Abstract
Objective
To investigate the association between the length of the polymorphic trinucleotide CAG microsatellite repeats in exon 1 of the AR gene and risk of prostate cancer containing TMPRSS2:ETS fusion genes.
Methods
This nested case-control study came from subjects enrolled in the Prostate Cancer Prevention Trial and included 195 biopsy-proven prostate cancer cases with known TMPRSS2:ETS status and 1344 matched controls.
Results
There was no association between CAG repeat length and risk of TMPRSS2:ETS-positive (OR=0.97, 95% CI, 0.91–1.04) or TMPRSS2:ETS-negative prostate cancer (OR=1.04, 95% CI, 0.97–1.11) and in patients with low- or high-grade disease.
Conclusion
Our findings suggested that AR CAG repeats are not associated with TMPRSS2:ETS formation in prostate cancer.
Keywords: androgen receptor, prostate cancer, CAG repeat, TMPRSS2:ETS
Introduction
The androgen receptor (AR) has been the focus of numerous studies in attempts to understand the role of this ligand-activated transcription factor in the development and progression of prostate cancer. It is a nearly ubiquitous protein that regulates tissue specific genes through androgens, particularly testosterone and dihydrotestosterone (DHT), controlling the regulation of prostate cellular proliferation and differentiation.
Residing within the large amino-terminal domain of the AR gene is a highly polymorphic CAG repeat sequence 1, that normally ranges from 8 to 31 repeats with an average of 20 repeats 2. Increased binding affinity for androgens and a higher transactivation activity of the AR, as well as a number of androgen-related clinical conditions including benign prostatic hyperplasia are associated with shorter AR CAG repeats 3, 4. Several studies have found no significant association between the AR CAG repeat length and the risk of prostate cancer 5–7, but there is evidence that genetic variation within the AR may influence the TMPRSS2:ETS fusions, which is detected in approximately half of all prostate cancers, and this gene fusion has been shown to be associated with unfavorable outcomes 8–10. Bastus et al., reporting on results from 40 prostate cancer clinical samples, showed that the CAG repeat length was shorter in TMPRSS2:ERG fusion positive samples than in fusion negative samples 8. Unfortunately the small sample size in that study was insufficient to find statistically significant results. In this report, we utilized tumor specimens collected within the Prostate Cancer Prevention Trial (PCPT), where all cases were biopsy detected and all pathology and TMPRSS2:ETS status were confirmed by central review, to further investigate the CAG repeat length – TMPRSS2:ETS relationship.
Methods
Study design, study population and data collection
We obtained biospecimen and study data from the PCPT, a randomized, placebo controlled trial that tested whether the 5α-reductase inhibitor finasteride would decrease the period prevalence of prostate cancer during a 7-year intervention. Institutional review boards at all participating institutions approved the study protocols and all participants provided informed consent. Details of the study design and participant characteristics were described previously 11, 12. Briefly, 18,882 men 55 years old or older with normal digital rectal exam (DRE), prostate specific antigen (PSA) 3 ng/ml or less and no history of prostate cancer or other clinically significant comorbid conditions that would have precluded successful completion of the study protocol were randomized to receive 5 mg finasteride daily or placebo daily for 7 years with enrollment completed between 1994 and 1997. During the course of the PCPT men underwent annual DRE and PSA measurement. Prostate biopsy was recommended in all with abnormal DRE or finasteride adjusted PSA greater than 4.0 ng/ml. All men without a prostate cancer diagnosis after seven years on study were recommended to undergo an ‘end of study’ prostate biopsy. Cases were men with biopsy determined prostate cancer identified by ‘for cause’ or ‘end of study’ biopsy and who had DNA available from white blood cells. Controls were selected from men with negative end-of-study biopsies, and they were frequency matched to cases by age (in 5-year increments), treatment arm (finasteride versus placebo) and family history of a first-degree relative with prostate cancer. Controls were oversampled on race to include all non-white subjects to increase power for subgroup analyses. We previously evaluated the AR CAG repeat polymorphisms in a case-control study and found no association between the AR CAG repeat length and risk of prostate cancer 7. For this study sample, we included 195 prostate cancer cases with archival tumor tissue available for characterization of TMPRSS2:ETS fusion status and 1,344 controls.
Details on age, race/ethnicity, family history, physical activity (type, frequency, duration, pace and intensity), usual alcohol consumption and smoking history were collected at baseline using self-administered questionnaires. Clinic staff measured height and weight at randomization and body mass index was calculated as weight in kg divided by height in m2. Tumors were graded and categorized; we retained the same low (Gleason less than 7) and high (Gleason 7 or greater) grade classifications as in the original trial report.
Genotyping and Characterization of TMPRSS2:ETS status
Blood collection, DNA extraction and genotyping for CAG repeat length have been described previously 7. Unstained 5 μm sections from all biopsy cores containing cancer were subjected to fluorescent in situ hybridization to determine TMPRSS2:ERG status. The 3′–5′ TMPRSS2 break-apart probe set comprised human DNA from two BAC clone, RP11-35C4 (labeled with SpectrumRed) just distal to TMPRSS2 5′-end and RP11-354C5 (labeled with SpectrumGreen) proximal to the TMPRSS2 3′-end. DNA-labeling and dual-target FISH assay were performed as previously described 13. Tumor areas were marked in the H&E stained reference slides by a pathologist. All scorable nuclei were analyzed in the lesions identified by the markings. Scorable nuclei have well-defined not disrupted borders, are not covered or involved with autofluorescent background particles, have at least one copy of the signal of interest, do not overlap with others and have at least the medium size of all tumor nuclei in the tumor section under screen. As negative controls, areas far away from the marked lesions and with round, epithelial-like nuclei about the same size as in the lesions were selected and scored. These controls were used for definition of the cut-offs for normal patterns. Specimens were classified as: (a) Negative for TMPRSS2 fusions when showing at least 70% of cells with only fused red/green signals; (b) Positive for TMPRSS2 fusions by deletion when showing more than 30% of cells with single reds; or (c) Positive for TMPRSS2 fusions by other mechanisms than deletion when showing more than 30% of cells with split red and green signals.
Statistical Analysis
The distribution of baseline participant characteristics for TMPRSS2:ETS positive and negative prostate cancer cases was compared using t-tests for continuous variables and chi-square tests for categorical variables. Odds ratios (OR) and 95% confidence intervals (CI) were calculated using unadjusted unconditional logistic regression to evaluate the association between CAG repeat length and prostate cancer risk. Polytomous logistic regression models were used to estimate the odds ratios and 95% confidence intervals for the association of CAG repeat length with risk of TMPRSS2:ETS-negative and TMPRSS2:ETS-positive prostate cancer. All analyses were for both all prostate cancer and stratified by grade. Because the transcriptional activity of the androgen receptor decreases linearly with CAG number 14, CAG length was evaluated both as a continuous variable and using categories from previously published studies (less than 19, 19 to 25 and 26 or greater) in relation to prostate cancer risk. Tests for linear trend across CAG length categories were based on an ordinal variable corresponding to rank. All p values were 2-sided and considered statistically significant at p < 0.05. SAS® (version 9.2) was used for all statistical analysis.
Results and Discussion
Baseline demographics and clinical characteristics of the case-control study population have been described previously 7. Characteristics of the men with prostate cancer by TMPRSS2:ETS tumor status are shown in Table 1. Overall, repeat length of the fusion-positive cases (mean=21.7) showed a trend to be shorter than fusion-negative cases (mean=22.3); however, this difference is not statistically significant (P=0.12). When comparing cases (n=195) with TMPRSS:ETS status to cases (n=1159) without TMPRSS:ETS status, men with TMPRSS:ETS status are younger (60.4 yrs vs. 63.4 yrs, respectively, p=<0.001) and have higher baseline PSA values (2.11 ng/dL vs. 1.59 ng/dL,, respectively, P<0.001) (data not shown).
Table 1.
Demographics and characteristics of men with prostate cancer by TMPRSS2:ETS status of the PCPT study participants.
| All (n=195) | TMPRSS2:ETS positive (n=97) | TMPRSS2:ETS Negative (n=98) | p-value | |
|---|---|---|---|---|
| Mean (SD) | ||||
| Age at Baseline (y) | 60.44 (4.00) | 60.02 (3.84) | 60.85 (4.14) | 0.15 |
| BMI (kg/m2) | 27.35 (3.63) | 27.35 (4.03) | 27.36 (3.21) | 0.99 |
| Alcohol intake (g/d) | 9.49 (14.74) | 8.85 (12.88) | 10.13 (16.42) | 0.54 |
| Baseline PSA (ng/dL) | 2.11 (1.66) | 2.06 (1.76) | 2.15 (1.55) | 0.72 |
| CAG repeat length | 22.0 (2.9) | 21.7 (2.5) | 22.3 (3.2) | 0.12 |
| n (%) | ||||
| Race | 0.15 | |||
| White | 182 (93.33) | 88 (90.72) | 94 (95.92) | |
| Non-white | 13 (6.67) | 9 (9.28) | 4 (4.08) | |
| Family history | 44 (22.56) | 25 (25.77) | 19 (19.39) | 0.29 |
| Diabetes | 10 (5.13) | 6 (6.19) | 4 (4.08) | 0.51 |
| Treatment Arm | 0.09 | |||
| Finasteride | 84 (43.08) | 36 (37.11) | 48 (48.98) | |
| Placebo | 111 (56.92) | 61 (62.89) | 50 (51.02) | |
| Smoking Status | 0.07 | |||
| Never Smoker | 65 (33.33) | 39 (40.21) | 26 (26.53) | |
| Current Smoker | 8 (4.10) | 5 (5.15) | 3 (3.06) | |
| Past Smoker | 122 (62.56) | 53 (54.64) | 69 (70.41) | |
| Physical Activity | 0.90 | |||
| Sedentary | 35 (18.04) | 19 (19.79) | 16 (16.33) | |
| Light | 86 (44.33) | 41 (42.71) | 45 (45.92) | |
| Moderate | 57 (29.38) | 27 (28.13) | 30 (30.61) | |
| Active | 16 (8.25) | 9 (9.38) | 7 (7.14) | |
| Gleason | <0.01 | |||
| Gleason 2–6 | 143 (73.71) | 81 (83.51) | 62 (63.92) | |
| Gleason 7–10 | 51 (26.29) | 16 (16.49) | 35 (36.08) | |
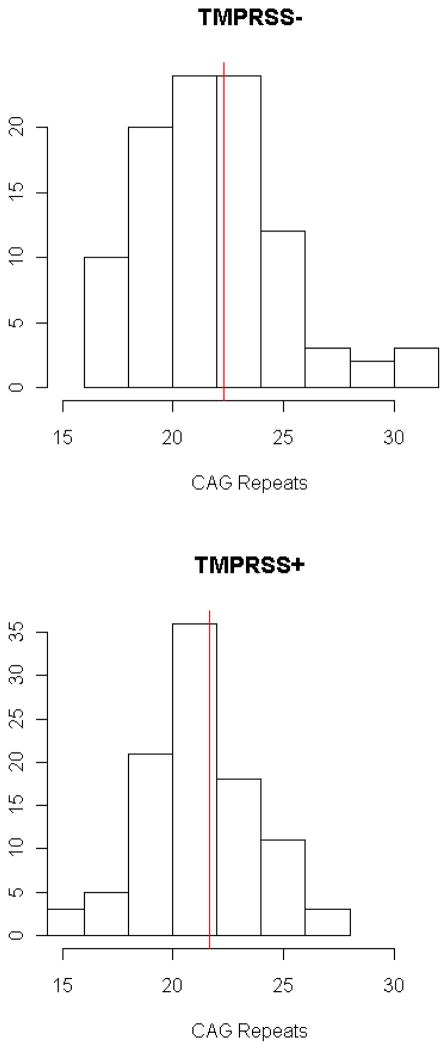
The frequency distributions of AR CAG repeat length in TMPRSS:ETS positive and negative cases are shown in Figure 1. There was no association between CAG repeat length and risk of TMPRSS2:ETS-positive (OR=0.97, 95% CI, 0.91–1.04) or TMPRSS2:ETS-negative prostate cancer (OR=1.04, 95% CI, 0.97–1.11) when CAG repeat length was analyzed as a continuous variable or categorized by cutoff points (Table 2). We also found no significant associations between AR CAG repeat length and risk of TMPRSS2:ETS-positive or TMPRSS2:ETS-negative prostate cancer in patients with low- or high-grade disease. Analysis of white-only race (in both cases and controls) also demonstrated no significant association between CAG repeats and TMPRSS2:ETS fusion status (data not shown).
Figure 1.
Frequency distribution of AR CAG repeat length in TMPRSS:ETS positive and negative cases.
Table 2.
AR CAG repeat length and risk of prostate cancer in PCPT controls and patients with low- and high-grade prostate cancer by TMPRSS2:ETS fusion status
| No. Controls | All prostate cancer with TMPRSS2:ETS status | TMPRSS2:ETS negative | TMPRSS2:ETS positive | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| No. Pts | OR (95% CI) | p-value | No. Pts | OR (95% CI) | p-value | No. Pts | OR (95% CI) | p-value | ||
| All prostate cancer | ||||||||||
| CAG categories | 0.99 | 0.48 | 0.46 | |||||||
| Less than 19 (ref) | 159 | 18 | ref | 10 | ref | 8 | ref | |||
| 19–25 | 1032 | 160 | 1.37 (0.82–2.92) | 75 | 1.16 (0.59–2.82) | 85 | 1.64 (0.78–3.44) | |||
| 26+ | 153 | 17 | 0.98 (0.49–1.98) | 13 | 1.35 (0.58–3.17) | 4 | 0.52 (0.15–1.76) | |||
| Per 1 CAG repeat increase | 1344 | 195 | 1.00 (0.96–1.05) | 0.91 | 98 | 1.04 (0.97–1.11) | 0.29 | 97 | 0.97 (0.91–1.04) | 0.37 |
| Gleason 2–6 | ||||||||||
| CAG categories | 0.82 | 0.55 | 0.41 | |||||||
| Less than 19 (ref) | 159 | 12 | ref | 6 | ref | 6 | ref | |||
| 19–25 | 1032 | 121 | 1.55 (0.84–2.88) | 48 | 1.23 (0.52–2.93) | 73 | 1.88 (0.80–4.38) | |||
| 26+ | 153 | 10 | 0.87 (0.36–2.06) | 8 | 1.39 (0.47–4.09) | 2 | 0.35 (0.07–1.74) | |||
| Per 1 CAG repeat increase | 1344 | 143 | 1.00 (0.94–1.05) | 0.91 | 62 | 1.04 (0.95–1.12) | 0.41 | 81 | 0.97 (0.90–1.04) | 0.38 |
| Gleason 7–10 | ||||||||||
| CAG categories | 0.73 | 0.69 | 0.97 | |||||||
| Less than 19 (ref) | 159 | 6 | ref | 4 | ref | 2 | ref | |||
| 19–25 | 1032 | 38 | 0.98 (0.41–2.35) | 26 | 1.00 (0.35–2.91) | 12 | 0.92 (0.21–4.17) | |||
| 26+ | 153 | 7 | 1.21 (0.40–3.69) | 5 | 1.30 (0.34–4.93) | 2 | 1.04 (0.15–7.47) | |||
| Per 1 CAG repeat increase | 1344 | 51 | 1.01 (0.93–1.11) | 0.76 | 35 | 1.03 (0.93–1.14) | 0.59 | 16 | 0.98 (0.84–1.15) | 0.81 |
Previous studies have shown that the length of CAG repeats is inversely correlated to AR transcriptional activity, increased AR activity induced TMPRSS2:ETS formation, and CAG repeat length of TMPRSS2:ETS fusion-positive tumors showed a trend to be shorter than fusion-negative cases 8, 14. We undertook the current study to better define the relationship of CAG repeat length and TMPRSS2:ETS fusion status with prostate cancer risk. To our knowledge, this is the first study to report that CAG repeats are not associated with the development of TMPRSS2:ETS fusion prostate cancer.
The precise effect of CAG repeats on AR function and the molecular pathways implicated in TMPRSS2-ERG gene fusion remain to be elucidated with the common defining factor being AR/AR-mediated signaling. In fact, changes in the homeostatic balance of AR and coregulator occupancy on target genes may explain the variable penetrance of the CAG repeat 15 and/or affect subsequent TMPRSS2-ETS gene fusion formation. Given the inconsistent findings on the relationship between CAG repeat length and prostate cancer risk across study populations coupled with the disparity in prevalence of these fusions across ethnic groups 16, future studies are warranted to further characterize the relationship between CAG repeats and TMPRSS2-ETS formation in prostate cancer.
Acknowledgments
This research was supported in part by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research; The Biology of the Prostate Cancer Prevention Trial (P01 CA108964); the University of Colorado Cancer Center Support Grant (P30 CA046934); the Cancer Therapy and Research Center Support Grant (P30 CA054174); and Public Health Service Grant (CA37429) from the National Cancer Institute, Division of Cancer Prevention.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Trapman J, Cleutjens KB. Androgen-regulated gene expression in prostate cancer. Semin Cancer Biol. 1997;8:29–36. doi: 10.1006/scbi.1997.0050. [DOI] [PubMed] [Google Scholar]
- 2.Edwards A, Hammond HA, Jin L, Caskey CT, Chakraborty R. Genetic variation at five trimeric and tetrameric tandem repeat loci in four human population groups. Genomics. 1992;12:241–53. doi: 10.1016/0888-7543(92)90371-x. [DOI] [PubMed] [Google Scholar]
- 3.Giovannucci E. Is the androgen receptor CAG repeat length significant for prostate cancer? Cancer Epidemiol Biomarkers Prev. 2002;11:985–6. [PubMed] [Google Scholar]
- 4.Rajender S, Singh L, Thangaraj K. Phenotypic heterogeneity of mutations in androgen receptor gene. Asian J Androl. 2007;9:147–79. doi: 10.1111/j.1745-7262.2007.00250.x. [DOI] [PubMed] [Google Scholar]
- 5.Freedman ML, Pearce CL, Penney KL, et al. Systematic evaluation of genetic variation at the androgen receptor locus and risk of prostate cancer in a multiethnic cohort study. Am J Hum Genet. 2005;76:82–90. doi: 10.1086/427224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Platz EA, Leitzmann MF, Rifai N, et al. Sex steroid hormones and the androgen receptor gene CAG repeat and subsequent risk of prostate cancer in the prostate-specific antigen era. Cancer Epidemiol Biomarkers Prev. 2005;14:1262–9. doi: 10.1158/1055-9965.EPI-04-0371. [DOI] [PubMed] [Google Scholar]
- 7.Price DK, Chau CH, Till C, et al. Androgen receptor CAG repeat length and association with prostate cancer risk: results from the prostate cancer prevention trial. J Urol. 2010;184:2297–302. doi: 10.1016/j.juro.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bastus NC, Boyd LK, Mao X, et al. Androgen-induced TMPRSS2:ERG fusion in nonmalignant prostate epithelial cells. Cancer Res. 2010;70:9544–8. doi: 10.1158/0008-5472.CAN-10-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hossain D, Bostwick DG. Significance of the TMPRSS2:ERG gene fusion in prostate cancer. BJU Int. 2013;111:834–5. doi: 10.1111/bju.12120. [DOI] [PubMed] [Google Scholar]
- 10.Mani RS, Tomlins SA, Callahan K, et al. Induced chromosomal proximity and gene fusions in prostate cancer. Science. 2009;326:1230. doi: 10.1126/science.1178124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feigl P, Blumenstein B, Thompson I, et al. Design of the Prostate Cancer Prevention Trial (PCPT) Control Clin Trials. 1995;16:150–63. doi: 10.1016/0197-2456(94)00xxx-m. [DOI] [PubMed] [Google Scholar]
- 12.Thompson IM, Goodman PJ, Tangen CM, et al. The influence of finasteride on the development of prostate cancer. N Engl J Med. 2003;349:215–24. doi: 10.1056/NEJMoa030660. [DOI] [PubMed] [Google Scholar]
- 13.Massion PP, Zou Y, Uner H, et al. Recurrent genomic gains in preinvasive lesions as a biomarker of risk for lung cancer. PLoS One. 2009;4:e5611. doi: 10.1371/journal.pone.0005611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buchanan G, Yang M, Cheong A, et al. Structural and functional consequences of glutamine tract variation in the androgen receptor. Hum Mol Genet. 2004;13:1677–92. doi: 10.1093/hmg/ddh181. [DOI] [PubMed] [Google Scholar]
- 15.Buchanan G, Need EF, Barrett JM, et al. Corepressor effect on androgen receptor activity varies with the length of the CAG encoded polyglutamine repeat and is dependent on receptor/corepressor ratio in prostate cancer cells. Mol Cell Endocrinol. 2011;342:20–31. doi: 10.1016/j.mce.2011.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Magi-Galluzzi C, Tsusuki T, Elson P, et al. TMPRSS2-ERG gene fusion prevalence and class are significantly different in prostate cancer of Caucasian, African-American and Japanese patients. Prostate. 2011;71:489–97. doi: 10.1002/pros.21265. [DOI] [PubMed] [Google Scholar]