Abstract
Metabolic reprogramming is a central hallmark of cancer, enabling tumor cells to obtain the macromolecular precursors and energy needed for rapid tumor growth. Understanding how oncogenes coordinate altered signaling with metabolic reprogramming and global transcription may yield new insights into tumor pathogenesis, and provide a new landscape of promising drug targets, while yielding important clues into mechanisms of resistance to the signal transduction inhibitors currently in use. Here we review the recently identified central regulatory role for mTORC2, a downstream effector of many cancer-causing mutations, in metabolic reprogramming and cancer drug resistance. We consider the impact of mTORC2-related metabolism on epigenetics and therapeutics, with a particular focus on the intractable malignant brain tumor, glioblastoma (GBM).
Keywords: mTORC2, c-Myc, metabolic reprogramming, epigenetics, drug resistance, glioblastoma
Metabolic reprogramming in cancer – a coordinated effort
Metabolic reprogramming is a central hallmark of cancer [1]. Nearly a hundred years ago, Otto Warburg demonstrated that cancer cells convert the majority of glucose they take up, into lactate even in the presence of sufficient oxygen to support oxidative phosphorylation. This biochemical adaptation, termed “the Warburg effect,” has once again assumed a central role in framing cancer as a metabolic disease, spurring considerable interest in trying to understand the survival advantages conferred by this adaptation and the signaling pathways that regulate it [2].
Cancer cells increase glucose uptake to meet the increased energetic and biosynthetic demands imposed by rapid tumor growth. However, ratcheting up glucose uptake is not without risk to the cell. If all of the glucose taken up by tumor cells were to be fully oxidized in the tricarboxylic acid (TCA) cycle, the levels of reactive oxygen species (ROS) generated could be catastrophic. The transfer of electrons from NADH and FADH2 to molecular oxygen through the cellular respiratory chain is energy efficient, yielding 36 ATP molecules per molecule of glucose, but superoxide anions are produced in this process, generating mitochondrial ROS [3-8]. Cancer cells have developed adaptations to allow them to: 1) utilize the glucose-derived carbons for lipid synthesis through the activity of ATP citrate lyase; 2) leverage the glucose-derived carbons for production of ribose, glycerol, serine and glycine and 3) secrete the excess glucose-derived carbons as lactate. These coordinated glycolytic adaptations enable tumor cells to meet their energetic and anabolic needs without suffering catastrophically high levels of ROS. However, they need to take up more glucose to do it, because only 2 molecules of ATP are yielded per molecule of glucose.
The Warburg effect alone cannot account for the full spectrum of metabolic changes required for tumor growth [2,9]. Glutaminolysis, the catabolism of glutamine to support tumor cell proliferation, is also a central feature of cancer metabolic reprogramming, providing: 1) a source of nitrogen for nucleotide and amino acid synthesis; 2) a mechanism to produce NADPH for lipid and nucleotide synthesis and 3) an alternative carbon source to supply TCA cycle intermediates [10]. Tumor cells also require large amounts of lipid for membrane biogenesis, signal transduction and potentially as an energy source. De novo lipogenesis is a metabolic hallmark of cancer, which can be augmented by uptake of exogenous lipids [11-14].
The Warburg effect, glutaminolysis and lipogenesis are not exclusive to cancer. They can all be activated in rapidly proliferating cells engaged in physiological processes such as immune response or wound repair [15,16]. This raises the question of whether cancer metabolic reprogramming simply represents the enhanced use of biochemical adaptations available to rapidly proliferating cell types, or whether the two differ in fundamental ways. One of the crucial differences between cancer cells and non-cancer cells lies in the inability of non-cancer cells to autonomously take up sufficient nutrients for anabolic metabolism [16]. In metazoans, the metabolism of individual cells is tightly regulated by balancing intrinsic and extrinsic molecular cues, thus instructing cells on how best to meet their demand for ATP generation, biosynthesis of macromolecules, and maintenance of redox in the context of a multicellular organism [9]. In contrast, cancer cells meet their metabolic demands in an entirely cell-intrinsic fashion, enabling cell-autonomous growth, a sine qua non of cancer [16]. The specificity of cancer metabolic reprogramming may therefore lie in the coordination of responses that enable tumor cells to do what non-neoplastic cells cannot; that is, to meet all of their needs in an entirely cell-autonomous fashion.
Understanding how cancer-causing mutations cause coordinated engagement of cellular signaling pathways, biochemical repertoires and global transcription ensembles may yield critical insights into the pathogenesis of cancer and shed new light on how tumor cells resist targeted therapies to which they should be vulnerable. In this light, it is not surprising that mutations in key regulators of PI3K-AKT/PKB-mTOR signaling and/or upstream receptor tyrosine kinases (RTKs) are found in the vast majority of cancers [17]. PI3K-AKT-mTOR signaling is the key mechanism that normal cells use to metabolize glucose in response to insulin [3]. Further, it is not surprising that c-Myc, a critical regulator of glutaminolysis, is also amplified or mutated in some types of cancer [18], although co-occurrence of PI3K-activating mutations and c-Myc amplification appears to be the exception [17]. Understanding how tumors with PI3K-AKT-mTOR activating mutations engage c-Myc signaling may provide important clues as to how tumor cells coordinate metabolic reprogramming to optimize growth. Mutations in metabolic enzymes such as isocitrate dehydrogenase 1/2 (IDH1/2) are highly informative because they provide a direct link between altered cellular metabolism and epigenetics [19,20]. How does metabolic reprogramming caused by more common cancer-causing mutations alter the epigenetic landscape of the cell? Does it do so through indirect regulation of enzymes that regulate histone acetylation and/or by regulating the level of intermediate metabolites such as acetyl-CoA whose levels directly influence epigenetic regulation [19]? This review focuses on a paradigmatic example, which may have broad implications for understanding cancer metabolic reprogramming. Epidermal growth factor receptor (EGFR) is the most commonly activated oncogene in GBM, the highly lethal form of adult brain cancer [21]. In particular, EGFRvIII (Box 1), a constitutively active gain-of-function mutation resulting from an extracellular in-frame genomic deletion, has recently been shown to reprogram tumor cell metabolism, driving the Warburg effect [22-24], glutaminolysis [22,24] and lipogenesis [25]. Here, we review a set of recent discoveries involving EGFR-mutant GBM that highlight the integration of altered signaling, metabolic reprogramming and epigenetic changes downstream of common cancer mutations, potentially providing new therapeutic opportunities.
mTORC1 and mTORC2 – essential partners in metabolic reprogramming
In many cancers, RTK amplification and mutations, PIK3CA mutations and PTEN loss conspire to constitutively activate PI3K-AKT-mTOR signaling [17] and thereby to reprogram cellular metabolism. EGFRvIII mutation and PTEN loss, a common cooccurrence in GBM, play a central role both in tumorigenesis and in metabolic reprogramming through PI3K-AKT-mTOR activation [21,26]. mTOR is a serine/threonine protein kinase that integrates growth factor receptor signaling with cellular growth, proliferation and survival through two distinct multi-protein complexes. mTORC1, a validated cancer drug target, regulates protein translation through its substrates S6K1 and 4E-BP1 as well as anabolic metabolism downstream of growth factor receptor-activated PI3K-AKT signaling and in response to amino acid nutrient levels [27-29].
mTORC2 is less well understood. mTORC2 has been considered to be insensitive to nutrient levels, but responsive to growth factor signaling and to function mainly through activating AKT by phosphorylating it on Ser473 [30]. It can also phosphorylate other AGC kinases. Recent studies, however, suggest that mTORC2 may have an unexpectedly important role in cancer pathogenesis, promoting tumor growth and chemotherapy resistance in cancer cells [31], as well as controlling genome stability in yeast [32]. These effects appear to occur through AKT-independent signaling [31,32]. Both mTORC1 and mTORC2 are also necessary for the formation of EGFR-PI3K driven gliomas in a Drosophila model [33], suggesting an important role for mTORC2 signaling, independent of AKT-mTORC1 activation.
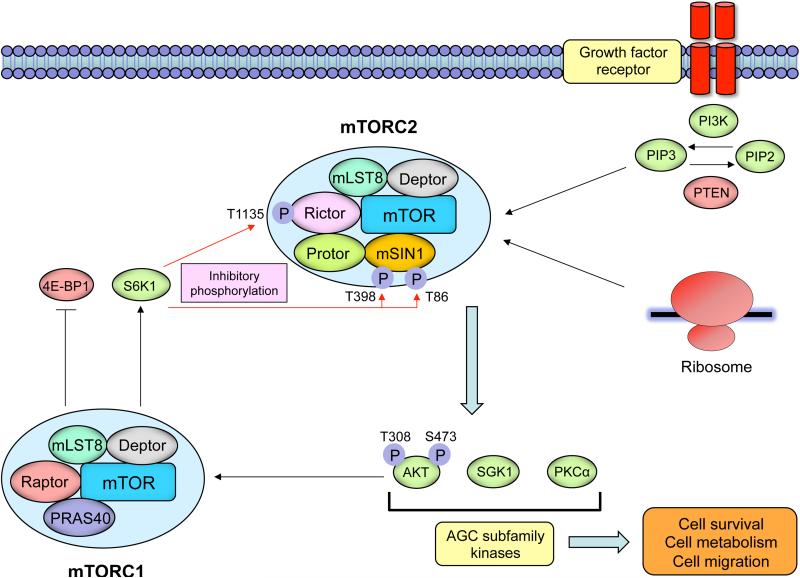
Structurally, both mTORC1 and mTORC2 contain mTOR, mLST8 and Deptor. The binding of Rictor to the HEAT repeats of the mTOR protein defines the mTORC2 complex, and the complex contains the additional proteins mSIN1 and Protor [29] (Figure 1). AKT is recruited to the plasma membrane enabling phosphorylation at Thr308 in the catalytic domain by phosphoinositide-dependent kinase 1 [34], and Rictor-mTOR complex phosphorylates AKT at Ser473 for its maximal activation [30,35]. mSIN1 also enables mTORC2 to phosphorylate and activate AKT. Protor, a Rictor-binding subunit, may function to activate serum and glucocorticoid regulated kinase 1 (SGK1) [36]. mTORC2 allosterically activates AGC kinase family members including AKT, PKCα, and SGK1 by phosphorylating their turn motif and hydrophobic motif sequences [37] (Figure 1).
Figure 1. The structure and signaling of the mTORC2 complex.
Growth factor receptor-dependent activation of PI3K promotes mTORC2's binding to ribosomes, which activates mTORC2 in a fashion that is still incompletely understood. mTORC2 phosphorylates conserved motifs in AGC kinases to promote their allosteric activation. mTORC2 is a critical node in growth factor receptor-PI3K signaling, phosphorylating Akt on Ser473 to promote its maximal activation. mTORC2 is negatively regulated by mTORC1. S6K1 downstream of mTORC1 phosphorylates Rictor on Thr1135 and mSIN1 on Thr86 and Thr398, inhibiting mTORC2-dependent phosphorylation of AKT.
mTORC2 is responsive to growth factor signaling, possibly by associating with translating ribosomes in response to growth factor receptor-PI3K activation [38,39]. mTORC2 activity may also be “tuned” in response to growth factor signaling. Persistent mTORC1 activity suppresses mTORC2 through a series of post-translational modifications [40-43] (Figure 1). Downstream of mTORC1, S6K1 phosphorylates Rictor on Thr1135, inhibiting mTORC2-dependent phosphorylation of AKT [40]. S6K1 also phosphorylates mSIN1 on Thr86 and Thr398, dissociating mSIN1 from the mTORC2 complex to suppress mTORC2-dependent AKT Ser473 phosphorylation [41]. Upstream of mTORC1, AKT itself may regulate mTORC2 activity, either by directly phosphorylating mSIN1 on Thr86, or indirectly through mTORC1/S6K1-dependent phosphorylation of mSIN1 [41, 42]. Interestingly, AKT's effect on mSIN1 phosphorylation and mTORC2 activity appear to be highly cell type-dependent, and cell context-dependent [41-43].
mTORC1 and mTORC2 converge on c-Myc to control metabolic reprogramming in cancer
C-Myc integrates cellular proliferation with metabolism in many cancers [3,44-46], coordinating nutrient uptake with biomass accumulation downstream of growth factor receptor signaling [4]. C-Myc is amplified in a relatively small subset of cancers, but its coordination with activating mutations in growth factor receptor-PI3K signaling appears to be important for cancer metabolic reprogramming, including through regulation of glucose transport, glycolysis, glutaminolysis, lipogenesis and nucleotide synthesis [47]. Thus, c-Myc may be a critical node of convergence through which growth factor receptor signaling mutations reprogram cellular metabolism.
Two complementary and interlacing mechanisms were recently identified by which mTORC1 and mTORC2 coordinately control tumor cell metabolism through c-Myc (Figure 2). In EGFRvIII-expressing GBMs, mTORC1 upregulates the hnRNPA1 splicing factor, catalyzing the splicing of the c-Myc interacting protein Max to generate Delta Max, thereby enhancing Myc-dependent glycolysis and tumor growth in vivo [22]. Concurrently, mTORC2 increases the level of c-Myc by inactivating class IIa histone deacetylases (HDACs), leading to inactivating acetylation of FoxO1 and FoxO3. Inactivation of FoxOs releases c-Myc from a suppressive miR-34c-dependent network which targets the 3’UTR of c-Myc mRNA and inhibits its translation [24,48]. Thus, mTORC1 and mTORC2 conspire to link growth factor receptor-PI3K signaling with c-Myc-dependent metabolic reprogramming by controlling both c-Myc levels and its activity (Figure 2). The role of mTORC1 in metabolic reprogramming has been recently comprehensively reviewed [27-29]; therefore, this review will focus on mTORC2.
Figure 2. mTORC2 plays a central role in reprogramming tumor cell metabolism through interlacing and synergistic mechanisms.
In GBM, the gain of function EGFR mutation, EGFRvIII, promotes glycolytic metabolism by activating hnRNPA1-dependent alternative splicing of a Myc-binding partner Delta Max downstream of AKT, thereby functionally augmenting the oncogenic activity of c-Myc (left panel). Concurrently, mTORC2 controls c-Myc transcription, translation and protein level through FoxO acetylation (right panel). These findings point to the central role for mTORC2 in linking EGFR mutation with c-Myc to reprogram cancer cell metabolism.
Metabolic reprogramming by mTORC2
Glycolytic metabolism
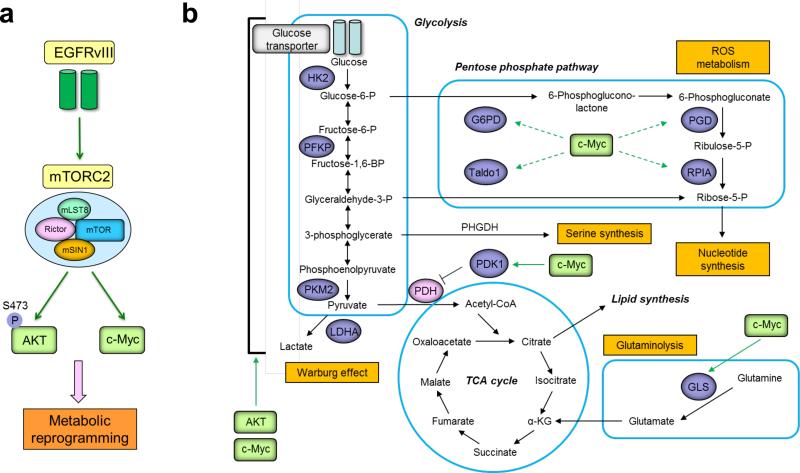
mTORC2 regulates glycolytic metabolism in cancer in at least three ways. First, mTORC2 phosphorylates AKT on Ser473 (Figure 3a), ensuring its full activation, which: 1) increases the expression of glucose transporters (GLUT); 2) phosphorylates and activate hexokinase 2 (HK2) to mediate the first step of glycolysis, and 3) allosterically activates phosphofructokinase-1 (PFK-1) to regulate the rate-limiting step of glycolysis [49-51] (Figure 3b). In the livers of Rictor knockout mice, loss of AKT Ser473 phosphorylation and glucokinase leads to constitutive gluconeogenesis and impaired glycolysis which alters whole-body glucose homeostasis [52]. This is consistent with the expected role for mTORC2 in regulating glucose metabolism through AKT. Secondly, mTORC2 regulates glycolytic metabolism in cancer by controlling c-Myc levels, as described above [24] (Figure 3a). C-Myc-regulates the expression of key regulatory genes that control glucose transport and glycolysis, including GLUT1, HK2, pyruvate kinase M2 isoform (PKM2), lactate dehydrogenase A (LDHA) and pyruvate dehydrogenase kinase isozyme 1 (PDK1) which inhibits pyruvate dehydrogenase (PDH). Thus, c-Myc controls the expression of a repertoire of genes that promotes the Warburg effect in cancer [53,54] (Figure 3b).Thirdly, mTORC2 can potentially decrease transcription of gluconeogenic genes in tumor cells in a FoxO-dependent fashion [24,55], resulting in the diversion of glucose-derived carbons to other metabolic pathways to enhance their metabolic flexibility.
Figure 3. mTORC2 signaling regulates metabolic reprogramming via AKT and c-Myc.
(a) EGFRvIII-mTORC2 axis promotes the activation of two independent downstream effectors AKT and c-Myc, which facilitate the metabolic reprogramming. (b) The chief metabolic pathways that contribute to the production of macromolecules and energy in rapidly dividing cells are glycolysis, TCA cycle, pentose phosphate pathway (PPP), and glutaminolysis. Green boxes denote the downstream effector of mTORC2: AGC subfamily kinases including AKT, and c-Myc up-regulated independently of AKT. AKT and c-Myc reprogram cellular metabolism by activating glycolytic enzymes (GLUT1, HK2, PFKP, PKM2, LDHA and PDK1), PPP enzymes (G6PD, PGD, RPIA and Taldo1), and glutaminase (GLS). Serine pathway, which plays a role in the cancer pathogenesis [107,108], may be regulated by EGFR signaling through the expression of phosphoglycerate dehydrogenase (PHGDH) [109].
Glutaminolysis
mTORC2 can regulate glutaminolysis by controlling glutamine uptake and glutaminase (GLS) levels (Figure 3b). First, mTORC2 can increase the uptake of glutamine by regulating its cell surface transporters through activation of AGC kinases. mTORC2-mediated activation of AKT and SGK1 controls amino acid transport through the modulation of the plasma membrane glutamine transporter isoforms including SNAT2 (SLC38A2) and LAT1 (SLC7A5) [56], and regulates the Na(+)-coupled glutamine transporter SN1 by inactivating the ubiquitin ligase Nedd4-2 which targets SN1 for degradation. [57]. Secondly, mTORC2, through c-Myc-dependent upregulation of GLS [18,24], can provide a source of nitrogen for amino acid and nucleotide synthesis, as well as carbons to replenish TCA cycle intermediates [10]. Thus, mTORC2-dependent glutaminolysis might fuel NADPH production and provide an alternative source for generation of acetyl-CoA through IDH1/2-dependent reductive carboxylation of glutamine-derived α-ketoglutarate [58-60]. Future studies are necessary, but it is speculated that mTORC2-dependent, c-Myc-dependent signaling might reprogram glutamine metabolism towards nucleotide synthesis and lipogenesis, particularly in hypoxic tumor tissue in which TCA-derived carbons may be in short supply.
Lipogenesis
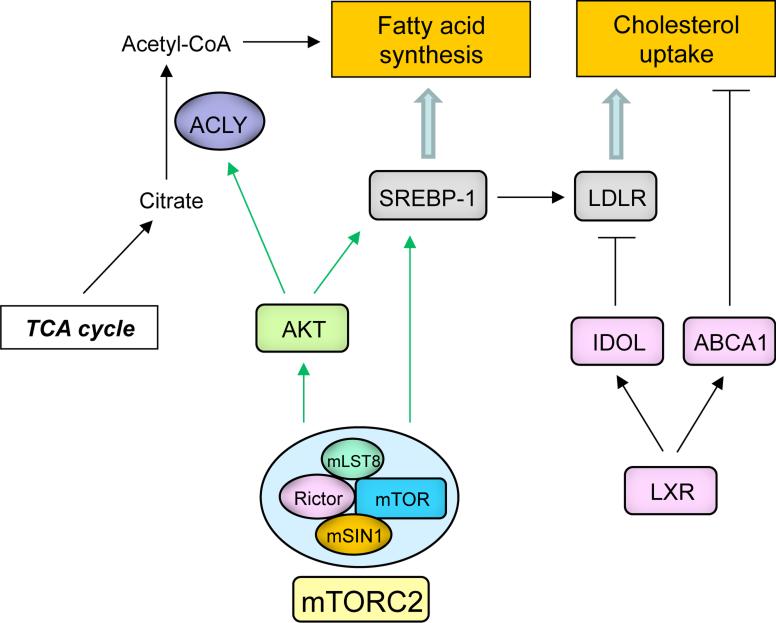
mTORC2 is also a critical regulator of lipid metabolism [61], regulating hepatic lipogenesis [52,62], lipolysis [63], adipogenesis [64] and lipid homeostasis [65,66], through AKT-dependent and AKT-independent mechanisms. Currently, the role of mTORC2 in tumor lipid metabolism is not well yet understood. Non-tumor cells regulate their intracellular lipids primarily by taking up free fatty acids and lipoproteins, ensuring that the lipogenic program is under tight systemic control. In contrast, cancer cells are thought to derive the bulk of their lipids through cell-autonomous de novo fatty acid and cholesterol synthesis, generating acetyl-CoA derived from glucose-derived citrate through the TCA cycle and ATP citrate lyase (ACLY) [11,67,68] as a source of structural lipids for membrane biogenesis (Figure 4). Lipids can be used for the generation of ATP through β-oxidation and as signaling molecules and post-translational modifiers of signaling proteins [67]. Tumor cells can also take up exogenous lipids, saving the high NADPH “cost” of de novo lipogenesis through mechanisms that may escape systemic regulation. Tumor cells of a variety of types take up low density lipoprotein (LDL), oxysterols or fatty acids to promote cell-autonomous tumor growth [69] and metastasis [70]. Ovarian tumor cells can also generate a source of their own lipids by stimulating neighboring adipocytes to release free fatty acids, thus fueling tumor growth and metastasis [14]. The mechanisms by which tumor cells increase their uptake of exogenous lipids, or stimulate their release from neighboring adipocytes through signaling cross talk, are only beginning to be understood. In GBM, EGFRvIII increases the expression of low density lipoprotein receptors (LDLRs), rendering tumor cells highly dependent on LDL for growth and survival. Importantly, these tumor cells have found a way to escape systemic regulatory feedback control [69], as will be described below. The ability of tumor cells to utilize multiple lipogenic pathways that skirt normal systemic control and optimize levels of fatty acids and cholesterol, while minimizing NADPH costs, may be central to cancer metabolic reprogramming.
Figure 4. mTORC2 may regulate tumor cell lipid metabolism in an AKT-dependent and AKT-independent manner.
mTORC2-dependent phosphorylation of AKT promotes activating cleavage of SREBP-1 to promote tumor cell lipogenesis. The possibility of mTORC2-dependent, AKT-independent mechanisms is currently being explored. mTORC2 may also facilitate the cholesterol uptake by LDLR in an EGFR-AKT-SREBP dependent manner. In non-cancerous cells when cholesterol levels rise, LXR-dependent transcription of IDOL, an E3 ubiquitin ligase that degrades LDLR, and ABCA1, a cholesterol efflux transporter, maintain a homeostatic level of cholesterol. Persistent SREBP-1 activation in response to mTORC2 signaling in cancer cells may subvert this feedback inhibition by LXR.
Sterol regulatory binding proteins (SREBPs) are the master transcriptional regulators of lipogenesis controlling expression of many of the key genes that regulate cholesterol and fatty acid synthesis [71]. SREBPs are responsive to growth factor receptor signaling through AKT and mTORC1 [72,73], and to nutrient and energy status [13], as well as cholesterol levels [71]. In GBM, activating cleavage of SREBP1 is downstream of mutant EGFR signaling through AKT, potently driving tumor growth in vivo [25]. Surprisingly, in GBM, SREBP1 cleavage is insensitive to rapamycin [25]. The failure of rapamycin to block SREBP1 cleavage may be a consequence of incomplete mTORC1 inhibition. Alternatively, mTORC2, which is not generally rapamycin-sensitive, could contribute to SREBP1 cleavage, either through AKT-dependent [25,52] or AKT-independent signaling [62], thus potentially nominating mTORC2 as an emerging key controller of lipid metabolism [61] (Figure 4).
SREBP1 also promotes GBM growth and survival by upregulating LDLR, ensuring sufficient levels of intracellular lipids to sustain rapid proliferation. In non-neoplastic cells, SREBP1 and the nuclear receptor LXR, maintain cholesterol homeostasis through complementary mechanisms of feedforward activation and feedback inhibition. Growth factor receptor signaling through PI3K-AKT, or low levels of cholesterol in the cell, activate SREBPs to promote expression of genes involved in cholesterol and fatty acid synthesis, including LDLR. When intracellular levels of cholesterol are high, LXRs promote expression of ABCA1 and ABCG1 cholesterol efflux transporters and induce transcription of IDOL, an E3 ubiquitin ligase that targets LDLR for degradation, thus lowering intracellular cholesterol levels to maintain homeostasis [74]. Remarkably, GBM cells have developed a mechanism to subvert feedback inhibition in the presence of high levels of cholesterol; EGFRvIII/EGFR signaling through PI3K/AKT promotes LDLR expression in an SREBP1-dependent manner, continuing to express abundant amounts of LDLR even when intracellular cholesterol levels are high (Figure 4). Perpetually elevated LDLR levels enable the tumor cells to escape the normal regulatory control system that should prevent relentless tumor growth [69].
Nucleotide and reactive oxygen species (ROS) metabolism
Tumor cells need an abundant supply of NADPH as a reducing agent for anabolic reactions such as liopogenesis and to maintain cellular redox balance by reducing glutathione disulfide (GSSG) to glutathione (GSH) to prevent damage from ROS [75]. Elevated levels of glycolysis in tumor cells divert glucose-6-phosphate into the pentose phosphate pathway (PPP), generating NADPH, as well as ribose for nucleotide synthesis [76] (Figure 3b). Knockdown of Rictor in GBM cells lowers cellular levels of c-Myc, decreasing expression of PPP oxidative (glucose-6-phosphate dehydrogenase: G6PD, phosphogluconate dehydrogenase: PGD) and non-oxidative (ribose 5-phosphate isomerase: RPIA, transaldolase 1: Taldo1) branch enzymes, thus suggesting a role for mTORC2 in regulating NADPH balance. The effects of mTORC2 inhibition are phenocopied by c-Myc knockdown, suggesting that the effects of mTORC2 on PPP flux and NADPH levels may be mediated through c-Myc [19] (Figure 3b). A recent chemical genetics study also supports the idea that TORC2 specifically interacts with PPP and balances the high energy demand required for ribosome biogenesis [77]. Cytoplasmic NADPH is also generated by malic enzyme, which converts α-ketoglutarate-derived malate into pyruvate, and by IDH1-dependent conversion of citrate into α-ketoglutarate [78]. Thus, mTORC2 appears to regulate NADPH levels in a c-Myc-dependent manner, generating NADPH through two α-ketoglutarate-related reactions, and by regulating PPP flux downstream of glycolysis.
Maintained activation of metabolic pathways can drive resistance to signal transduction inhibitors – a therapeutic role for mTORC2 inhibition
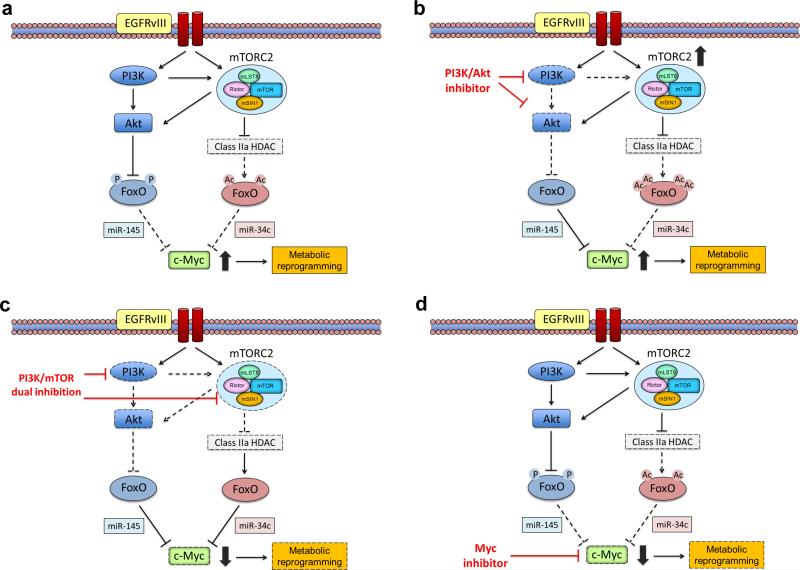
The FoxO-Myc axis is regulated via complementary posttranslational mechanisms and microRNA networks - PI3K/AKT-dependent inactivating phosphorylation of FoxO which relieves miR-145-mediated suppression of c-Myc [18,79-83] and mTORC2-dependent acetylation of FoxO releasing c-Myc from miR-34c suppression [24] (Figure 5). An important consequence of this dual-pronged regulation may be that the PI3K, AKT, and/or mTORC1 inhibitors, including combinations, that are being tested in early to mid-phase clinical trials, may not be sufficient to suppress cancer metabolic reprogramming through c-Myc, thus promoting clinical resistance. mTORC2 inhibition may also be required to abrogate c-Myc expression and metabolic reprogramming in order to achieve clinical remission (Figure 5). For reasons that are as yet unclear, mTORC2 appears much more difficult to suppress than mTORC1 with ATP-competitive mTOR kinase inhibitors, and mTORC2 specific inhibitors, which would likely need to be allosteric inhibitors, have yet to be developed. Another potentially important consequence of this type of dual regulation of signaling pathways converging on metabolic outputs, is that Positron Emission Tomography (PET), especially 18F-fluorodeoxy-glucose (FDG)-PET, may prove to be an invaluable tool for assessing target inhibition and predicting response in the clinic, because it may enable quantitative assessment of the effect of signal transduction inhibitors on metabolic pathways in patients through non-invasive imaging [78,84]. Further, new types of PET radiotracers may be a prospective candidate for identifying molecular subtypes and therapeutic effects in metabolically-active tumors stimulated by mTORC2/c-Myc signaling, including 89Zr-transferrin which binds to Myc-regulated transferrin receptor 1 [85], L-[5-11C]-glutamine for glutaminolytic tumors [86] and 11C-acetate for non 18F-FDG-avid neoplasms [87] as well as 18F-FDG for Myc-activated tumors [88]. In addition, the convergence of multiple upstream signaling pathways on c-Myc, raises the possibility that small molecule inhibitors which disrupt Myc/Max dimerization [89], or bromodomain and extraterminal (BET) bromodomain inhibitors that interfere with BET family protein binding to lysine-acetylated histone tails to suppress c-Myc-dependent target gene expression [90,91], may have a role in solid tumors with PI3K and mTORC2 activation (Figure 5).
Figure 5. mTORC2-c-Myc-dependent tumor cell metabolism can cause resistance to PI3K and AKT-targeted therapies.
(a) C-Myc levels in tumor cells are subject to dual-pronged regulation. Persistent PI3K/AKT signaling in tumor cells phosphorylates and inhibits FoxO, releasing c-Myc from miR-145-dependent suppression. Concurrently, persistent mTORC2 signaling independent of AKT acetylates and inhibits FoxO, de-repressing c-Myc from miR-34c-dependent regulation. (b) PI3K and AKT-targeted therapies suppress FoxO phosphorylation, but may paradoxically elevate mTORC2-dependent FoxO acetylation, maintaining high levels of c-Myc in tumor cells, potentially causing drug resistance. (c) Combined inhibition of PI3K and mTORC2 potently suppresses both FoxO-dependent pathways, lowering tumor cell levels of c-Myc and causing tumor cell death. (d) C-Myc inhibition by small molecular drugs could also possibly be effective in suppressing mTORC2-mediated cancer metabolism.
Metabolic influence on epigenetics – the next frontier
Metabolic reprogramming may exert some of its most important consequences by globally altering gene transcription [19,92]. Although the mechanisms underlying metabolic control of epigenetics are not well understood, a number of studies focusing on GBM have begun to shed light on the interconnection between these fundamental processes.
Mutations in IDH1/2 play an important role in gliomas through their effects on global transcription [20,93]. R132H IDH1 mutations cause the enzyme to acquire a neomorphic activity that converts α-ketoglutarate to 2-hydroxyglutarate [94], which inhibits the TET2 and the Jumonji-domain-containing protein 2A (JMJD2A/KDM4A) DNA demethylases, thus maintaining DNA in a hypermethylated state [95,96]. One current model posits that in the presence of IDH mutations, differentiation-related genes become “locked” in an inactive state, contributing to tumorigenesis [20,94,97]. IDH1 mutations occur largely in lower grade gliomas and in only a small fraction of adult de novo GBMs [21]. Also, the histone 3.3 mutations and the chromatin modifier mutations detected in pediatric gliomas are not common in adult de novo GBMs [21]. Taken together, these findings suggest that most adult GBMs, which are driven by RTK mutations and PI3K-AKT-mTOR pathway activation, utilize an alternative mechanism to hijack the epigenetic machinery.
In EGFR-mutant GBMs, constitutive PI3K activation could potentially engage the epigenetic machinery through a number of complementary routes. First, EGFR activation causes the glycolytic enzyme PKM2 to translocate to the nucleus where it phosphorylates histone 3 at Thr11, causing dissociation of HDAC3 to regulate transcription [98]. Acetylation on the N-terminal lysine tail of histones leads to an open chromatin configuration facilitating transcription [99]. Conversely, deacetylation of histones is associated with condensed chromatin and reduction of transcriptional activity. Thus, the balance between histone acetyltransferases (HATs), which transfer the acetyl group from acetyl-CoA to histone N-terminal lysines, and histone deacetylases (HDACs), including at specific genes, contribute to epigenetic regulation.
We, and others [19,92], speculate that the level of acetyl-CoA derived from metabolic reactions could have a profound effect on epigenetic regulation. Coordinated flux in the levels of acetyl-CoA, histone acetylation and global transcription with the yeast metabolic cycle suggest that HAT and HDAC activities are highly responsive to acetyl-CoA levels, ensuring that cells are appropriately responsive to their biochemical environment [19]. In cancer cells, glucose and other nutrients drive the production of acetyl-CoA through the TCA [100,101]. Further, cancer cells convert citrate into acetyl-CoA via the ATP citrate lyase to drive histone acetylation [102], suggesting that tumors may increase the production of acetyl-CoA as well as its demand. Therefore, it is possible that EGFR mutant GBMs may control epigenetic regulation by elevating the level of acetyl-CoA, while coordinately shifting the balance of HATs and HDACs towards one that supports tumor growth. This review has outlined three ways in which EGFR signaling could elevate acetyl-CoA levels in GBM through mTORC2: 1) by increasing glycolysis to generate pyruvate-derived acetyl-CoA; 2) through glutaminolysis to generate glutamine-derived acetyl-CoA, and 3) by uptake and β-oxidation of exogenous lipids to generate acetyl-CoA. Third, EGFR-PI3K-AKT-mTOR signaling may also alter global transcription in GBM through mTORC2-dependent upregulation of c-Myc, which has recently been shown to globally amplify transcripts from active genes. Taken together, these diverse pieces of evidence suggest a series of complementary routes by which EGFR-PI3K-AKT-mTOR signaling in tumors controls the epigenetic machinery. In the future, it will be critically important to dissect the molecular mechanisms tumors use to link metabolic reactions downstream of altered signaling with epigenetic control, enabling cells to rapidly adapt to a changing environment to ensure maximum tumor growth.
Concluding remarks and future perspectives
The specificity of cancer metabolic reprogramming lies in the coordination of responses that enable tumor cells to meet all of their needs in an entirely cell-autonomous fashion. Signal transduction inhibitors hold out the promise of much more effective, much less toxic treatments for cancer patients. However, that promise is unlikely to be realized until the consequences of cancer-causing mutations on metabolic reprogramming and epigenetic regulation are understood, including the flexible ways in which tumor cells adapt to changing conditions (such as drug treatment) to coordinately maintain the activity of downstream effectors necessary for tumor growth. Here, we have briefly reviewed the recent literature pointing to an unexpectedly important role for mTORC2 in cancer metabolic reprogramming, where it coordinates altered signal transduction with biochemical repertoires and potential transcriptional consequences that drive tumor growth and drug resistance. We have also highlighted a central role for c-Myc in that process. Understanding how cancer-causing mutations engage signaling pathways to coordinate repertoires of biochemical reactions linked to global transcription ensembles may yield critical insights into the pathogenesis of cancer, shed new light on how tumor cells resist targeted therapies to which they should be vulnerable, and possibly point the way towards more effective targeted cancer treatments. Much remains to be learned about how these cellular modules are flexibly integrated to maximize tumor growth, but the door is opening.
Box 1. Epidermal growth factor receptor variant III (EGFRvIII).
Among numerous studies to decipher the interactions between growth factors and their cognate receptors, four members of the ErbB family receptors, particularly ErbB1 (epidermal growth factor receptor or EGFR), have been the most vigorously investigated. EGFR is a membrane-spanning glycoprotein consisting of an extracellular domain (ECD) and a cytoplasmic domain with multiple tyrosine residues which are phosphorylated upon ligand binding and receptor activation. EGFR is a chief regulator of epithelial cell growth and its deregulation, often leading to the tumor formation, is the result of overexpression which is commonly associated with gene amplification and/or mutation [103]. Among the several reported tumorigenic mutations of EGFR, the most common, EGFRvIII (also known as de2-7 EGFR and ΔEGFR) which is characterized by an in-frame deletion of exons 2-7 and results in a constitutively active oncogenic form, occurs in the ECD [104]. As a result of the removal of 801 base pairs and subsequent 267 amino acids from the ECD, EGFRvIII exhibits a molecular weight of 145 kDa compared with that of 170 kDa for wild type EGFR [103], and can be detected by an antibody specific for EGFRvIII or PCR including an RT-PCR technique developed for EGFRvIII quantification in formalin-fixed paraffin-embedded samples [104]. EGFR amplification (or copy number increases of chromosome 7p12, the site of the EGFR gene) is a hallmark of several cancers including primary GBM, and about 50% of EGFR-amplified GBM express the ligand-independent truncated variant EGFRvIII [21,104]. The ensuing strong and persistent activation of downstream PI3K/AKT signaling provides advantages for cell survival, proliferation and motility. The prooncogenic effects of EGFRvIII are also mediated by several signaling pathways including Ras/MAPK and STAT3 [103]. Recently, EGFRvIII has been shown to activate mTORC2, which in turn activates NF-kB independently of AKT, causing resistance to chemotherapy [31]. The expression of EGFRvIII can affect the efficacy of cancer targeted therapies such as tyrosine kinase inhibitor (TKI). Expression of the constitutively active mutant EGFRvIII sensitizes tumors to EGFR inhibitors, but only if the PTEN tumor suppressor protein is intact because PI3K signal flux is sustained by PTEN deficiency [105]. Recent single-cell analyses using GBM patient-derived models and clinical samples revealed that resistance to EGFR TKI occurs by a surprisingly dynamic elimination and re-emergence of mutant EGFR (EGFRvIII) from extrachromosomal DNA (episomes), indicating a highly adaptive route by which cancers can circumvent therapies which target oncogenes [106].
Highlights.
Oncogenes reprogram cellular metabolism needed for rapid tumor growth
mTORC2 is a central regulator of cancer metabolic reprogramming
mTORC2 coordinates cancer metabolism through AKT-dependent and AKT-independent mechanisms
c-Myc is a critical effector of mTORC2-dependent metabolic reprogramming
Acknowledgements
This work is supported by grants from National Institute for Neurological Diseases and Stroke (NS73831), the National Cancer Institute (CA151819), The Ben and Catherine Ivy Foundation, Defeat GBM Research Collaborative, a subsidiary of National Brain Tumor Society, and generous donations from the Ziering Family Foundation in memory of Sigi Ziering. WKC is a Fellow of the National Foundation for Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vander Heiden MG, et al. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dang CV. Links between metabolism and cancer. Genes Dev. 2012;26:877–90. doi: 10.1101/gad.189365.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ray PD, et al. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell. Signal. 2012;24:981–990. doi: 10.1016/j.cellsig.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balaban RS, et al. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 7.García-Jiménez C, et al. A new link between diabetes and cancer: enhanced WNT/β-catenin signaling by high glucose. J. Mol. Endocrinol. 2014;52:R51–R66. doi: 10.1530/JME-13-0152. [DOI] [PubMed] [Google Scholar]
- 8.Turturro F, et al. Hyperglycemia regulates thioredoxin-ROS activity through induction of thioredoxin-interacting protein (TXNIP) in metastatic breast cancer-derived cells MDA-MB-231. BMC Cancer. 2007;7:96. doi: 10.1186/1471-2407-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cairns RA, et al. Regulation of cancer cell metabolism. Nat. Rev. Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 10.DeBerardinis RJ, Cheng T. Q's next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29:313–24. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baenke F, et al. Hooked on fat: the role of lipid synthesis in cancer metabolism and tumour development. Dis. Model Mech. 2013;6:1353–63. doi: 10.1242/dmm.011338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Currie E, et al. Cellular fatty acid metabolism and cancer. Cell Metab. 2013;18:153–61. doi: 10.1016/j.cmet.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffiths B, et al. Sterol regulatory element binding protein-dependent regulation of lipid synthesis supports cell survival and tumor growth. Cancer Metab. 2013;1:3. doi: 10.1186/2049-3002-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nieman KM, et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat Med. 2011;17:1498–503. doi: 10.1038/nm.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kidani Y, et al. Sterol regulatory element-binding proteins are essential for the metabolic programming of effector T cells and adaptive immunity. Nat. Immunol. 2013;14:489–99. doi: 10.1038/ni.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson CB. Rethinking the regulation of cellular metabolism. Cold Spring Harb. Symp. Quant. Biol. 2011;76:23–9. doi: 10.1101/sqb.2012.76.010496. [DOI] [PubMed] [Google Scholar]
- 17.Ciriello G, et al. Emerging landscape of oncogenic signatures across human cancers. Nat. Genet. 2013;45:1127–1133. doi: 10.1038/ng.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaelin WG, Jr., McKnight SL. Influence of metabolism on epigenetics and disease. Cell. 2013;153:56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu C, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–8. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brennan CW, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–77. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Babic I, et al. EGFR mutation-induced alternative splicing of Max contributes to growth of glycolytic tumors in brain cancer. Cell Metab. 2013;17:1000–8. doi: 10.1016/j.cmet.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo D, et al. The AMPK agonist AICAR inhibits the growth of EGFRvIII-expressing glioblastomas by inhibiting lipogenesis. Proc. Natl. Acad. Sci. U.S.A. 2009;106:12932–7. doi: 10.1073/pnas.0906606106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masui K, et al. mTOR complex 2 controls glycolytic metabolism in glioblastoma through FoxO acetylation and upregulation of c-Myc. Cell Metab. 2013;18:726–39. doi: 10.1016/j.cmet.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo D, et al. EGFR signaling through an Akt-SREBP-1-dependent, rapamycin-resistant pathway sensitizes glioblastomas to antilipogenic therapy. Sci. Signal. 2009;2:Ra82. doi: 10.1126/scisignal.2000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cloughesy TF, et al. Glioblastoma: from molecular pathology to targeted treatment. Annu. Rev. Pathol. 2014;9:1–25. doi: 10.1146/annurev-pathol-011110-130324. [DOI] [PubMed] [Google Scholar]
- 27.Cornu M, et al. mTOR in aging, metabolism, and cancer. Curr. Opin. Genet. Dev. 2013;23:53–62. doi: 10.1016/j.gde.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 28.Howell JJ, et al. A growing role for mTOR in promoting anabolic metabolism. Biochem. Soc. Trans. 2013;41:906–12. doi: 10.1042/BST20130041. [DOI] [PubMed] [Google Scholar]
- 29.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–93. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sarbassov DD, et al. Phosphorylation and regulation of Akt/PKB by the rictor–mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 31.Tanaka K, et al. Oncogenic EGFR signaling activates an mTORC2-NF-kB pathway that promotes chemotherapy resistance. Cancer Discov. 2011;1:524–538. doi: 10.1158/2159-8290.CD-11-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimada K, et al. TORC2 signaling pathway guarantees genome stability in the face of DNA strand breaks. Mol Cell. 2013;51:829–39. doi: 10.1016/j.molcel.2013.08.019. [DOI] [PubMed] [Google Scholar]
- 33.Read RD, et al. A drosophila model for EGFR- Ras and PI3K-dependent human glioma. PLoS Genet. 2009;5:e1000374. doi: 10.1371/journal.pgen.1000374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alessi DR, et al. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Balpha. Curr. Biol. 1997;7:261–9. doi: 10.1016/s0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- 35.Iwanami A, et al. Striking the balance between PTEN and PDK1: it all depends on the cell context. Genes. Dev. 1997;23:1699–704. doi: 10.1101/gad.1832909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pearce LR, et al. Protor-1 is required for efficient mTORC2-mediated activation of SGK1 in the kidney. Biochem. J. 2011;436:169–179. doi: 10.1042/BJ20102103. [DOI] [PubMed] [Google Scholar]
- 37.Oh WJ, Jacinto E. mTOR complex 2 signaling and functions. Cell Cycle. 2011;10:2305–16. doi: 10.4161/cc.10.14.16586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oh WJ, et al. mTORC2 can associate with ribosomes to promote cotranslational phosphorylation and stability of nascent Akt polypeptide. EMBO J. 2010;29:3939–51. doi: 10.1038/emboj.2010.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zinzalla V, et al. Activation of mTORC2 by Association with the Ribosome. Cell. 2011;144:757–68. doi: 10.1016/j.cell.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 40.Julien LA, et al. mTORC1-activated S6K1 phosphorylates Rictor on threonine 1135 and regulates mTORC2 signaling. Mol. Cell. Biol. 2010;30:908–21. doi: 10.1128/MCB.00601-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu P, et al. Sin1 phosphorylation impairs mTORC2 complex integrity and inhibits downstream Akt signaling to suppress tumorigenesis. Nat. Cell Biol. 2013;15:1340–1350. doi: 10.1038/ncb2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Humphrey SJ, et al. Dynamic adipocyte phosphoproteome reveals that Akt directly regulates mTORC2. Cell Metab. 2013;17:1009–1020. doi: 10.1016/j.cmet.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu P, et al. Dual phosphorylation of Sin1 at T86 and T398 negatively regulates mTORC2 complex integrity and activity. Protein. Cell. 2014;5:171–7. doi: 10.1007/s13238-014-0021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DeBerardinis RJ, et al. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 45.Koppenol WH, et al. Otto Warburg's contributions to current concepts of cancer metabolism. Nat. Rev. Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 46.Levine AJ, Puzio-Kuter AM. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science. 2010;330:1340–1344. doi: 10.1126/science.1193494. [DOI] [PubMed] [Google Scholar]
- 47.Dang CV. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harb. Perspect. Med. 2013;3 doi: 10.1101/cshperspect.a014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kress TR, et al. The MK5/PRAK kinase and Myc form a negative feedback loop that is disrupted during colorectal tumorigenesis. Mol. Cell. 2011;41:445–57. doi: 10.1016/j.molcel.2011.01.023. [DOI] [PubMed] [Google Scholar]
- 49.Deprez J, et al. Phosphorylation and activation of heart 6-phosphofructo-2-kinase by protein kinase B and other protein kinases of the insulin signaling cascades. J. Biol. Chem. 1997;272:17269–17275. doi: 10.1074/jbc.272.28.17269. [DOI] [PubMed] [Google Scholar]
- 50.Gottlob K, et al. Inhibition of early apoptotic events by Akt/PKB is dependent on the first committed step of glycolysis and mitochondrial hexokinase. Genes Dev. 2001;15:1406–1418. doi: 10.1101/gad.889901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kohn AD, et al. Expression of a constitutively active Akt Ser/Thr kinase in 3T3-L1 adipocytes stimulates glucose uptake and glucose transporter 4 translocation. J. Biol. Chem. 1996;271:31372–31378. doi: 10.1074/jbc.271.49.31372. [DOI] [PubMed] [Google Scholar]
- 52.Hagiwara A, et al. Hepatic mTORC2 activates glycolysis and lipogenesis through Akt, glucokinase, and SREBP1c. Cell Metab. 2012;15:725–38. doi: 10.1016/j.cmet.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 53.Dang CV, et al. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin. Cancer Res. 2009;15:6479–83. doi: 10.1158/1078-0432.CCR-09-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McFate T, et al. Pyruvate dehydrogenase complex activity controls metabolic and malignant phenotype in cancer cells. J. Biol. Chem. 2008;283:22700–8. doi: 10.1074/jbc.M801765200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang RH, et al. Hepatic Sirt1 deficiency in mice impairs mTorc2/Akt signaling and results in hyperglycemia, oxidative damage, and insulin resistance. J. Clin. Invest. 2011;121:4477–90. doi: 10.1172/JCI46243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boehmer C, et al. Properties and regulation of glutamine transporter SN1 by protein kinases SGK and PKB. Biochem. Biophys. Res. Commun. 2003;306:156–62. doi: 10.1016/s0006-291x(03)00921-5. [DOI] [PubMed] [Google Scholar]
- 57.Rosario FJ, et al. Mammalian target of rapamycin signalling modulates amino acid uptake by regulating transporter cell surface abundance in primary human trophoblast cells. J. Physiol. 2013;591:609–25. doi: 10.1113/jphysiol.2012.238014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Metallo CM, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2011;481:380–4. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mullen AR. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2011;481:385–8. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wise DR, et al. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of α-ketoglutarate to citrate to support cell growth and viability. Proc. Natl. Acad. Sci. U.S.A. 2011;108:19611–6. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lamming DW, Sabatini DM. A Central role for mTOR in lipid homeostasis. Cell Metab. 18:465–9. doi: 10.1016/j.cmet.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yuan M, et al. Identification of Akt-independent regulation of hepatic lipogenesis by mammalian target of rapamycin (mTOR) complex 2. J. Biol. Chem. 2012;287:29579–88. doi: 10.1074/jbc.M112.386854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cybulski N, et al. mTOR complex 2 in adipose tissue negatively controls whole-body growth. Proc. Natl. Acad. Sci. U. S. A. 2009;106:9902–7. doi: 10.1073/pnas.0811321106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yao Y, et al. BSTA promotes mTORC2-mediated phosphorylation of Akt1 to suppress expression of FoxC2 and stimulate adipocyte differentiation. Sci. Signal. 2013;6:ra2. doi: 10.1126/scisignal.2003295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jones KT, et al. Rictor/TORC2 regulates Caenorhabditis elegans fat storage, body size, and development through sgk-1. PLoS Biol. 2009;7:e60. doi: 10.1371/journal.pbio.1000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Soukas AA, et al. Rictor/TORC2 regulates fat metabolism, feeding, growth, and life span in Caenorhabditis elegans. Genes. Dev. 2009;23:496–511. doi: 10.1101/gad.1775409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Santos CR, Schulze A. Lipid metabolism in cancer. FEBS J. 2012;279:2610–2623. doi: 10.1111/j.1742-4658.2012.08644.x. [DOI] [PubMed] [Google Scholar]
- 68.Migita T, et al. ATP citrate lyase: activation and therapeutic implications in non-small cell lung cancer. Cancer Res. 2008;68:8547–54. doi: 10.1158/0008-5472.CAN-08-1235. [DOI] [PubMed] [Google Scholar]
- 69.Guo D, et al. An LXR agonist promotes glioblastoma cell death through inhibition of an EGFR/AKT/SREBP-1/LDLRdependent pathway. Cancer Discov. 2011;2:290–1. doi: 10.1158/2159-8290.CD-11-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nelson ER, et al. 27-Hydroxycholesterol links hypercholesterolemia and breast cancer pathophysiology. Science. 2013;342:1094–8. doi: 10.1126/science.1241908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brown MS, Goldstein JL. Cholesterol feedback: from Schoenheimer's bottle to Scap's MELADL. J. Lipid Res. 50:S15–27. doi: 10.1194/jlr.R800054-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Düvel K, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol. Cell. 2010;39:171–83. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Porstmann T, et al. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 2008;8:224–36. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zelcer N, et al. LXR regulates cholesterol uptake through Idol-dependent ubiquitination of the LDL receptor. Science. 2009;325:100–4. doi: 10.1126/science.1168974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- 76.Ramos-Montoya A, et al. Pentose phosphate cycle oxidative and nonoxidative balance: a new vulnerable target for overcoming drug resistance in cancer. Int. J. Cancer. 2006;119:2733–2741. doi: 10.1002/ijc.22227. [DOI] [PubMed] [Google Scholar]
- 77.Kliegman JI, et al. Chemical genetics of rapamycin-insensitive TORC2 in S. cerevisiae. Cell Rep. 2013;5:1725–36. doi: 10.1016/j.celrep.2013.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schulze A, Harris AL. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature. 2012;491:364–73. doi: 10.1038/nature11706. [DOI] [PubMed] [Google Scholar]
- 79.Biggs WH, 3rd, et al. Protein kinase B/Akt-mediated phosphorylation promotes nuclear exclusion of the winged helix transcription factor FKHR1. Proc. Natl. Acad. Sci. U.S.A. 1999;96:7421–7426. doi: 10.1073/pnas.96.13.7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bouchard C, et al. Myc-induced proliferation and transformation require Akt-mediated phosphorylation of FoxO proteins. EMBO J. 2004;23:2830–40. doi: 10.1038/sj.emboj.7600279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Delpuech O, et al. Induction of Mxi1-SR alpha by FOXO3a contributes to repression of Myc-dependent gene expression. Mol. Cell. Biol. 2007;27:4917–4930. doi: 10.1128/MCB.01789-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gan B, et al. FoxOs enforce a progression checkpoint to constrain mTORC1-activated renal tumorigenesis. Cancer Cell. 2010;18:472–84. doi: 10.1016/j.ccr.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Peck B, et al. Antagonism between FOXO and MYC Regulates Cellular Powerhouse. Front. Oncol. 2013;3:96. doi: 10.3389/fonc.2013.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen Z, et al. A murine lung cancer co-clinical trial identifies genetic modifiers of therapeutic response. Nature. 2012;483:613–7. doi: 10.1038/nature10937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Holland JP, et al. Annotating MYC status with 89Zr-transferrin imaging. Nat. Med. 2012;18:1586–91. doi: 10.1038/nm.2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Qu W, et al. Preparation and characterization of L-[5-11C]-glutamine for metabolic imaging of tumors. J. Nucl. Med. 2012;53:98–105. doi: 10.2967/jnumed.111.093831. [DOI] [PubMed] [Google Scholar]
- 87.Grassi I, et al. The clinical use of PET with (11)C-acetate. Am. J. Nucl. Med. Mol. Imaging. 2012;2:33–47. [PMC free article] [PubMed] [Google Scholar]
- 88.Palaskas N, et al. 18F-fluorodeoxy-glucose positron emission tomography marks MYC-overexpressing human basal-like breast cancers. Cancer Res. 2011;71:5164–74. doi: 10.1158/0008-5472.CAN-10-4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shi J, et al. Small molecule inhibitors of Myc/Max dimerization and Myc-induced cell transformation. Bioorg. Med. Chem. Lett. 2009;19:6038–41. doi: 10.1016/j.bmcl.2009.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Alderton GK. Targeting MYC? You BET. Nat. Rev. Cancer. 2011;11:693. doi: 10.1038/nrc3147. [DOI] [PubMed] [Google Scholar]
- 91.Delmore JE, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–17. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lu C, Thompson CB. Metabolic regulation of epigenetics. Cell Metab. 2012;16:9–17. doi: 10.1016/j.cmet.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Turcan S, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–83. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xu W, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chowdhury R, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12:463–469. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Venneti S, Thompson CB. Metabolic modulation of epigenetics in gliomas. Brain Pathol. 2013;23:217–21. doi: 10.1111/bpa.12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yang W, et al. PKM2phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell. 2012;150:685–96. doi: 10.1016/j.cell.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Clayton AL, et al. Enhanced histone acetylation and transcription: a dynamic perspective. Mol. Cell. 2006;23:289–296. doi: 10.1016/j.molcel.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 100.Maher EA, et al. Metabolism of [U-13 C]glucose in human brain tumors in vivo. NMR Biomed. 2012;25:1234–44. doi: 10.1002/nbm.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Marin-Valencia I, et al. Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell Metab. 2012;15:827–37. doi: 10.1016/j.cmet.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wellen KE, et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–80. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gan HK, et al. The epidermal growth factor receptor variant III (EGFRvIII): where wild things are altered. FEBS J. 2013;280:5350–70. doi: 10.1111/febs.12393. [DOI] [PubMed] [Google Scholar]
- 104.Masui K, et al. Review: molecular pathology in adult high-grade gliomas: from molecular diagnostics to target therapies. Neuropathol. Appl. Neurobiol. 2012;38:271–91. doi: 10.1111/j.1365-2990.2011.01238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mellinghoff IK, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N. Engl. J. Med. 2005;353:2012–24. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 106.Nathanson DA, et al. Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science. 2014;343:72–6. doi: 10.1126/science.1241328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Locasale JW, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nature Genet. 2011;43:869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Possemato R, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Noh S, et al. Expression levels of serine/glycine metabolism-related proteins in triple negative breast cancer tissues. Tumour Biol. 2014 doi: 10.1007/s13277-013-1588-z. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]