Abstract
Metabolic homeostasis is achieved through coordinated regulation across several tissues. Studies using mouse genetic models have shown that perturbation of specific pathways of lipid metabolism in metabolically active tissues impacts systemic metabolic homeostasis. The use of metabolomic technologies combined with genetic models has helped identify several potential lipid mediators that serve as metabolic messengers to communicate energy status and modulate substrate utilization among tissues. When provided exogenously, these lipid metabolites exhibit biological effects on glucose and lipid metabolism, implicating a therapeutic potential for treating metabolic diseases. In this review, we will summarize recent advances in inter-organ communication through novel mechanisms with a focus on lipid mediators synthesized de novo or derived from dietary sources and discuss challenges and future directions.
Keywords: Lipid metabolites, lipogenesis, Tregs, PPARs, metabolic flexibility, BAIBA
Metabolic flexibility in energy metabolism
Obesity is a growing health issue that is associated with a collection of metabolic disorders, such as dyslipidemia, insulin resistance and cardiovascular diseases. The underlying mechanism for how the obese state is causative to metabolic diseases is multifaceted and has been reviewed extensively [1-4]. Significant effort has focused on the pathological effect of obesity-induced ectopic fat deposition, or lipotoxicity, characterized by accumulation of lipid intermediates in non-adipose tissue [3, 4]. This leads to cellular dysfunction and diminished efficiency and flexibility in energy metabolism, thereby establishing a metabolic vicious cycle.
Metabolic flexibility can be described as the body’s capacity to switch between carbohydrates and lipids as the predominant source of energy substrates [5]. After a meal, elevated blood glucose level triggers insulin release by pancreatic beta cells. The anabolic action of insulin promotes glucose uptake and utilization by insulin-sensitive tissues while suppressing fatty acid release from white adipose tissue. Surplus glucose is converted to fat via de novo lipogenesis in the liver and exported to white adipose tissue for long-term storage. Conversely, a decline in the blood glucose level during the fasted state dampens insulin secretion and enhances the action of counter-regulatory hormones that facilitate fatty acid release from adipose tissue to be used as a major energy source [6].
While it is indisputable that insulin is a key regulator of substrate switching as seen in the fasting/feeding cycle, metabolic flexibility appears to be applicable to a broader range of physiology. As discussed in detail below, after a period of caloric restriction, food intake leads to a transient up-regulation of adipose tissue lipogenesis and suppression of hepatic lipogenesis [7]. On the other hand, endurance exercise not only enhances fatty acid burning in the muscle but also in adipose tissue and the liver [8]. How these metabolic states are achieved is under active investigation. Nevertheless, cumulative evidence suggests the existence of additional metabolic signals that serve as messengers among metabolically active tissues to orchestrate coordinated control of energy metabolism. The seminal work by Randle et al. (Box 1), which demonstrated altered glucose uptake in isolated rat muscle upon fatty acid perfusion in the absence of insulin, suggests that fatty acids or their derivatives could be potential regulators of metabolic flexibility [9].
Box 1. De novo lipogenesis, lipotoxicity and Randle hypothesis.
Fatty acids are a major component of dietary macronutrients, normally stored in the form of triacylglyerols, which provide energy substrates for cellular metabolism. With the exception of a few essential fatty acids, such as linoleic acid, the human body can synthesize fatty acids de novo from carbohydrates. Products of de novo lipogenesis are building blocks for complex lipids such as triglycerides and phospholipids. The breakdown intermediates of these lipids accumulate in obesity to generate several classes of lipid metabolites implicated in mediating lipotoxicity. Randle and colleagues proposed a glucose-fatty acid cycle hypothesis, in which fat oxidation in the mitochondria generates metabolic signals that block glucose usage through inhibition of glycolytic enzymes [9]. This basic concept of lipotoxicity has been expanded and several underlying mechanisms have been proposed [3]. For example, fatty acids have been shown to activate c-Jun N-terminal kinases, which suppress insulin signaling through inhibitory phosphorylation of insulin receptor substrates 1/2. The metabolites of fatty acids, including diacylglycerol, ceramides and acyl-carnitines, have all been suggested to decrease insulin sensitivity via different signaling pathways [3, 4].
Here we describe recent discoveries of novel mechanisms mediated by lipid metabolites that are capable of modulating metabolic flexibility and other key metabolic pathways. They are proposed to serve as long-range hormones but can act in a paracrine manner to integrate metabolic processes between tissues or cells. The therapeutic potential of these lipid mediators and the challenges in studying their functions in physiological contexts will also be discussed.
Lipogenic pathways and systemic metabolic homeostasis
While animals can usually acquire a sufficient amount of fatty acids through dietary sources [10], mouse genetic models have suggested an essential role of the de novo lipogenic pathway. Whole body knockout of acetyl-CoA carboxylase 1 (Acc1, or Acaca) or fatty acid synthase (Fasn), two rate-limiting enzymes of de novo lipogenesis (Figure 1), is embryonic lethal [11, 12]. On the other hand, essential fatty acids are required for normal physiology by virtue of being precursors of signaling molecules such as eicosanoids [4], as will be discussed later.
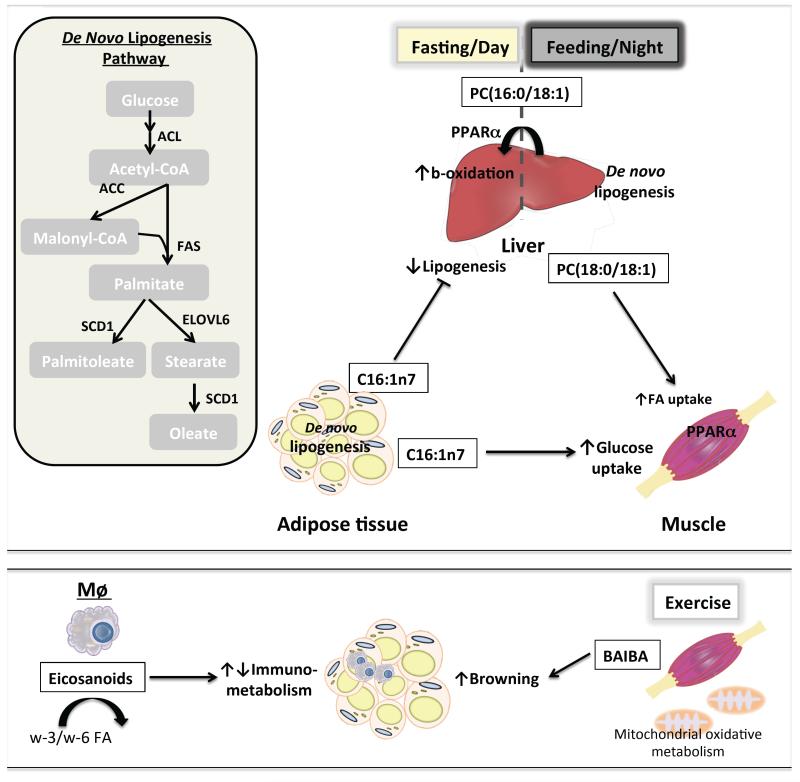
Figure 1. Lipid metabolites function as long-range hormones to regulate systemic energy metabolism.
Top panel: de novo lipogenesis is controlled by several rate-limiting enzymes that convert acetyl-CoA into palmitate (C16:0), palmitoleate (C16:1), stearate (C18:0) and oleate (C18:1). These fatty acids provide building blocks for complex lipids such as triglycerides and phospholipids. In the liver, de novo lipogenesis increases circulating concentrations of PC(18:0/18:1) following feeding to promote fatty acid uptake by the muscle, an activity requiring intact PPARα signaling. Furthermore, hepatic synthesis of PC(16:0/18:1) during the dark cycle may later act as a PPARα ligand during the light cycle to increase oxidation of fatty acids. De novo synthesis of palmitoleate by adipose is conducive to increased glucose uptake by muscle and decreased lipogenesis in the liver. FA: fatty acid; ACL: ATP citrate lyase; ACC: acetyl-CoA carboxylase; FAS: fatty acid synthase; SCD1: stearoyl-CoA desaturase 1; ELOVL6: elongation of long-chain fatty acids family member 6. Lower panel: eicosanoids, which include both pro- and anti-inflammatory lipid mediators, are synthesized from essential fatty acids (e.g., ω-3 and ω-6 fatty acids). They are primarily produced by immune cells such as tissue resident macrophages (Mϕ). Resolvins RvD1 and RvD2 have been shown to enhance adiponectin expression and suppress meta-inflammation in white adipose tissue. Additionally, β-aminoisobutyric acid (BAIBA) is a by-product of the valine degradation and the fatty acid oxidation pathway in muscle. Its circulating level is increased during endurance exercise. BAIBA has been implicated as an exercise-induced myokine that promotes white adipose tissue browning.
Although excessive lipid synthesis is causative to metabolic diseases such as hepatic steatosis [13], de novo lipogenesis is not always associated with pathology. In the liver, de novo lipogenesis promotes the sequestration of detrimental lipid species in lipid droplets, thereby limiting hepatic lipotoxicity [14]. The insulin-sensitizing effect of the thiazolidinedione class of drugs, high affinity ligands for the nuclear receptor peroxisome proliferator-activated receptor gamma (PPARγ), is in part mediated by the lipogenic effect of PPARγ activation in adipose tissue (Box 2) [15]. While these localized beneficial activities of de novo lipogenesis are well documented, studies derived from mouse genetic models seem to suggest that the effects of de novo lipogenic programs extend beyond the tissue boundary.
Box 2. PPARs and lipid metabolism.
Peroxisome proliferator-activated receptors (PPARs) belong to the nuclear receptor superfamily. They are transcription factors whose activities can be modulated by dietary lipids, notably fatty acids. The three PPARs, PPARα, PPARβ/δ and PPARγ, show distinct tissue distributions and regulate various aspects of lipid metabolism [71]. The best-described functions for these receptors include the adipogenic and insulin-sensitizing effects of PPARγ and regulation of fatty acid catabolism/mitochondrial oxidative metabolism by PPARα and PPARδ in the liver and muscle, respectively. In the liver, PPARα and PPARδ exhibit opposing activities in the control of diurnal lipid metabolism. PPARα is up-regulated in the fasted state to regulate fat catabolism. By contrast, PPARδ is most active at the fed state and controls the transcription of lipogenic genes.
Forced glucose uptake by adipose tissue through fat-specific over-expression of the glucose transporter Glut4 (Slc2a4) activates the transcription factor carbohydrate response element binding protein β (ChREBP-β, also called Mlxipl), a protein that, in a glucose-dependent manner, increases the transcription of lipogenic genes in adipose tissue. This leads to an improvement not only in the metabolic profile of adipose tissue but also in peripheral insulin-stimulated glucose uptake [16]. On the other hand, adipose tissue-specific deletion of Glut4 impairs insulin sensitivity in both muscle and liver [17]. Similarly, mice with a double knockout of fatty acid binding proteins Fabp4 and Fabp5 exhibit altered cellular and systemic lipid transport and composition with enhanced adipose tissue lipogenesis and protection from diet-induced obesity [18]. Activation of de novo lipogenesis in the liver also promotes metabolic changes in other tissues. For example, hepatic overexpression of glucokinase or 6-phosphofructo-2-kinase, which drives glucose flux for hepatic lipogenesis, concomitantly promotes muscle fatty acid oxidation [19]. In addition, liver-specific knockout of stearic-CoA desaturase 1 (Scd1), an enzyme that catalyzes a rate-limiting step in the synthesis of unsaturated fatty acids, protects mice from high carbohydrate diet-induced metabolic disorders as a consequence of suppressed hepatic lipid accumulation and gluconeogenesis and reduced adipose tissue weight [20].
These early studies provide clues for a link between the lipogenic program and systemic metabolic responses, although how tissue crosstalk in these genetic models is mediated is mostly unclear. The common feature of perturbed de novo lipogenesis underlying these mice models points to an unexpected role for lipogenic pathways in the maintenance of systemic metabolic homeostasis. It also raises the possibility that blood-borne factors, likely lipid metabolites, can serve as signaling molecules for inter-organ communication to achieve coordinated energy substrate utilization. In fact, metabolic improvements, including reduced hepatic lipogenesis and enhanced insulin actions in muscle in Fabp4/5 double knockout mice was shown to be stimulated by a lipogenic product originated from adipose tissue (discussed below) [21]. In the following section, we discuss recent studies that identify bioactive lipids synthesized de novo or derived from dietary fats implicated in such communication.
Lipid metabolites as signaling molecules mediating tissue crosstalk
Liver
Despite ample genetic evidence implicating the hepatic lipogenic pathway in metabolic homeostasis, few lipid mediators have been identified. A notable observation came from hepatic Fasn knockout mice. These mice developed hypoglycemia and fatty liver under a fat free diet, phenotypes resembling nuclear receptor PPARα deficiency in the liver. Furthermore, these metabolic defects can be rescued with a synthetic PPARα agonist, suggesting that Fasn is required to produce endogenous PPARα ligands under fat-free conditions [22]. In a follow-up study, mass spectrometry profiling of liver extracts from wild type or Fasn knockout mice was performed to screen for lipids bound to PPARα [23]. Specifically, phosphocholine PC(16:0/18:1) was identified as a putative PPARα ligand (Figure 1). In fact, PC(16:0/18:1) infusion through the portal vein increased fatty acid oxidation in a PPARα-dependent manner. However, it is interesting to note that in mice, Fasn and lipogenesis are most active in the dark (feeding) cycle, while PPARα is known to control fat catabolism in the light cycle. Thus, further investigation is needed with regard to the role of PC(16:0/18:1) as a PPARα ligand in physiologic feeding/fasting cycles.
To explore how hepatic de novo lipogenesis is able to affect systemic metabolic homeostasis, Liu et al. [24, 25] utilized genetic models perturbing the activity of PPARδ, which is identified as a key regulator of diurnal de novo lipogenesis in the liver partly through transcriptional regulation of Acc1. Overexpression of PPARδ improved whole-body metabolic homeostasis and insulin sensitivity. While suppression of hepatic gluconeogenesis via AMP-activated protein kinase is partly responsible for the metabolic benefits, the authors also found a paradoxical reduction of serum lipids, corresponding to enhanced muscle fatty acid burning. To investigate whether this is a result of direct signaling events rather than changes in serum lipid contents, transient knockdown of the PPARδ target Acc1 was performed, which resulted in reduced fatty acid uptake in the muscle despite elevated serum lipid levels. This suggests that changes in total serum lipids are a consequence of direct modulation of muscle fatty acid metabolism. Consequently, muscle fatty acid uptake was shown to exhibit a diurnal rhythm with increased uptake in the dark cycle when PPARδ-controlled lipogenesis in the liver is most active. This diurnal activity was abolished in liver-specific PPARδ knockout mice. Through unbiased lipidomic profiling (Box 3) of serum or liver samples from mice with hepatic PPARδ activation/deletion or Acc1 knockdown, the authors identified PC(18:0/18:1), or SOPC, to be most significantly correlated with altered muscle fatty acid uptake phenotypes. In vitro or in vivo delivery of this lipid enhanced muscle cell fatty acid uptake. Interestingly, this effect requires intact PPARα signaling in the muscle, indicating that PC(16:0/18:1) and PC(18:0/18:1) share similar biological effects on PPARα activation. Therefore, functional identification of PC(16:0/18:1) and PC(18:0/18:1) suggests that these lipogenic products may provide signals for postprandial fat utilization in the liver and muscle, respectively, when excess glucose has been converted to fats.
Box 3. Metabolomic approaches to identify bioactive lipid metabolites.
The quest to identify lipid signaling molecules has proven to be challenging in the past, mainly due to the lack of methods to survey and annotate lipid molecules from complex biological samples. The rapid development of lipidomics and metabolomics in the past decade has enabled researchers to start understanding the role of specific lipids in metabolism and physiology. A branch of lipidomics mainly focuses on the fatty acyl-chain composition of major lipid classes in a targeted manner, using enzymatic digestion products or direct fragmentation in mass spectrometer. These methods can unequivocally detect and accurately quantify fatty acids but lack the ability to identify unknown lipid species. Liquid chromatography mass spectrometry (LC-MS) based unbiased lipidomics takes unfractionated lipid extracts from biological samples and assigns a retention time and an accurate mass to each species, allowing a global survey of lipids with known and unknown identities. The limitations of this approach include the lack of definitive identification of lipid species, limited sensitivity owing to detection of a broad spectrum of lipid species and the need to develop additional methods to validate and quantify identified lipids
Adipose Tissue
It has long been recognized that adipose tissue is an endocrine organ capable of producing a number of adipokines with beneficial (e.g. leptin, adiponectin and interleukin-13) or detrimental (e.g. Tumor necrosis factor alpha and resistin) effects [26, 27]. Primarily viewed as an organ specialized to take up fatty acids from diet or synthesized in the liver, adipose tissue de novo lipogenesis is qualitatively less important in ad libitum feeding. However, it is significantly up-regulated under caloric restriction, suggesting a role in caloric restriction-associated metabolic benefits [7].
The aforementioned genetic models that exhibit altered adipose tissue lipogenesis present intriguing new opportunities to identify lipid mediators important for metabolic flexibility. Based on the observation that Fabp4 and Fabp5 double knockout mice have improved systemic metabolic homeostasis [18], Cao et al. [21] asked whether specific lipids mediated the beneficial effects of these mice. They quantified the fatty acyl chain composition of major lipid classes from the adipose tissue, muscle, liver and serum of wild type and the Fabp4/Fabp5 double knockout mice. Extensive and detailed bioinformatics analysis revealed that the fatty acid palmitoleate (C16:1n7), either in its free form or incorporated into other lipid classes, was the most statistically significant fatty acid down-regulated by high fat diet and up-regulated by genetic ablation of Fabp4/5. The authors suggest that palmitoleate is a lipid signaling molecule and term it a lipokine. Mechanistically, palmitoleate suppresses hepatic lipogenesis and enhances insulin action in the muscle, thereby contributing to the improved whole-body glucose disposal rate of Fabp4/5 double knockout mice on high fat diet. The molecular mechanism leading to enhanced insulin sensitivity in the muscle by palmitoleate remains unclear. Interestingly, palmitoleate alone does not enhance insulin action. Instead, it counters the deleterious effects of palmitate on insulin signaling. These data raise the possibility that lipid composition as a whole rather than a particular lipid constitutes a signal to modulate insulin response. A recent study found that altered plasma membrane fluidity and enhanced lipid raft-associated src family kinase activation play a role in saturated fatty acid induced insulin resistance, which can be suppressed by monounsaturated fatty acids such as palmitoleate and oleate [28]. Whether a similar mechanism is responsible for crosstalk between adipose tissue and muscle through the lipokine requires further study.
Immune cells
Among the most classically described bioactive lipid signaling molecules are the eicosanoids, a class of autocoid regulators of inflammatory response secreted primarily by immune cells including tissue macrophages, dendritic cells, neutrophils and mast cells [29]. Eicosanoids are synthesized from essential fatty acids rather than from products of de novo lipogenesis. Prostaglandins and leukotrienes belong to the “classical eicosanoids” and are derived from the omega-6 fatty acid arachidonic acid via the cyclooxygenase pathway. Although eicosanoids are often short-lived and act locally in a paracrine manner, they play an integral role in the intercommunication of immune cell populations and the vasculature to coordinate inflammatory response and leukocyte trafficking [30-32].
By contrast, pro-resolving/anti-inflammatory lipid mediators include lipoxins, which are similarly derived from arachidonic acid, and two novel classes of lipids, the resolvins and protectins, derived from either of the omega-3 polyunsaturated fatty acids eicosapentaenoic acid and docosahexanoic acid (DHA) [33, 34]. Interestingly, a deficit in pro-resolving lipid signaling has been reported in chronic low-grade inflammation associated with obesity. Lipidomic analyses of adipose tissue samples taken from humans and mice have identified a reduction in the synthesis of the DHA-derived resolvins RvD1 and RvD2 in obese fat [35]. Treatment of murine adipose tissue explants with either RvD1 or RvD2 enhanced adiponectin expression and secretion and attenuated the production of proinflammatory cytokines including leptin, TNFα, IL-6 and IL-1β. RvD1 and RvD2 are additionally capable of blunting murine monocyte migration and adhesion to adipocytes in response to the chemoattractant molecules monocyte chemotactic protein 1 and leukotriene B4 in vitro. While these observations are in line with the notion that resident immune cells regulate the metabolic set point of adipose tissues [36], mechanisms dictating the conversion of essential fatty acids to pro- or anti-inflammatory lipid mediators in the context of energy metabolism are not clear.
Muscle
Although the current review primarily focuses on the products of lipogenesis, it is noteworthy that their metabolites can also serve as signaling molecules. Using metabolomics, Roberts et al. [37] identified muscle derived β-aminoisobutyric acid (BAIBA), a product of the valine degradation and the fatty acid oxidation pathway, as a myokine that may convey signals to other tissues during exercise. BAIBA is enriched in the muscle of mice overexpressing peroxisome proliferator-activated receptor-gamma coactivator-1α (PGC-1α), a transcriptional co-activator that regulates metabolism and muscle fiber type switching in skeletal muscle and contributes to the response of muscle to exercise. Intramuscular and circulating BAIBA levels are increased after endurance exercise in humans and rodents. In vitro and in vivo delivery of BAIBA to adipocytes or in mice enhances white adipose tissue browning as evident by increased expression of brown adipocyte-specific genes. In addition, BAIBA stimulated fatty acid oxidation in hepatocytes. Collectively, these data identify BAIBA as a muscle-derived factor and a potential mechanism through which endurance exercise may improve whole-body metabolic homeostasis.
Gut
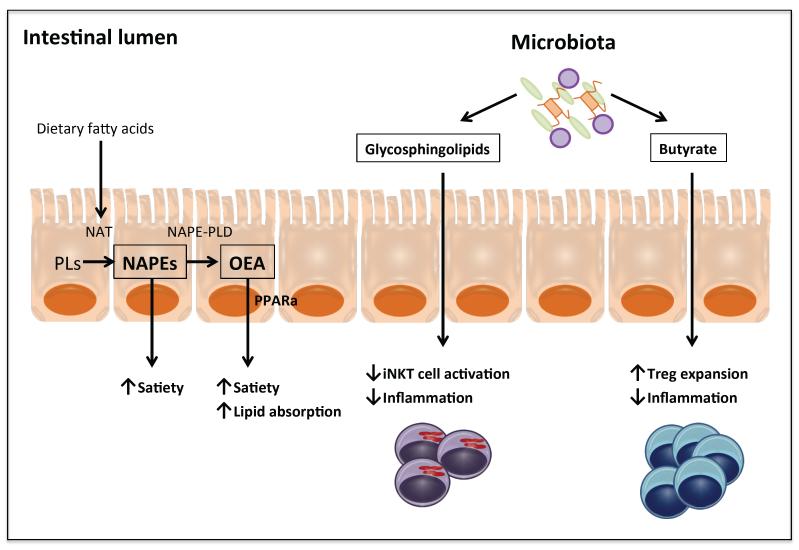
The gut-adipose-brain axis is central in balancing energy intake with expenditure. Several gut-derived hormones such as glucagon-like peptide 1 and cholecystokinin are known to facilitate digestion and absorption of food components or act as a feedback mechanism to control food intake [38]. The brain can also directly sense glucose and amino acids levels to regulate feeding behavior [39, 40]. Ingestion of fat induces satiation. However, plasma fatty acid levels do not affect feeding behavior, suggesting that dedicated lipid hormones may be responsible for these effects. This notion has led to the discovery of two related lipid species: oleylethanolamide (OEA) and N-acylphosphatidylethanolamine (NAPE) (Figure 2). Uptake of dietary fatty acids by enterocytes activates N-acyltransferases that convert lipids into NAPEs [41]. The subsequent action of NAPE phospholipase D (NAPE-PLD) converts NAPEs into fatty acyl ethanolamides, such as OEA [42]. OEA has been shown to enhance across-meal satiety and promote intestinal lipid absorption [42]. It acts locally through PPARα [43]. A secondary signaling cascade mediated by the neuropeptide oxytocin is also proposed to induce satiation in the brain [44]. By contrast, NAPEs suppress food intake through direct actions in the hypothalamus, as centrally administered NAPE is effective to suppress food intake and its effect remains intact in vagotomized rats [45]. Interestingly, upon intralipid infusion the most increased NAPE is palmitoyl-NAPE, despite palmitate-containing lipids not being the most enriched in the intralipid emulsion [46]. The selectivity of NAPE synthesis suggests either palmitate-containing lipids are preferentially taken up or the presentation of lipids per se induces de novo synthesis of palmitate.
Figure 2. Gut/microbiota-derived lipid mediators.
N-acylphosphatidylethanolamines (NAPEs) and oleylethanolamide (OEA) are gut-derived lipid metabolites shown to promote satiety. Uptake of dietary fatty acids by enterocytes activates N-acyltransferases (NAT) to convert phospholipids (PLs) into NAPEs. The subsequent action of NAPE phospholipase D (NAPE-PLD) converts NAPEs into fatty acyl ethanolamides, such as OEA. OEA has been shown to promote satiety and intestinal lipid absorption. It acts locally through PPARα. By contrast, NAPEs suppress food intake through direct actions in the hypothalamus. Gut microbiota can also produce lipid metabolites that have immunomodulatory activities. Butyrate and glycosphingolipids produced by gut bacteria have been shown to induce colonic regulatory T (Treg) cells and repress natural killer T (NKT) cells, respectively, to modulate colonic inflammation. Both Treg and NKT cells have been found in white adipose tissues and are believed to modulate meta-inflammation and progression of metabolic diseases.
Although it may not seem to fit the scope of inter-organ communication, the gut microbiome is arguably an integral component of human body. The health of microbiota affects metabolism [47-50], inflammation [51], cancer [52, 53] and circadian rhythms [54]. Recent studies demonstrate that microbe-derived lipids are capable of modulating host immune responses. Notably, butyrate and glycosphingolipids produced by gut bacteria have been shown to induce colonic regulatory T (Treg) cells and repress natural killer T (NKT) cells, respectively, to modulate colonic inflammation [51, 55-57]. Both Treg and NKT cells are recruited to white adipose tissues and modulate meta-inflammation and progression of metabolic diseases [58-60].
Exploring the therapeutic potential of novel lipid mediators
To understand the therapeutic potentials of the lipid molecules discussed above, important questions have to be addressed regarding their synthesis, delivery and mechanism of action. While de novo lipogenesis appears to be a common upstream pathway for many of the signaling molecules discussed, activation or suppression of this pathway leads to changes in a few fatty acid species [24, 25]. How the seemingly generic perturbation results in changes of a specific molecule but not related species remains unclear. In the case of PC(18:0/18:1), no enzyme is known to specifically synthesize this lipid molecule. PC can be synthesized de novo through the Kennedy pathway using diacylglycerols (DAGs) [61]. This pathway preferentially accommodates several substrates including DAG(16:1/16:1), DAG(18:1/18:1) and DAG(16:0/18:1) [62]. The diverse side chain composition of PCs is mainly generated through a remodeling process, termed the Lands cycle [63], in which the sn-2 position is removed by phospholipases and a new acyl chain is added by lysophosphocholine acyltransferases (LPCATs). In the liver, the major LPCAT activity is believed to come from Lpcat3. Lpcat3 prefers to add arachidonic, linoleic and oleic acid side chain to the sn-2 position [64]. It is possible that the combination of a particular lipid profile with these limited but not strict substrate preferences enriches the production of selective lipid species. In addition, genetic ablation of different hepatic lipogenic genes induces similar but not identical global metabolic changes, suggesting the existence of perhaps several lipid signaling pathways.
Charged lipophilic molecules rarely circulate freely in the blood. How some of the aforementioned lipid metabolites are delivered to target tissues has not been examined. It is tempting to speculate that uncharacterized chaperones are responsible for the delivery of these lipid mediators. What should not be overlooked, however, is the possibility that they simply incorporate into known deliverable forms such as albumin-fatty acids complex and lipoprotein particles. The release of fatty acids from adipose tissue or the production of lipoproteins from liver is associated with a particular metabolic state, in which context these lipid mediators may be effective. The molecular actions of several lipid mediators seem to converge at the activation of PPARα. However, it is unlikely that all these lipid mediators are direct PPARα ligands. Although empirical evidence shows a rapid enrichment of exogenously added lipids in the nucleus [23], PCs and fatty acids are not known to cross plasma membrane through passive diffusion. Unlike classical lipophilic hormones such as glucocorticoids, the baseline concentration is high and the dynamic range is small for some of these lipids [24, 45]. Our knowledge for how small changes can elicit strong physiological effects is limited. The mechanisms for the apparent tissue specificity of these lipid mediators are also not known. Identifying the cell surface receptors or transporters for lipid mediators, if they indeed exist, will shed light into mechanistic insights and provide therapeutic targets for developing drugs to treat metabolic disorders.
Concluding remarks and future perspectives
The few examples summarized here represent exciting new developments in our understanding of how tissues talk to each other to achieve metabolic homeostasis. The use of genetic models with strong phenotypes is key to uncovering underlying lipid signaling molecules. The rationale is that they mimic certain physiological states. Whether the identified lipid molecules indeed function within such physiological contexts requires rigorous investigation. Unlike gene function that can be revealed with genetic models, in vivo validation of tissue origins and metabolic functions of putative lipid signaling molecules remains challenging. Nevertheless, the development of bioinformatics tools that facilitate the annotation of metabolomics profiling has made it easier to identify novel lipid mediators [65-68]. Recently, several peptide signaling molecules have been shown to modulate specific tissue crosstalk [69, 70]. With the discovery of new circulating peptides and lipids, we will soon have a fuller picture of how metabolic flexibility in energy substrate utilization is communicated among metabolically active tissues.
Highlights.
Metabolites of key lipogenic pathways generate long-range metabolic messengers
Several lipid metabolites are shown to coordinate energy metabolism among tissues
Novel mechanisms of tissue crosstalk are identified through these lipid metabolites
Lipid mediators provide new therapeutic opportunities to treat metabolic diseases
Table 1.
List of putative lipid mediators implicated in inter-tissue communication
| Tissue source | Lipid mediator | Mechanism | Target tissue | Metabolic function | REF |
|---|---|---|---|---|---|
| Liver | PC(16:0/18:1) |
de novo lipogenesis |
liver | promote FA oxidation |
[23] |
| Liver | PC(18:0/18:1) |
de novo lipogenesis |
muscle | promote FA uptake/oxidation |
[24] |
| Adipocyte | C16:1n7 |
de novo lipogenesis |
liver, muscle | improve insulin sensitivity |
[21] |
| Immune cells | eicosanoids | derived from essential FA |
various tissues | modulate immuno- metabolism |
|
| Muscle | β-aminoisobutyric acid |
valine degradation/ β-oxidation pathway |
white adipose tissue, liver |
promote adipocyte browning, FA oxidation |
[37] |
| Gut | NAPEs, OEA | gut, brain | suppress food intake |
[42- 45] |
|
| Gut/Microbiota | short-chain FA (e.g., butyrate) |
immune cells | Treg cell differentiation |
[51, 56-77] |
|
| Gut/Microbiota | sphingolipids | immune cells | modulate NKT cell activity |
[55] |
Acknowledgments
Work in the laboratory of the authors is supported by National Institutes of Health grant R01DK075046, American Diabetes Association grant 1-14-BS-122 and American Heart Association.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Vernochet C, Kahn CR. Mitochondria, obesity and aging. Aging. 2012;4:859–860. doi: 10.18632/aging.100518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gregor MF, Hotamisligil GS. Inflammatory mechanisms in obesity. Annual review of immunology. 2011;29:415–445. doi: 10.1146/annurev-immunol-031210-101322. [DOI] [PubMed] [Google Scholar]
- 3.Samuel VT, Shulman GI. Mechanisms for insulin resistance: common threads and missing links. Cell. 2012;148:852–871. doi: 10.1016/j.cell.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glass CK, Olefsky JM. Inflammation and lipid signaling in the etiology of insulin resistance. Cell metabolism. 2012;15:635–645. doi: 10.1016/j.cmet.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galgani JE, et al. Metabolic flexibility and insulin resistance. American journal of physiology. Endocrinology and metabolism. 2008;295:E1009–1017. doi: 10.1152/ajpendo.90558.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nussey S, Whitehead S. Endocrinology: An Integrated Approach. 2001 [PubMed] [Google Scholar]
- 7.Bruss MD, et al. Calorie restriction increases fatty acid synthesis and whole body fat oxidation rates. American journal of physiology. Endocrinology and metabolism. 2010;298:E108–116. doi: 10.1152/ajpendo.00524.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pedersen BK, Febbraio MA. Muscles, exercise and obesity: skeletal muscle as a secretory organ. Nature reviews. Endocrinology. 2012;8:457–465. doi: 10.1038/nrendo.2012.49. [DOI] [PubMed] [Google Scholar]
- 9.Randle PJ, et al. The glucose fatty-acid cycle. Its role in insulin sensitivity and the metabolic disturbances of diabetes mellitus. Lancet. 1963;1:785–789. doi: 10.1016/s0140-6736(63)91500-9. [DOI] [PubMed] [Google Scholar]
- 10.Parks EJ, Hellerstein MK. Carbohydrate-induced hypertriacylglycerolemia: historical perspective and review of biological mechanisms. The American journal of clinical nutrition. 2000;71:412–433. doi: 10.1093/ajcn/71.2.412. [DOI] [PubMed] [Google Scholar]
- 11.Abu-Elheiga L, et al. Mutant mice lacking acetyl-CoA carboxylase 1 are embryonically lethal. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:12011–12016. doi: 10.1073/pnas.0505714102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chirala SS, et al. Fatty acid synthesis is essential in embryonic development: fatty acid synthase null mutants and most of the heterozygotes die in utero. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:6358–6363. doi: 10.1073/pnas.0931394100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown MS, Goldstein JL. Selective versus total insulin resistance: a pathogenic paradox. Cell metabolism. 2008;7:95–96. doi: 10.1016/j.cmet.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 14.Sun Z, Lazar MA. Dissociating fatty liver and diabetes. Trends Endocrinol Metab. 2013;24:4–12. doi: 10.1016/j.tem.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahmadian M, et al. PPARgamma signaling and metabolism: the good, the bad and the future. Nature medicine. 2013;19:557–566. doi: 10.1038/nm.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herman MA, et al. A novel ChREBP isoform in adipose tissue regulates systemic glucose metabolism. Nature. 2012;484:333–338. doi: 10.1038/nature10986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abel ED, et al. Adipose-selective targeting of the GLUT4 gene impairs insulin action in muscle and liver. Nature. 2001;409:729–733. doi: 10.1038/35055575. [DOI] [PubMed] [Google Scholar]
- 18.Maeda K, et al. Adipocyte/macrophage fatty acid binding proteins control integrated metabolic responses in obesity and diabetes. Cell metabolism. 2005;1:107–119. doi: 10.1016/j.cmet.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 19.Wu C, et al. Enhancing hepatic glycolysis reduces obesity: differential effects on lipogenesis depend on site of glycolytic modulation. Cell metabolism. 2005;2:131–140. doi: 10.1016/j.cmet.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 20.Miyazaki M, et al. Hepatic stearoyl-CoA desaturase-1 deficiency protects mice from carbohydrate-induced adiposity and hepatic steatosis. Cell metabolism. 2007;6:484–496. doi: 10.1016/j.cmet.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 21.Cao H, et al. Identification of a lipokine, a lipid hormone linking adipose tissue to systemic metabolism. Cell. 2008;134:933–944. doi: 10.1016/j.cell.2008.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chakravarthy MV, et al. “New” hepatic fat activates PPARalpha to maintain glucose, lipid, and cholesterol homeostasis. Cell metabolism. 2005;1:309–322. doi: 10.1016/j.cmet.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 23.Chakravarthy MV, et al. Identification of a physiologically relevant endogenous ligand for PPARalpha in liver. Cell. 2009;138:476–488. doi: 10.1016/j.cell.2009.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu S, et al. A diurnal serum lipid integrates hepatic lipogenesis and peripheral fatty acid use. Nature. 2013;502:550–554. doi: 10.1038/nature12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu S, et al. Role of peroxisome proliferator-activated receptor {delta}/{beta} in hepatic metabolic regulation. The Journal of biological chemistry. 2011;286:1237–1247. doi: 10.1074/jbc.M110.138115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao H. Adipocytokines in obesity and metabolic disease. The Journal of endocrinology. 2014;220:T47–59. doi: 10.1530/JOE-13-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacobi D, et al. Adipose tissue signaling by nuclear receptors in metabolic complications of obesity. Adipocyte. 2012;1:4–12. doi: 10.4161/adip.19036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holzer RG, et al. Saturated fatty acids induce c-Src clustering within membrane subdomains, leading to JNK activation. Cell. 2011;147:173–184. doi: 10.1016/j.cell.2011.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harizi H, et al. Arachidonic-acid-derived eicosanoids: roles in biology and immunopathology. Trends Mol Med. 2008;14:461–469. doi: 10.1016/j.molmed.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 30.Ricciotti E, FitzGerald GA. Prostaglandins and inflammation. Arterioscler Thromb Vasc Biol. 2011;31:986–1000. doi: 10.1161/ATVBAHA.110.207449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okunishi K, Peters-Golden M. Leukotrienes and airway inflammation. Biochimica et biophysica acta. 2011;1810:1096–1102. doi: 10.1016/j.bbagen.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sadik CD, Luster AD. Lipid-cytokine-chemokine cascades orchestrate leukocyte recruitment in inflammation. J Leukoc Biol. 2012;91:207–215. doi: 10.1189/jlb.0811402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levy BD, et al. Lipid mediator class switching during acute inflammation: signals in resolution. Nature immunology. 2001;2:612–619. doi: 10.1038/89759. [DOI] [PubMed] [Google Scholar]
- 34.Schwab JM, Serhan CN. Lipoxins and new lipid mediators in the resolution of inflammation. Curr Opin Pharmacol. 2006;6:414–420. doi: 10.1016/j.coph.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 35.Claria J, et al. Resolvin D1 and resolvin D2 govern local inflammatory tone in obese fat. J Immunol. 2012;189:2597–2605. doi: 10.4049/jimmunol.1201272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Medzhitov R. Origin and physiological roles of inflammation. Nature. 2008;454:428–435. doi: 10.1038/nature07201. [DOI] [PubMed] [Google Scholar]
- 37.Roberts LD, et al. beta-Aminoisobutyric acid induces browning of white fat and hepatic beta-oxidation and is inversely correlated with cardiometabolic risk factors. Cell metabolism. 2014;19:96–108. doi: 10.1016/j.cmet.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coll AP, et al. The hormonal control of food intake. Cell. 2007;129:251–262. doi: 10.1016/j.cell.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cota D, et al. Hypothalamic mTOR signaling regulates food intake. Science. 2006;312:927–930. doi: 10.1126/science.1124147. [DOI] [PubMed] [Google Scholar]
- 40.Wolfgang MJ, et al. Regulation of hypothalamic malonyl-CoA by central glucose and leptin. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:19285–19290. doi: 10.1073/pnas.0709778104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fu J, et al. Food intake regulates oleoylethanolamide formation and degradation in the proximal small intestine. The Journal of biological chemistry. 2007;282:1518–1528. doi: 10.1074/jbc.M607809200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwartz GJ, et al. The lipid messenger OEA links dietary fat intake to satiety. Cell metabolism. 2008;8:281–288. doi: 10.1016/j.cmet.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fu J, et al. Oleylethanolamide regulates feeding and body weight through activation of the nuclear receptor PPAR-alpha. Nature. 2003;425:90–93. doi: 10.1038/nature01921. [DOI] [PubMed] [Google Scholar]
- 44.Gaetani S, et al. The fat-induced satiety factor oleoylethanolamide suppresses feeding through central release of oxytocin. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30:8096–8101. doi: 10.1523/JNEUROSCI.0036-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gillum MP, et al. N-acylphosphatidylethanolamine, a gut- derived circulating factor induced by fat ingestion, inhibits food intake. Cell. 2008;135:813–824. doi: 10.1016/j.cell.2008.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morris S, et al. Characterization of fatty acid clearance in premature neonates during intralipid infusion. Pediatric research. 1998;43:245–249. doi: 10.1203/00006450-199802000-00015. [DOI] [PubMed] [Google Scholar]
- 47.Ridaura VK, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koeth RA, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nature medicine. 2013;19:576–585. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parks BW, et al. Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell metabolism. 2013;17:141–152. doi: 10.1016/j.cmet.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Henao-Mejia J, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–185. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith PM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iida N, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342:967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Viaud S, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342:971–976. doi: 10.1126/science.1240537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mukherji A, et al. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell. 2013;153:812–827. doi: 10.1016/j.cell.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 55.An D, et al. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell. 2014;156:123–133. doi: 10.1016/j.cell.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arpaia N, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Furusawa Y, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 58.Cipolletta D, et al. PPAR-gamma is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature. 2012;486:549–553. doi: 10.1038/nature11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ji Y, et al. Activation of natural killer T cells promotes M2 Macrophage polarization in adipose tissue and improves systemic glucose tolerance via interleukin-4 (IL-4)/STAT6 protein signaling axis in obesity. The Journal of biological chemistry. 2012;287:13561–13571. doi: 10.1074/jbc.M112.350066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Osborn O, Olefsky JM. The cellular and signaling networks linking the immune system and metabolism in disease. Nature medicine. 2012;18:363–374. doi: 10.1038/nm.2627. [DOI] [PubMed] [Google Scholar]
- 61.Kennedy EP, Weiss SB. The function of cytidine coenzymes in the biosynthesis of phospholipides. The Journal of biological chemistry. 1956;222:193–214. [PubMed] [Google Scholar]
- 62.Wright MM, McMaster CR. PC and PE synthesis: mixed micellar analysis of the cholinephosphotransferase and ethanolaminephosphotransferase activities of human choline/ethanolamine phosphotransferase 1 (CEPT1) Lipids. 2002;37:663–672. doi: 10.1007/s11745-002-0947-6. [DOI] [PubMed] [Google Scholar]
- 63.Lands WE. Metabolism of glycerolipids. 2. The enzymatic acylation of lysolecithin. The Journal of biological chemistry. 1960;235:2233–2237. [PubMed] [Google Scholar]
- 64.Zhao Y, et al. Identification and characterization of a major liver lysophosphatidylcholine acyltransferase. The Journal of biological chemistry. 2008;283:8258–8265. doi: 10.1074/jbc.M710422200. [DOI] [PubMed] [Google Scholar]
- 65.Kind T, et al. LipidBlast in silico tandem mass spectrometry database for lipid identification. Nature methods. 2013;10:755–758. doi: 10.1038/nmeth.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brown M, et al. Automated workflows for accurate mass-based putative metabolite identification in LC/MS-derived metabolomic datasets. Bioinformatics. 2011;27:1108–1112. doi: 10.1093/bioinformatics/btr079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang X, et al. X(13)CMS: Global Tracking of Isotopic Labels in Untargeted Metabolomics. Analytical chemistry. 2014;86:1632–1639. doi: 10.1021/ac403384n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xia J, et al. MetaboAnalyst 2.0--a comprehensive server for metabolomic data analysis. Nucleic acids research. 2012;40:W127–133. doi: 10.1093/nar/gks374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bostrom P, et al. A PGC1-alpha-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature. 2012;481:463–468. doi: 10.1038/nature10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yi P, et al. Betatrophin: a hormone that controls pancreatic beta cell proliferation. Cell. 2013;153:747–758. doi: 10.1016/j.cell.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 71.Lee CH, et al. Minireview: lipid metabolism, metabolic diseases, and peroxisome proliferator-activated receptors. Endocrinology. 2003;144:2201–2207. doi: 10.1210/en.2003-0288. [DOI] [PubMed] [Google Scholar]