Abstract
Biliary atresia is a progressive fibroinflammatory obstruction of extrahepatic bile ducts that presents as neonatal cholestasis. Due to the overlap in clinical, biochemical, and histological features with other causes of cholestasis, the diagnosis requires an intraoperative cholangiogram. Thus, we determined whether diseased livers express a gene expression signature unique to biliary atresia. Applying stringent statistical analysis to a genome-wide liver expression platform of 64 infants with biliary atresia at the time of diagnosis, 14 age-appropriate subjects with intrahepatic cholestasis as diseased controls, and 7 normal controls, we identified 15 genes uniquely expressed in biliary atresia with an accuracy of 92.3%. Among these genes, IL8 and LAMC2 were sufficient to classify subjects with biliary atresia distinctly from diseased controls with an area under the curve of 0.934 (95%CI: 0.84-1.03), sensitivity of 96.9%, and specificity of 85.7% using their combined first principal component. Direct measurement of IL8 protein in the serum, however, was not different between the two groups. To investigate whether the liver-restricted increase in IL8 was relevant to disease pathogenesis, we inactivated the signaling of IL8 homologs by genetic targeting of the Cxcr2 receptor in a murine model of experimental biliary atresia. Disruption of Cxcr2 shortened the duration of cholestasis, decreased the incidence of bile duct obstruction, and improved survival above wild-type neonatal mice. Conclusion: The hepatic expression of IL8 and LAMC2 has high sensitivity for biliary atresia at diagnosis and may serve as a biomarker of disease, with an important role for the IL8 signaling in experimental biliary atresia.
Keywords: Cholestasis, Kasai portoenterostomy, Cxcr2
INTRODUCTION
Biliary atresia results from a segmental or complete obstruction of extrahepatic bile ducts of devastating clinical consequences to the child. Despite advances in understanding the multifactorial nature of pathogenesis of disease, there is no hepatic or serologic biomarker that distinguishes the disease from other causes of neonatal cholestasis. Analyzing a large number of biopsies from neonates with cholestasis, pathologists reported that the presence of ductular proliferation, bile plugs and other histological features have a predictive value of 90.7% to identify biliary obstruction, but with a variability of 79-98% among individual pathologists (1). As a consequence, the diagnosis requires intraoperative cholangiogram to evaluate for duct obstruction.
Notwithstanding the histological limitations, studies of liver biopsies have provided important clues regarding the role of the innate and adaptive immune systems in pathogenesis of disease (2-4), as well as staging of the liver pathology at diagnosis (5). Therefore, we investigated the hepatic transcriptome in search for the molecular differentiation between biliary atresia and other causes of neonatal cholestasis, and to identify biomarkers of disease. Here, we report that a survey of a genome-wide expression platform identified 15 genes whose expression levels were elevated in biliary atresia, among which the genes encoding interleukin-8 (IL8) and laminin gamma-2 (LAMC2) were sufficient to identify the disease with an accuracy of 94.9%. Interestingly, while serum concentrations of IL8 did not support its use as a biomarker with similar accuracy, complementary studies in a mouse model of disease demonstrated that circuits involving IL8 homologs play a mechanistic role in the pathogenesis of bile duct injury and phenotypic expression of experimental biliary atresia.
PATIENTS AND METHODS
Patients
Liver biopsies, serum samples and clinical data were obtained from infants with cholestasis enrolled into a prospective study (ClinicalTrials.gov Identifier: NCT00061828) of the NIDDK-funded Childhood Liver Disease Research and Education Network (www.childrennetwork.org) or from infants evaluated at Cincinnati Children's Hospital Medical Center. Information on tissues, diagnosis and ages for subjects with biliary atresia (BA), intrahepatic cholestasis (as diseased controls and named non-BA), and normal controls (NC) is provided as “Supplementary – Patients and Methods.” The study protocols were approved by the human research review boards of all participating institutions.
Microarray analyses
Genome-wide expression datasets of human liver samples were generated for individual subjects using pools of biotinylated cRNAs synthesized from frozen liver samples and the GeneChip® Human Gene 1.0 ST Array (Affymetrix, Santa Clara, CA), as described previously (2, 5-7). Detailed information on handling of biopsy samples, assay protocols, signal curation and data analysis is deposited in the Gene Expression Omnibus [GEO: GSE46995].
Mouse model of experimental biliary atresia
Newborn BALB/c mice with or without the genetic inactivation of Cxcr2 (C.129S2[B6]-Cxcr2tm1Mwm/J, Cxcr2−/− mice) were injected with 1.5×106 fluorescent-forming units of rhesus rotavirus (RRV) or 0.9% NaCl (saline) to induce experimental biliary atresia, and phenotyped according to protocols published by us previously (6, 8). Genome-wide expression datasets for extrahepatic bile ducts were generated as described above (data deposited in GEO: GSE46995). Details for the mouse model and protocols for assays of PCR and cytokine levels are provided as “Supplementary Patients and Methods.”
Statistical analyses
Conventional statistical procedures were applied for the gene array experiments using GeneSpring GX11.5 platform, beginning with a selection of genes whose expression differed by ≥2 fold among the groups of BA, non-BA, and NC according to Welch One-way ANOVA with a significance of 0.05 and Benjamini-Hochberg multiple testing correction (false discovery rate [FDR] 0.05) with Tukey's Honestly Significant difference test. Selected genes were subjected to functional analyses using ToppFun application in the ToppGene Suite (http://toppgene.cchmc.org/) with a right-tailed Fisher's exact test (FDR 0.05) (9), and the ToppNet application of the ToppGene Suite, which allows to build protein interaction network with selected neighborhood distance (level one in this case, i.e., immediate interactants) (9), using only interactions from the Human Protein Reference Database (HPRD; http://www.hprd.org, HPRD Release 9) (10). The final group of genes was subjected to random forest analysis and principal component analysis using R package to evaluate the predictive ability of the genes to differentiate biliary atresia samples from diseased controls. Receiver operating characteristic (ROC) analysis was performed using Prism 5.0c (GraphPad Software, San Diego, CA).
PCR data and serum concentrations of human IL8 were analyzed with Student's t-test and Kruskal-Wallis test with Dunn's multiple comparison test; P<0.05 was regarded as significant.
RESULTS
Gene expression signature for biliary atresia at diagnosis
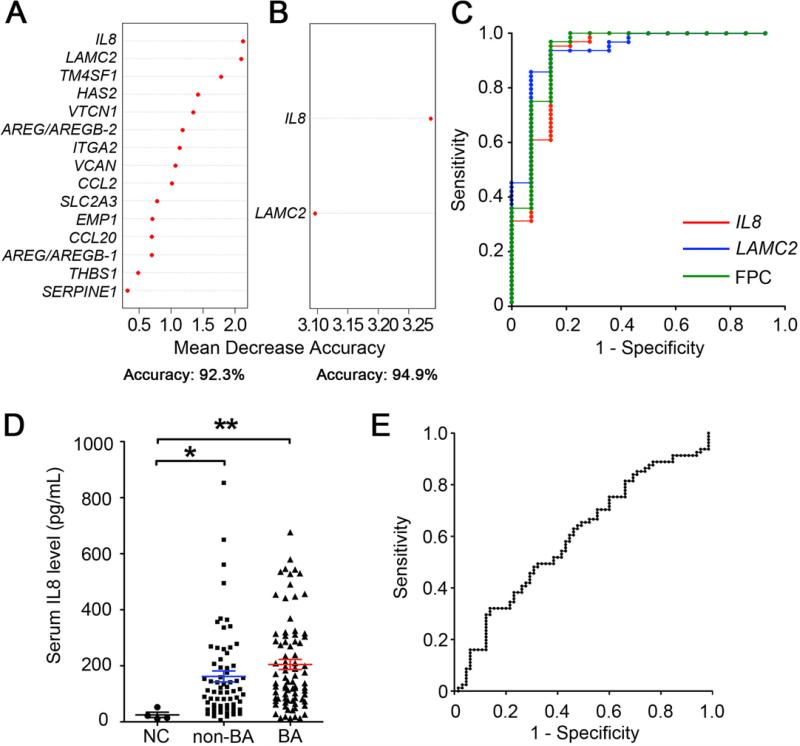
From a total of 33,297 probe sets covering 21,014 genes for each of the three conditions (BA, non-BA, and NC), we identified 574 probe sets differentially expressed by at least 2 fold between BA and NC, 32 between BA and non-BA, and 544 between the two control groups (non-BA and NC; Supplementary Table 1). To identify genes uniquely expressed in livers from subjects with biliary atresia, we searched for genes that were differentially expressed both in BA versus NC and BA versus non-BA, and identified 24 probe sets shared by both groups (Figure 1A, top Venn diagram). A conditional cluster analysis of this gene list formed an expression profile highly enriched for biliary atresia (Figure 1B), with each gene uniformly showing higher levels of expression in BA above the levels seen in NC and non-BA (Supplementary Figure 1). Next, we removed 9 from the 24 probe sets because they were also differentially expressed in the comparison between the two control groups (non-BA versus NC; Figure 1A, bottom Venn diagram), identifying a final list of 15 probe sets corresponding to 15 unique genes linked exclusively to differential expression in BA (Table 1 and Supplementary Figure 2). Thus, the unique gene overexpression in BA and the enriched bundling of subjects in each group suggested that the signature might be able to distinguish BA from the other two groups.
Figure 1. Molecular signature of biliary atresia.
(A) Venn diagram showing the initial selection of 24 probe sets based on the shared overexpression in biliary atresia (BA) over both normal control (NC) and non-biliary atresia (non-BA) as diseased controls. From these probe sets, 9 were excluded based on their increased expression in comparative analysis between the two control groups (NC and non-BA). (B) Hierarchical clustering of the 24 probe sets. Each column represents signal intensities in each subject, and the expression level is depicted by color variation from red (high expression) to blue (low expression); yellow indicates expression level of the median of 7 normal controls.
Table 1.
List of genes specifically regulated in the biliary atresia samples over both disease control (DC) and normal control (NC).
| Gene symbol | Gene description | Fold change | |
|---|---|---|---|
| BA to NC | BA to DC | ||
| EMP1 | epithelial membrane protein 1 | 5.28 | 2.66 |
| HAS2 | hyaluronan synthase 2 | 4.85 | 2.94 |
| VCAN | versican | 4.36 | 2.50 |
| IL8 | interleukin 8 | 3.81 | 2.29 |
| CCL20 | chemokine (C-C motif) ligand 20 | 3.66 | 2.37 |
| VTCN1 | V-set domain containing T cell activation inhibitor 1 | 3.51 | 2.12 |
| ITGA2 | integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) | 3.21 | 2.15 |
| SERPINE1 | serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 | 2.88 | 2.56 |
| THBS1 | thrombospondin 1 | 2.86 | 2.00 |
| CCL2 | chemokine (C-C motif) ligand 2 | 2.61 | 2.59 |
| TM4SF1 | transmembrane 4 L six family member 1 | 2.60 | 3.27 |
| LAMC2 | laminin, gamma 2 | 2.55 | 2.03 |
| AREG|AREGB | amphiregulin | amphiregulin B | 2.53 | 2.42 |
| SLC2A3 | solute carrier family 2 (facilitated glucose transporter), member 3 | 2.22 | 2.23 |
| AREG|AREGB | amphiregulin | amphiregulin B | 2.09 | 2.00 |
All of the genes were upregulated in biliary atresia samples compared to control
Hepatic expression of IL8 and LAMC2 as biomarkers of biliary atresia
To examine whether this signature of 15 genes differentiates patients in the BA from non-BA groups, we applied random forest analyses to determine the relative contribution of individual genes to the accuracy of differentiation (11). In this analysis, we excluded the NC group because clinical examination alone (or combined with standard laboratory tests) is enough for practitioners to differentiate normal asymptomatic infants from symptomatic infants at risk for biliary atresia. Random forest analysis using all 15 genes showed an accuracy of 92.3% (Figure 2A, Supplementary Table 2). Surprisingly, a progressive withdrawal of genes that appeared to minimally influence accuracy revealed that IL8 and LAMC2 alone were sufficient to distinguish BA from non-BA with an increased accuracy of 94.9% (Figure 2B, Supplementary Table 2), with an area under the curve (AUC) of 0.917 (95%CI: 0.81-1.02), sensitivity of 95.3% and specificity of 85.7% using an optimal cutoff for IL8; the AUC using an optimal cutoff for LAMC2 was 0.935 (95%CI: 0.86-1.01), with a sensitivity of 93.8% and specificity of 85.7%. Combining the two genes to generate their first principal component, ROC-AUC was 0.934 (95%CI: 0.84-1.03), with an even greater sensitivity of 96.9% and specificity of 85.7% (Figure 2C). This implied that although the initial list of 15 genes differentiated BA from non-BA, this could also be accomplished with the combined expression levels of only IL8 and LAMC2 while maintaining high sensitivity and specificity.
Figure 2. Levels of discriminatory power of hepatic genes and serum IL8 to differentiate biliary atresia from diseased controls.
Random forest analysis with expression levels of all 15 probe sets (panel A) shows the relative impact of individual genes on the accuracy when differentiating biliary atresia from diseased controls. In panel B, the accuracy improves for IL8 and LAMC2 as a subgroup. In panel C, subjecting IL8 and LAMC2 as individual values or combined as a first principal component (FPC) to ROC curves identifies livers of subjects with biliary atresia at the specified values. Considering IL8 and LAMC2 combined as FPC (green line in [C]), the area under curve is 0.934, sensitivity is 96.9%, and specificity is 85.7%. In panel D, scatter plots show the serum concentration of IL8 for infants at the time of diagnosis of biliary atresia (BA; N=81) compared to non-biliary atresia (non-BA; N=66) and normal control (NC; N=5). The average concentrations for BA and non-BA are not different. Values are expressed as mean ± SEM; *P<0.05, **P<0.01. In panel E, ROC curves of serum IL8 to differentiate samples from biliary atresia and diseased controls has sensitivity of 63.0% and specificity of 53.0%, with the ideal cutoff value as 117.8 pg/mL.
Similar levels of serum IL8 in subjects in the BA and non-BA groups
The high hepatic expression of IL8 and LAMC2 raised the possibility that their serum concentration could serve as a non-invasive biomarker of disease. Here, we focused on IL8 levels because antibodies were not available to determine the immunostaining or concentration of LAMC2. Immunostaining of liver sections showed that the portal tracts of livers from BA subjects contained several nonparenchymal mononuclear cells expressing IL8, which were infrequent in non-BA livers, and not detected in NC livers (Supplementary Figure 3). In the serum, we quantified IL8 in 81 serum samples obtained at the time of diagnosis of biliary atresia, in comparison with 66 samples from subjects with other causes of neonatal intrahepatic cholestasis who were younger than 6 months of age (serving as NON-BA), and 5 healthy infants at 2-4 months of age (serving as NC); these subjects differed from the previous subjects analyzed for liver gene expression. Serum IL8 levels were significantly higher in subjects in the BA (204.8±17.9 pg/mL) and non-BA (163.2±19.5 pg/mL) groups when compared to normal controls (24.9±9.5 pg/mL; Figure 2D), but were not different between BA and NON-BA. Using ROC to distinguish BA from non-BA, the area under the curve was 0.59 (95% CI: 0.50-0.69; Figure 2E). The lack of translation of high hepatic IL8 mRNA expression into high levels of IL8 concentration suggested the existence of a post-transcriptional regulation and/or a liver-restricted activation of IL8 signaling (not reflected systemically). This raised the possibility that expression of IL8 and/or other genes in the molecular signature could play a role in pathogenesis of liver disease.
Biological relatedness of the genes THBS1, SERPINE1, CCL2, ITGA2 and IL8
To explore whether the genes forming the molecular signature for biliary atresia are mechanistically linked to pathogenic mechanisms of the disease, we examined their relatedness through shared biological properties. Functional enrichment analysis using a right-tailed Fisher's exact test with false discovery rate correction (0.05) showed that the 15 genes were linked to 75 biological processes, which could be combined into regeneration or wound healing, inflammation, cell movement and other processes (Figure 3A, Supplementary Table 3). Subjecting the 15 genes to 2-way clustering analysis, we found that THBS1, SERPINE1, CCL2, ITGA2 and IL8 segregated as a group, implying some degree of shared biological relatedness (shown in Figure 3A as common red areas). We explored this further by searching for potential sub-networks based on curated protein-protein interactions among the encoded gene products. This resulted in a network consisting of 114 proteins (12 of 15 seed proteins and 102 direct interactants; 3 of 15 seed proteins did not have any known protein interactions) and 283 direct interactions (Figure 3B, Supplementary Table 4). The largest cluster in this interactome containing 77 proteins with the highest levels of connection to THBS1, SERPINE1, VCAN, and ITGA2, primarily encompassed biological processes that regulate tissue development/morphology and extracellular matrix remodeling (Supplementary Table 5). The second cluster contained 21 proteins with CCL2 and IL8, both related to immunity (Supplementary Table 6). These analyses supported a functional relationship among THBS1, SERPINE1, CCL2, ITGA2 and IL8, and pointed to a potential role of this group of genes in the pathogenesis of bile duct injury.
Figure 3. Functional analyses of the 15 genes highly enriched for biliary atresia.
(A) Functional enrichment analysis of the 15 genes depicted as a 2-way condition tree for individual genes and biological processes (from Gene Ontology), classified into 4 functional categories. The red area in the heatmap indicates the closely related biological processes that are shared by the subgroup of genes shown in the horizontal axis. (B) Protein-protein interaction network of the 15 genes and their accessory proteins, with red nodes indicating original seed proteins and green nodes indicating accessory proteins that are known to directly bind to one of the original 15 proteins. The size of the nodes is proportional to the number of edges connected to the node. A detailed description of (A) and (B) is included in Supplementary Figures 4 and 5, and Supplementary Tables 3 and 4.
Regulation of the 15 genes in extrahepatic bile ducts from RRV-induced mouse model of biliary atresia
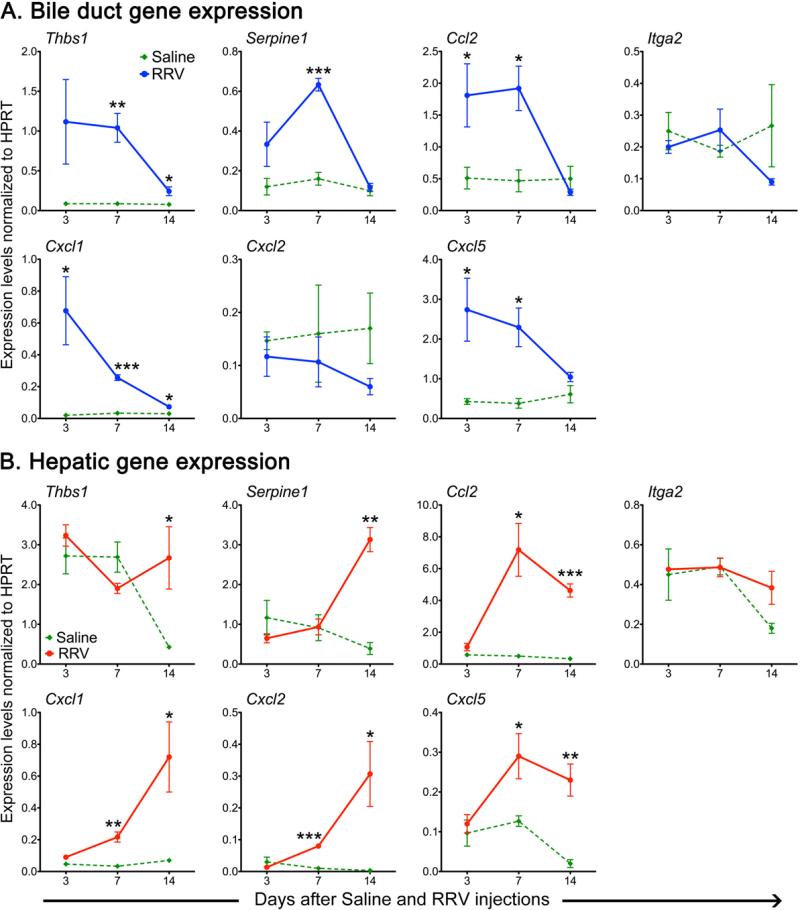
To circumvent obvious experimental challenges in performing mechanistic experiments in humans, we used the experimental model of RRV-induced biliary atresia to investigate the role of these genes in pathogenesis of duct injury. First we examined whether extrahepatic bile ducts from newborn mice have a similar regulation of expression for Thbs1, Serpine1, Ccl2 and Itga2, and for Cxcl1, Cxcl2 and Cxcl5 (representing the murine orthologs of human IL8) after RRV injection. The expression of Thbs1, Serpine1, and Ccl2, but not Itga2, increased 2-7 fold above saline-injected controls at the times of epithelial injury (3 days) and duct obstruction (7 days), and decreased to near baseline levels at the time of atresia (14 days; Figure 4A). Both Cxcl1 and Cxcl5 had a similar pattern, with no consistent changes in Cxcl2 (Figure 4A). The pattern was largely reproduced for Ccl2, Cxcl1, 2 and 5 when mRNA was expressed in the liver, but there was no change in the other genes (Figure 4B). Thus, the increased expression profile of 4 of 5 genes and related genes was reproduced in extrahepatic bile ducts, the main site of RRV-induced injury in experimental biliary atresia, as described previously (6). Among these genes, we hypothesized that signaling linked to Cxcl1, 2 and 5 plays a regulatory role in determining the phenotype of experimental biliary atresia. The decision to focus on this signaling pathway was based on the increased levels of all three murine orthologs of IL8, on their functional relatedness to the biological activity of myeloid cells previously linked to mechanisms of duct obstruction in experimental atresia (12, 13), and on previous reports of increased hepatic IL8 mRNA expression in smaller groups of children with biliary atresia (2, 14).
Figure 4. Expression levels of murine orthologs of functionally enriched genes in experimental biliary atresia.
mRNA expression for Thbs1, Serpine1, Ccl2, Itga2 and 3 murine orthologues of IL8 (Cxcl1, Cxcl2, Cxcl5; B) in extrahepatic bile ducts (A) and liver (B) at 3, 7 and 14 days after injection of RRV or saline in the first day of life. N=4 samples per group and per time point; *P<0.05, **P<0.01, ***P<0.001 between the 2 groups at each time point. mRNA expression is normalized to internal Hprt control; values are expressed as mean ± SEM.
Inactivation of Cxcr2 suppresses the phenotype of experimental atresia
To directly determine whether cellular signaling triggered by Cxcl1, 2 and 5 plays a mechanistic role in the pathogenesis of bile duct injury, we subjected Balb/c mice carrying the inactivation of Cxcr2 (Cxcr2−/− mice) to RRV challenge. Cxcr2 is one of the two receptors shared by Cxcl1, 2 and 5, and was selected because the quantification of mRNA expression in extrahepatic bile ducts for Cxcr2, but not its functional homologue Cxcr1, increased after RRV injection by 2-4 fold above saline controls (Figure 5A). In wild-type (WT) mice, RRV infection induced the onset of jaundice and acholic stools within 5-6 days and 100% mortality by 15 days (Figure 5B,C), similar to findings reported previously (8). RRV infection of Cxcr2−/− mice also produced the timely development of jaundice and acholic stools, but these symptoms were transient and cleared in the majority of the mice, and survival improved substantially from 0% to 48% well beyond 21 days of age (Figure 5B,C).
Figure 5. Overexpression of Cxcr2 in wild type mice after RRV infection and suppression of hepatobiliary injury in Cxcr2−/− mice.
(A) Expression levels of Cxcr1 and Cxcr2 in extrahepatic bile ducts from wild type mice at different time points after normal saline or RRV injection. N=4 samples per group; **P<0.01, ***P<0.001 between the 2 groups at each time point. Development of symptoms (B), and survival (C) of wild type (WT) and Cxcr2−/− neonatal mice after RRV injection. N=21 and 56 samples for (B) and 21 and 40 samples for (C) in each group, respectively. H&E staining of longitudinal sections of murine extrahepatic bile ducts (D) and livers (E) at different time points after RRV challenge. Asterisk: bile duct lumen; PV: portal vein; arrow: infiltration of immune cells; arrowhead: necrotic area in the parenchyma. Scale bars: 50 μm (D) and 100 μm (E).
To investigate the tissue basis for improved symptoms and survival, we examined hematoxylin/eosin-stained histological sections of extrahepatic bile ducts from WT and Cxcr2−/− mice. WT bile ducts had mild inflammation and mild epithelial injury 3 days after RRV infection, which progressed to complete loss of epithelium at day 7 and obstruction at day 14 (Figure 5D). In contrast, Cxcr2−/− bile ducts had largely normal histology, except for transient cholangitis at day 7 (Figure 5D). A similar pattern of injury was present in the liver, with qualitatively greater expansion of portal space with inflammation and necrotic area in the parenchyma in WT mice when compared to milder portal inflammation and no necrosis in Cxcr2−/− mice (Figure 5E).
Decreased activation of NK and dendritic cells in Cxcr2−/− livers
To explore the potential cellular and molecular mechanisms promoting the improved phenotype in Cxcr2−/− mice, we quantified the number and activation status of hepatic immune cells by FACS. There were no differences in the population of the liver by neutrophils, macrophages, CD4+ or CD8+ T cells between WT and Cxcr2−/− mice at any time point after RRV challenge, but NK and dendritic cells decreased in number (Figure 6A-C) and had lower expression of the activation markers Nkg2d and Rae1 (Figure 6D, E) 14 days after RRV. Examining the impact of Cxcr2 inactivation on the expression of cytokines/chemokines, we found decreases in mRNA expression in extrahepatic bile ducts for its ligand Cxcl1 (but not for Cxcl2 and 5) and for Ifng, Tnf-alpha, and IL10 at different times after RRV infection when compared to RRV-infected WT mice (Figure 7A); in the liver, the levels of expression were suppressed only for Cxcl1, 2 and 5 and for IL10 (Figure 7B). Interestingly, the hepatic expression of its functional homolog Cxcr1 increased in Cxcr2−/− mice at all time points in both tissues (Figure 7A and B). This gene expression pattern is consistent with our previous report that greater magnitudes of changes are present when the measures of mRNA expression is done in extrahepatic bile ducts, the primary site of tissue injury in experimental biliary atresia (6).
Figure 6. Population of neonatal livers by inflammatory cells after RRV challenge.
Flow cytometry-based quantification of hepatic plasmacytoid dendritic cells (CD11c+B220+PDCA1+), myeloid dendritic cells (CD11c+CD11b+), natural killer cells (CD49b+), neutrophils (CD11b+Gr1+), macrophages (CD11b+F4/80+), CD4+ (CD3+CD4+) and CD8+ T cells (CD3+CD8+) at 3 (A), 7 (B) and 14 (C) days after RRV challenge. Values are expressed as total number of cells per liver. (D) shows the percentage of NK cells expressing the NKg2d receptor and (E) of mDC expressing its ligand Rae-1 at 14 days after RRV challenge. Data are shown as an average of 4 different experiments. *P<0.05, **P<0.01; values are expressed as mean ± SEM.
Figure 7. Hepatic expression of cytokines, chemokines and their receptors in wild-type and Cxcr2−/− mice.
Graphs display mRNA expression for genes encoding individual cytokines in extrahepatic bile ducts (A) and livers (B) of WT and Cxcr2−/− mice at 3, 7 and 14 days after RRV infection. N = 4-8 livers per group and time point. *P <0.05; **P<0.01, ***P<0.001 between the 2 groups at each time point. mRNA expression is normalized to internal Hprt control; Values are expressed as mean ± SEM.
DISCUSSION
Analyzing the hepatic transcriptome, we identified a gene expression signature that identifies infants with biliary atresia as a unique group. The signature contains a relatively short number of 15 genes that are overexpressed individually and collectively when compared to infants with other causes of neonatal cholestasis and non-diseased control livers. Among the genes, IL8 and LAMC2 alone identified patients with biliary atresia at a sensitivity of 96.9% and specificity of 85.7%. The highly enriched hepatic expression pattern for IL8 was not reflected in the serum, as evidenced by the lack of significant increases in the concentrations of IL8 in biliary atresia compared to diseased controls. Exploring the role of the gene expression signature in pathogenesis of disease, we found a substantial overlap in biological functions for THBS1, SERPINE1, CCL2, ITGA2 and IL8, which collectively support the activation of processes regulating tissue development/morphology (organogenesis), extracellular matrix remodeling (wound healing), and inflammation, processes previously linked to pathogenesis of biliary atresia (5, 15). Based on the reproducibility of this signature in extrahepatic bile ducts from neonatal mice subjected to RRV infection, we directly investigated the mechanistic role of IL8-signaling in pathogenesis of bile duct injury. Disruption of the Cxcr2 gene, encoding one of the receptors of the IL8 murine orthologs Cxcl1, 2, and 5, suppressed the disease phenotype, with a decrease in epithelial injury, prevention of duct obstruction, and increased long-term survival. Altogether, these findings identify a molecular signature that is specific and seemingly restricted to the diseased liver of biliary atresia, not reflected systemically, and relevant to pathogenesis of biliary injury in an experimental model of the disease.
Tissue analysis of infants with biliary atresia has been invaluable in the search for pathogenic mechanisms of disease. An earlier survey of liver gene expression profile identified a coordinated over- and under-expression of lymphocyte-enriched genes at diagnosis (2). Although this study was limited in scope (expression of about half of the human genes) and in size of cohort (14 livers with biliary atresia), the relevance of its findings was supported by the prevention of the phenotypic expression of experimental biliary atresia in mice carrying the targeted inactivation of the gene encoding the lymphocyte-dependent cytokine interferon-gamma (8). Our current study overcomes these limitations by the inclusion of a much larger cohort and analyzes the expression of the entire human genome. Not surprisingly, there is reproducibility of gene expression results between the two studies, as supported by the differential expression of IL8, SPP1, MMP7, CFTR, DDR1, but it is notable that the application of stringent statistical approaches narrowed the final profile to 15 genes that were uniformly over-expressed in biliary atresia when compared to age-matched diseased controls.
One potential limitation of the study is that the use of livers from children at 22-42 months of age as normal controls may not adequately display unique features of the immune function that may be typical of young infants, although data from an infant at 75 days of age showed a similar pattern of expression for the gene signature. One particular benefit from the inclusion of the normal controls was that it allowed for the exclusion of genes that increased in response to cholestasis or liver injury (by comparing non-BA and NC livers). One example to support this approach is the findings of increased SPP1 in biliary atresia, but also in diseased controls over non-diseased controls, which led to the elimination of SPP1 in our final list. Interestingly, SPP1 was previously reported to be over-expressed in patients with biliary atresia (2, 16-18), but its inactivation did not influence the development of the phenotype in experimental biliary atresia (19), suggesting that SPP1 may be involved in a generalized physiological response of the liver to a cholestatic injury, but not necessarily linked to pathogenesis of biliary atresia.
The identification of a molecular signature of biliary atresia has potential implications for the development of biomarkers of disease. In the liver, the 15 genes formed a signature with high sensitivity and specificity. Surprisingly, an analysis of the relative contribution of individual genes to identify the disease revealed that the combination of IL8 and LAMC2 already provides a sensitivity of 96.9% and specificity of 85.7%. This level of sensitivity is similar to or slightly exceeds the levels reported in studies assessing the accuracy of histopathological examination of liver biopsies to diagnose biliary atresia (20-23). Based on the degree of variability among pathologists to accurately identify histological features in the liver that are diagnostic of extrahepatic obstruction (20), the availability of an operator-independent test, such as a molecular profiling reported here, may aid in the selection of those infants most likely to benefit from an exploratory laparotomy and hepatoportoenterostomy. Future investigations of this possibility could include studies that add the expression of IL8 and LAMC2 to histopathological scoring by pathologists to determine whether they would reach 100% sensitivity and specificity for biliary atresia if used together. If such outcome were demonstrated, their use in clinical practice would improve patient care by reliably identifying those infants that do not need to undergo exploratory laparotomy, thus avoiding morbidity and health care costs.
The availability of biomarkers of disease may improve patient care by incorporating objectivity to diagnostic and treatment algorithms, and are particularly valuable if it prevents operative procedures. Based on this premise and on the high hepatic expression of IL8, we quantified serum IL8 concentrations but found them to have lower sensitivity and specificity. In agreement with previous reports (24-26), serum IL8 was higher in subjects with biliary atresia when compared to age-matched healthy controls; however, the direct comparison between biliary atresia and diseased controls did not differ in our study, which is similar to a previous study (24) and conflicts with another study that reported higher levels in subjects with biliary atresia (26). The apparent conflict among the previous studies probably results from small cohort sizes or to a lack in uniformity in the timing of sample collection. Our study design attempted to control for these variables by matching all subjects by age and by the inclusion of a substantial cohort size. It is interesting that the hepatic expression of IL8 (at the mRNA and protein levels) was much higher in biliary atresia, but the levels of circulating IL8 were similar between biliary atresia and diseased controls. While our studies do not directly address the cellular basis for these findings, the high serum levels and low hepatic expression of IL8 in diseased controls suggest that the circulating levels reflect a site of origin outside the liver, perhaps in circulating mononuclear cells that may be activated in response to the cholestatic liver of non-BA diseases. This biological scenario is consistent with a previous report of high serum IL8 in adult patients with several liver disorders and at different stages of disease (including cirrhosis), suggesting that IL8 expression is linked to cholestatic conditions, cirrhosis and circulating mononuclear cells (27).
We used the RRV model of biliary injury in newborn mice to directly explore whether IL8 signaling is relevant to mechanisms of disease. In this experimental model, the constitutional disruption of signaling by IL8 orthologs in Cxcr2−/− mice resulted in transient cholestasis and improved survival. Cxcr2 and its functional counterpart Cxcr1 mediate antimicrobial host defense by rendering cells responsive to chemotaxis- and activation-inducing signals of their ligands Cxcl1, 2 and 5 (28). Some of the Cxcr1- and Cxcr2-bearing cells, namely NK cells, macrophages, and dendritic cells have been shown to play regulatory roles in the phenotypic expression of RRV-induced bile duct injury (13, 29-32). Consistent with these roles, NK and dendritic cells had decreased number and activation markers in Cxcr2−/− mice injected with RRV. Interestingly, the hepatic population of neutrophils and macrophages, cells known to have high expression of Cxcr2, did not differ between Cxcr2−/− and wild-type mice. This is unexpected based on previous reports that Cxcr2 expression is required for the infiltration of neutrophils and macrophages in peripheral tissues in response to an injury (33-35). Based on the inability of myeloid cells from Cxcr2−/− mice to leave the bone marrow (32), it is possible that the high number of these cells in the livers of Cxcr2−/− mice may reflect the well-described role of the newborn liver in extramedullary hematopoiesis. Another possibility is an increase in the expression of its functional counterpart Cxcr1, which may occur as a compensatory response to RRV in mice that are unable to express the Cxcr2 gene, thus creating an alternate receptor for the Cxcl1, 2, 5 ligands, as supported by an increased Cxcr1 expression in livers of Cxcr2−/− mice above the levels seen in RRV-injected wild-type mice. The inability of this receptor to completely abolish jaundice and mortality suggests that, to be effective, therapeutic strategies targeting IL8 signaling must be broad enough to prevent the compensatory activation of related pathways. The experimental data in Cxcr2−/− mice are aligned with the previous report that its ligand, Cxcl2, is released upon infection of macrophages and serves as chemoattractant to neutrophils (12). This early signaling between macrophages and neutrophils is probably one of the important initiating events, perhaps simultaneously to the activation of dendritic cells and NK lymphocytes following RRV infection, later to be followed by the release of pro-inflammatory cytokines and obstruction of the duct lumen to produce the phenotype of experimental biliary atresia [reviewed in (15)].
In summary, we report an expression signature of 15 genes at the time of presentation of neonatal cholestasis that is highly enriched for biliary atresia. Among these genes, IL8 and LAMC2 are uniformly over-expressed and produce an abbreviated signature with high sensitivity and specificity when compared to the levels in the livers of infants with other causes of neonatal cholestasis. At the mechanistic level, the disruption of IL8 signaling in newborn mice suppressed, but did not abolish, the disease phenotype in experimental biliary atresia. Despite these findings, the serum concentration of IL8 does not have discriminatory power and cannot be used as a minimally invasive biomarker of disease. This notwithstanding, future studies to validate the expression profile in the liver and to explore whether its addition to liver histopathology raises sensitivity and specificity to 100% have the potential to improve diagnostic algorithms and the care of infants with biliary atresia.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the Data Coordinating Center of the NIDDK-funded Childhood Liver Disease Research and Education Network (ChiLDREN) and the Principal Investigators and Clinical Research Coordinators of individual ChiLDREN Centers for patient recruitment and acquisition of tissue and data. The contents of the article do not necessarily reflect the opinions or views of the NIDDK, ChiLDREN, or ChiLDREN investigators.
Financial Support:
This work was supported by the NIH grant R01 DK83781 and DK62497 (to J.A.B.), a Liver Scholar Research Award from the American Association for Studies of Liver Disease (to P.S.), NIH grant U01 DK062456 (to J.C.M.), and the NIH grant DK78392 (Integrative Morphology Core of the Digestive Disease Research Core Center in Cincinnati). This work was also supported by the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through grant 8 UL1 TR000077-04
List of Abbreviations
- BA
biliary atresia
- non-BA
diseased controls
- NC
normal controls
- ROC
receiver operating characteristic
- WT
wild type
- RRV
rhesus rotavirus
REFERENCES
- 1.Moyer V, Freese DK, Whitington PF, Olson AD, Brewer F, Colletti RB, Heyman MB. Guideline for the evaluation of cholestatic jaundice in infants: recommendations of the North American Society for Pediatric Gastroenterology, Hepatology and Nutrition. J Pediatr Gastroenterol Nutr. 2004;39:115–128. doi: 10.1097/00005176-200408000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Bezerra JA, Tiao G, Ryckman FC, Alonso M, Sabla GE, Shneider B, Sokol RJ, et al. Genetic induction of proinflammatory immunity in children with biliary atresia. Lancet. 2002;360:1653–1659. doi: 10.1016/S0140-6736(02)11603-5. [DOI] [PubMed] [Google Scholar]
- 3.Mack CL, Falta MT, Sullivan AK, Karrer F, Sokol RJ, Freed BM, Fontenot AP. Oligoclonal expansions of CD4+ and CD8+ T-cells in the target organ of patients with biliary atresia. Gastroenterology. 2007;133:278–287. doi: 10.1053/j.gastro.2007.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohya T, Fujimoto T, Shimomura H, Miyano T. Degeneration of intrahepatic bile duct with lymphocyte infiltration into biliary epithelial cells in biliary atresia. J Pediatr Surg. 1995;30:515–518. doi: 10.1016/0022-3468(95)90120-5. [DOI] [PubMed] [Google Scholar]
- 5.Moyer K, Kaimal V, Pacheco C, Mourya R, Xu H, Shivakumar P, Chakraborty R, et al. Staging of biliary atresia at diagnosis by molecular profiling of the liver. Genome Med. 2010;2:33. doi: 10.1186/gm154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carvalho E, Liu C, Shivakumar P, Sabla G, Aronow B, Bezerra JA. Analysis of the Biliary Transcriptome in Experimental Biliary Atresia. Gastroenterology. 2005;129:713–717. doi: 10.1016/j.gastro.2005.05.052. [DOI] [PubMed] [Google Scholar]
- 7.Zhang DY, Sabla G, Shivakumar P, Tiao G, Sokol RJ, Mack C, Shneider BL, et al. Coordinate expression of regulatory genes differentiates embryonic and perinatal forms of biliary atresia. Hepatology. 2004;39:954–962. doi: 10.1002/hep.20135. [DOI] [PubMed] [Google Scholar]
- 8.Shivakumar P, Campbell KM, Sabla GE, Miethke A, Tiao G, McNeal MM, Ward RL, et al. Obstruction of extrahepatic bile ducts by lymphocytes is regulated by IFN-gamma in experimental biliary atresia. J Clin Invest. 2004;114:322–329. doi: 10.1172/JCI21153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nuc Acid Res. 2009;37:W305–311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, et al. Human Protein Reference Database--2009 update. Nuc Acid Res. 2009;37:D767–772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Breiman L. Random forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- 12.Mohanty SK, Ivantes CA, Mourya R, Pacheco C, Bezerra JA. Macrophages are targeted by rotavirus in experimental biliary atresia and induce neutrophil chemotaxis by mip2/cxcl2. Pediatr Res. 2010;67:345–351. doi: 10.1203/PDR.0b013e3181d22a73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J, Bessho K, Shivakumar P, Mourya R, Mohanty SK, Dos Santos JL, Miura IK, et al. Th2 signals induce epithelial injury in mice and are compatible with the biliary atresia phenotype. J Clin Invest. 2011;121:4244–4256. doi: 10.1172/JCI57728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang YH, Chou MH, Du YY, Huang CC, Wu CL, Chen CL, Chuang JH. Expression of toll-like receptors and type 1 interferon specific protein MxA in biliary atresia. Lab Invest. 2007;87:66–74. doi: 10.1038/labinvest.3700490. [DOI] [PubMed] [Google Scholar]
- 15.Bessho K, Bezerra JA. Biliary atresia: will blocking inflammation tame the disease? Ann Rev Med. 2011;62:171–185. doi: 10.1146/annurev-med-042909-093734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whitington PF, Malladi P, Melin-Aldana H, Azzam R, Mack CL, Sahai A. Expression of osteopontin correlates with portal biliary proliferation and fibrosis in biliary atresia. Pediatr Res. 2005;57:837–844. doi: 10.1203/01.PDR.0000161414.99181.61. [DOI] [PubMed] [Google Scholar]
- 17.Huang L, Wei MF, Feng JX. Abnormal activation of OPN inflammation pathway in livers of children with biliary atresia and relationship to hepatic fibrosis. Eur J Pediatr Surg. 2008;18:224–229. doi: 10.1055/s-2008-1038483. [DOI] [PubMed] [Google Scholar]
- 18.Honsawek S, Vejchapipat P, Chongsrisawat V, Thawornsuk N, Poovorawan Y. Association of circulating osteopontin levels with clinical outcomes in postoperative biliary atresia. Pediatr Sur Int. 2011;27:283–288. doi: 10.1007/s00383-010-2799-7. [DOI] [PubMed] [Google Scholar]
- 19.Hertel PM, Crawford SE, Finegold MJ, Estes MK. Osteopontin upregulation in rotavirus-induced murine biliary atresia requires replicating virus but is not necessary for development of biliary atresia. Virology. 2011;417:281–292. doi: 10.1016/j.virol.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Russo P, Magee JC, Boitnott J, Bove KE, Raghunathan T, Finegold M, Haas J, et al. Design and validation of the biliary atresia research consortium histologic assessment system for cholestasis in infancy. Clin Gastroenterol Hepatol. 2011;9:357–362. e352. doi: 10.1016/j.cgh.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brough AJ, Bernstein J. Conjugated hyperbilirubinemia in early infancy. A reassessment of liver biopsy. Hum Pathol. 1974;5:507–516. doi: 10.1016/s0046-8177(74)80003-1. [DOI] [PubMed] [Google Scholar]
- 22.Ferry GD, Selby ML, Udall J, Finegold M, Nichols B. Guide to early diagnosis of biliary obstruction in infancy. Review of 143 cases. Clin Pediatr. 1985;24:305–311. doi: 10.1177/000992288502400601. [DOI] [PubMed] [Google Scholar]
- 23.Lee WS, Looi LM. Usefulness of a scoring system in the interpretation of histology in neonatal cholestasis. World J Gastroenterol. 2009;15:5326–5333. doi: 10.3748/wjg.15.5326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nobili V, Marcellini M, Giovannelli L, Girolami E, Muratori F, Giannone G, Devito R, et al. Association of serum interleukin-8 levels with the degree of fibrosis in infants with chronic liver disease. J Pediatr Gastroenterol Nutri. 2004;39:540–544. doi: 10.1097/00005176-200411000-00017. [DOI] [PubMed] [Google Scholar]
- 25.El-Faramawy AA, El-Shazly LB, Abbass AA, Ismail HA. Serum IL-6 and IL-8 in infants with biliary atresia in comparison to intrahepatic cholestasis. Trop Gastroenterol. 2011;32:50–55. [PubMed] [Google Scholar]
- 26.Honsawek S, Chongsrisawat V, Vejchapipat P, Thawornsuk N, Tangkijvanich P, Poovorawan Y. Serum interleukin-8 in children with biliary atresia: relationship with disease stage and biochemical parameters. Pediatr Surg Int. 2005;21:73–77. doi: 10.1007/s00383-004-1329-x. [DOI] [PubMed] [Google Scholar]
- 27.Zimmermann HW, Seidler S, Gassler N, Nattermann J, Luedde T, Trautwein C, Tacke F. Interleukin-8 is activated in patients with chronic liver diseases and associated with hepatic macrophage accumulation in human liver fibrosis. PLoS One. 2011;6:e21381. doi: 10.1371/journal.pone.0021381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roth I, Hebert C. CXCR1 and CXCR2. In: Oppenheim JJ, Feldmann M, Durum SK, editors. Cytokine Reference: a Compendium of Cytokines and Other Mediators of Host Defense. Academic Press; San Diego: 2001. pp. 1982–2002. [Google Scholar]
- 29.Shivakumar P, Sabla GE, Whitington P, Chougnet CA, Bezerra JA. Neonatal NK cells target the mouse duct epithelium via Nkg2d and drive tissue-specific injury in experimental biliary atresia. J Clin Invest. 2009;119:2281–2290. doi: 10.1172/JCI38879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saxena V, Shivakumar P, Sabla G, Mourya R, Chougnet C, Bezerra JA. Dendritic cells regulate natural killer cell activation and epithelial injury in experimental biliary atresia. Science Transl Med. 2011;3:102ra194. doi: 10.1126/scitranslmed.3002069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lages CS, Simmons J, Chougnet CA, Miethke AG. Regulatory T cells control the CD8 adaptive immune response at the time of ductal obstruction in experimental biliary atresia. Hepatology. 2012;56:219–227. doi: 10.1002/hep.25662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eash KJ, Greenbaum AM, Gopalan PK, Link DC. CXCR2 and CXCR4 antagonistically regulate neutrophil trafficking from murine bone marrow. J Clin Invest. 2010;120:2423–2431. doi: 10.1172/JCI41649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Devalaraja RM, Nanney LB, Du J, Qian Q, Yu Y, Devalaraja MN, Richmond A. Delayed wound healing in CXCR2 knockout mice. J Invest Derm. 2000;115:234–244. doi: 10.1046/j.1523-1747.2000.00034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsai WC, Strieter RM, Mehrad B, Newstead MW, Zeng X, Standiford TJ. CXC chemokine receptor CXCR2 is essential for protective innate host response in murine Pseudomonas aeruginosa pneumonia. Inf Imun. 2000;68:4289–4296. doi: 10.1128/iai.68.7.4289-4296.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tateda K, Moore TA, Newstead MW, Tsai WC, Zeng X, Deng JC, Chen G, et al. Chemokine-dependent neutrophil recruitment in a murine model of Legionella pneumonia: potential role of neutrophils as immunoregulatory cells. Inf Imun. 2001;69:2017–2024. doi: 10.1128/IAI.69.4.2017-2024.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.