Abstract
BACKGROUND
Immuno-spin trapping (IST) is based on the reaction of a spin trap with a free radical to form a stable nitrone adduct, followed by the use of antibodies, rather than traditional electron paramagnetic resonance spectroscopy, to detect the nitrone adduct. IST has been successfully applied to mechanistic in vitro studies, and recently, macromolecule-centered radicals have been detected in models of drug-induced agranulocytosis, hepatotoxicity, cardiotoxicity, and ischemia/reperfusion, as well as in models of neurological, metabolic and immunological diseases.
SCOPE OF THE REVIEW
To critically evaluate advances, challenges, and pitfalls as well as the scientific opportunities of IST as applied to the study of protein-centered free radicals generated in stressed organelles, cells, tissues and animal models of disease and exposure.
MAJOR CONCLUSIONS
Because the spin trap has to be present at high enough concentrations in the microenvironment where the radical is formed, the possible effects of the spin trap on gene expression, metabolism and cell physiology have to be considered in the use of IST and in the interpretation of results. These factors have not yet been thoroughly dealt with in the literature.
GENERAL SIGNIFICANCE
The identification of radicalized proteins during cell/tissue response to stressors will help define their role in the complex cellular response to stressors and pathogenesis; however, the fidelity of spin trapping/ immuno-detection and the effects of the spin trap on the biological system should be considered.
Keywords: reactive chemical species, protein radical, spin trap, anti-DMPO, immuno-spin trapping, disease/exposure model
1. Introduction
The “gold-standard” technique that allows the unambiguous detection of free radicals is electron spin resonance spectroscopy (ESR, also called electron paramagnetic resonance (EPR)) because this technique is based on fundamental physics and makes no assumptions [1–3] (see also [4] in this issue). There is no doubt that ESR has a number of undisputed advantages over other methods of studying free radicals [5]. However, the greatest limitation of ESR to the study of free radicals in cells and tissues is its poor sensitivity in relation to the steady-state concentrations of free radicals under physiological conditions, or even in the response to severe stress. Typically, the steady-state concentration of free radicals under normal conditions is less than 1 nM or even 1 pM, which is far below the best sensitivity of ESR of 3 nM. These limitations of ESR have led to an intensive search for alternatives for the investigation of free radicals in biological systems.
The limitations of direct ESR in the study of short-lived free radicals led, in the 1960’s, to the development of spin trapping [6], in which a free radical adds to the carbon end of the nitrone function of a spin trap (Fig. 1). This reaction produces a much more stable free radical, a nitroxide radical adduct or radical adduct that can be seen by ESR spectroscopy [6]. The greatest advantage of using spin trapping in the study of free radicals is the increased stability leading to increased radical adduct concentrations, which are often above the 3 nM limit of ESR detection [2]. This has led to a renaissance of ESR-spin trapping in the study of oxidative processes in biological systems [7–8]. Indeed, a number of free radical metabolites, proteins and nucleic acids have been detected by ESR-spin trapping both in vitro and in vivo [2].
Figure 1. Spin trapping and fate of protein-DMPO adducts.
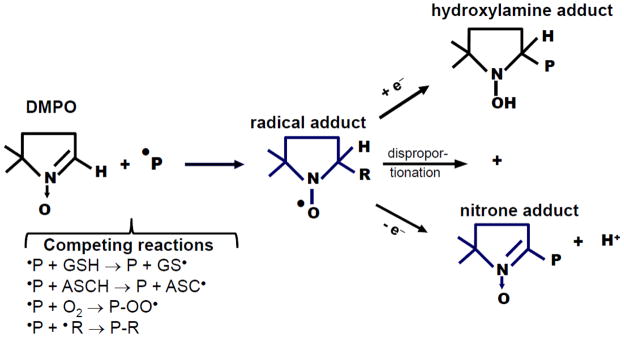
A protein radical (a radical site in a protein) reacts with DMPO to form a radical adduct. Depending on microenvironment conditions and structural characteristics of the target protein, the radical adduct can be reduced to hydroxylamine or oxidized to a stable nitrone adduct. It can also disproportionate to generate both the hydroxylamine and nitrone adducts. In cells and in vivo there are a number of competing reactions that can affect the yield of DMPO-protein adducts. Reduced glutathoine (GSH) and L-ascorbate (ASCH) [28] can react with protein radicals faster than the rate of reaction with DMPO, resulting in reduced yield of protein-DMPO nitrone adducts, a repaired protein and a less reactive radical (i.e., GS• and ASC•). Oxygen, the best spin trap in nature, can also react with protein radicals to form protein-peroxyl radicals, which very slowly react with DMPO. Protein radicals can react with other radical sites in the same or different proteins to form cross links (e.g., Tyr-Tyr, His-His or Trp-Trp). Protein radicals can also react with lipid radicals, •NO2/•NO or drug/toxicant radicals, thus resulting in protein-lipid or protein-drug/toxicant adducts (P-R).
Many spin traps, such as nitroso and nitrone compounds, have been used to study free radicals in biological systems [7, 9]. Because of their physico-chemical properties, membrane permeability [10–11], effectiveness at trapping free radicals [9], and low toxicity [12], nitrone spin traps have been employed both as reagents to detect radicals using ESR spectroscopy [7, 9] and as pharmacological agents against oxidative stress-mediated injury [13–14]. The most popular of these spin traps is 5,5-dimethyl-1-pyrroline N-oxide (DMPO) [15], which has been cited in Medline over 1,300 times. In a radicalized macromolecule, for example a radicalized protein, the spin trap adds to an atom in a solvent-exposed site with high electron spin density [6]. The greater the stability of the radical adduct, the higher the concentration of the radical adduct for a given rate of free radical generation [7]. The lifetime of the radical adduct is usually the most important factor in deciding the success of an ESR-spin trapping experiment [2]. In addition, it is important to note that unlike direct ESR, the spin trapping methodology depends on the absolute fidelity of the spin trapping reaction [16]. Importantly, nitrone spin traps are known to react with free radicals and nonradicals via electrophilic and nucleophilic addition reactions [17]. Two alternate mechanisms of radical adduct artifacts with DMPO have been recently investigated and discussed [16, 18]: inverted spin trapping (one-electron oxidation of the spin trap) and the Forrester-Hepburn (nucleophilic addition of the spin trap) mechanisms. In biological systems the Forrester-Hepburn mechanism, which is initiated by a nucleophilic addition of a nucleophile to the spin trap, would be the major mechanism of generation of potential artifactual DMPO-molecule adducts. See [16, 18] for a comprehensive chemical discussion of these two sources of artifacts in spin trapping.
The specificity of the reactions of nitrone spin traps with free radicals has already made spin trapping with ESR detection the most universal and specific tool for the detection of free radicals in biochemical systems as well as in cells, tissues and animals [19]. Unfortunately, ESR-spin trapping of protein radicals in vivo has severe limitations. Some of them are: 1) the cost of acquiring the instrument; 2) the instability of radical adducts in tissues, which may compromise its reproducibility; 3) the lossiness of the dielectric sample, and 4) the low radical adduct concentration due to the presence of antioxidants that can compete with DMPO.
2. Principle of Immuno-spin Trapping
To an ESR spectroscopist, the conservation of the unpaired electron is the most important aspect of the reaction of a protein-centered radical with a spin trap to form a radical adduct. To an organic chemist, the most unique feature of the reaction is the formation of a new covalent bond between the DMPO and the free radical in a reaction that is specific for free radicals. To an immunologist, the reaction of a free radical with a spin trap marks the creation of a novel epitope; and to a biochemist this is a novel way to identify a protein target of oxidation in states of stress that may lead to an understanding of the chemical basis of a free radical process in biologically-relevant scenarios.
DMPO is very stable and nearly redox inert, being reduced to the hydroxylamine only at the very low potential of -1.68 volts and oxidized only at the very high potential of 1.87 volts versus normal hydrogen electrode. Once formed DMPO-protein adducts can exist in three redox forms: (1) the nitroxide radical adduct, (2) the corresponding hydroxylamine formed by a one-electron reduction of the radical adduct, and (3) the corresponding nitrone formed by a one-electron oxidation of the radical adduct [20] (see Fig. 1). The nitroxide radical adducts and their corresponding hydroxylamine adducts are all unstable and decay over time. The nitrone adduct is the most thermodynamically stable product of the reaction of a free radical with a nitrone spin trap that can be studied in tissues excised from an experimental animal treated with the spin trap.
These basic concepts brought our attention to the development of an immunoassay for the DMPO motif in protein-DMPO nitrone adducts (Fig. 2), thus bringing the power of immunological techniques to the growing field of free radical biochemistry in medicine [21]. We called this technique immuno-spin trapping (IST) [22]. The basic idea of IST arose from the observation in Mason’s laboratory that when the DMPO/ hoMb (horse heart myoglobin) radical adduct decayed, the unique chemical bond formed by the reaction of the hoMb[Tyr103] radical and DMPO persisted [23]. MS analysis showed that this ESR-silent species was the one-electron oxidation product of the radical adduct, the nitrone adduct. Once the nitrone adduct is formed, it is a specific marker for the radical and for any analytical method that can be used to detect it (e.g., immunochemistry, high-performance liquid chromatography (HPLC), mass spectrometry (MS), MS/MS (tandem-MS), NMR, and magnetic resonance imaging (MRI)) [21]. See in this issue [24] for the use of MS to detect other posttranslational modifications in proteins, which may be applicable to study DMPO adducted to proteins. The impressive sensitivity, power and relative ease of immunological techniques led to the production of antibodies to DMPO as a specific radical probe.
Figure 2. Principle of immuno-spin trapping of protein radicals.
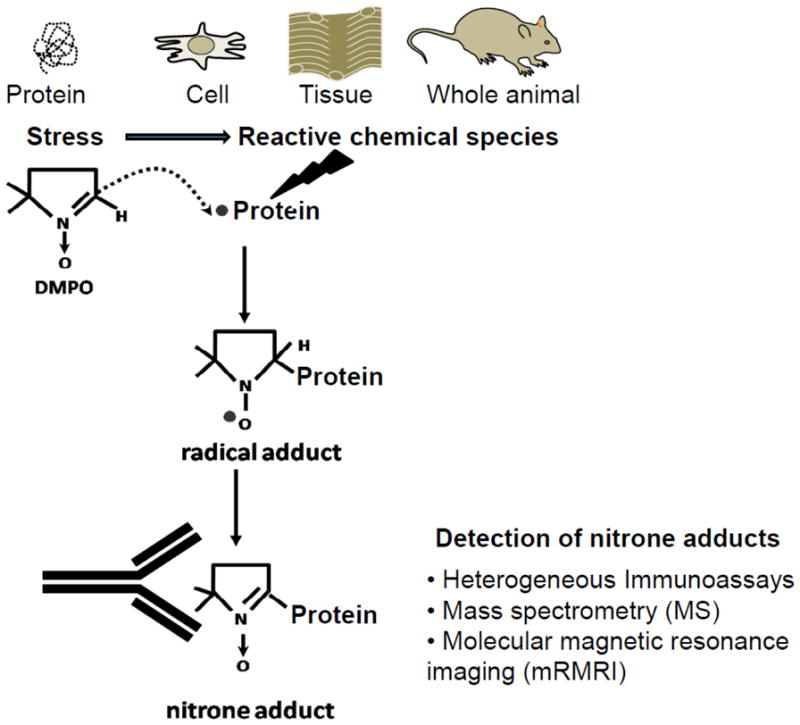
Immuno-spin trapping can be applied to investigate protein radicals in purified systems, cells, tissues and in the whole animal. The production of protein radicals is caused by one-electron-mediated oxidation of specific residues in a protein. These residues can be primary or secondary targets. Protein radicals are trapped in situ by DMPO to form protein-DMPO radical adducts. With time a radical adduct decays to form a stable DMPO-protein nitrone adduct. The DMPO motif of a protein-DMPO nitrone adduct can be detected with an anti-DMPO antibody using immunoassays. Protein-DMPO nitrone adducts can be pulled down by immunoprecipitation from complex mixtures such as homogenates of organelles, whole cells or tissues and then characterized using MS. To preserve tissue architecture, protein-nitrone adducts can be observed using the anti-DMPO antibody and fluorescent or immunogold techniques. Protein-DMPO nitrone adducts can also be detected using non-invasive techniques such as molecular magnetic resonance imaging (mMRI).
3. Anti-DMPO antibodies detect the DMPO motif of protein-DMPO nitrone adducts
Polyclonal and monoclonal antibodies have now been produced against the nitrone spin trap DMPO [23]. Because of the hapten characteristics of DMPO (i.e., its low molecular weight), to raise an antiserum that specifically recognizes DMPO, it was necessary to synthesize a DMPO-protein immunogen [23]. The production of the immunogen required the synthesis of a DMPO-linker compound, in this case octanoic acid (OA), followed by its reaction with a carrier protein. The nitrone group does not occur in nature and has proven to be highly antigenic, as is the related nitro group [20]. The compound 5,5-dimethyl-2-(8-octanoic acid)-1-pyrroline N-oxide (DMPO-OA) was chosen as a hapten because it mimics a nitrone-protein adduct and provides more access for antigen presentation during the immune response. Originally, the DMPO-OA hapten was conjugated to chicken egg albumin to produce the immunogen, which was further used to immunize rabbits and produce the anti-DMPO polyclonal antiserum. More recently a monoclonal anti-DMPO (hybridoma clone N1664A) has been developed from the original DMPO conjugated to chicken egg albumin [25]. Both polyclonal and monoclonal anti-DMPO immunoreagents are commercially available.
In the original experiments anti-DMPO bound to Mb-DMPO adducts was 50% displaced by 30 nM DMPO-OA, which showed the high affinity of the DMPO motif-antibody interaction. The sensitivity advantage of IST over traditional ESR-spin trapping is that a stable product, not a transient adduct, is being measured [20]. Recently, Summers et al. produced a chicken IgY antiserum against a DMPO-OA-BSA immunogen and used it to detect DNA-DMPO adducts [26]. Importantly, anti-DMPO IgY binds nonspecifically to agarose beads commonly used for immunoprecipitation experiments, therefore the same authors [26] suggested the need for a precleaning step to eliminate this nonspecific binding by incubating the IgY with 1% agarose in a blocking buffer for 2 h at room temperature. Moreover, rabbit monoclonal IgG and goat anti-DMPO antibodies against DMPO-OA-BSA and DMPO-OA-keyhole limpet hemocyanin have been produced and are awaiting validation (Ramirez Laboratory, unpublished data).
4. Considerations in the use of DMPO to study protein radicals in cells, tissues and whole animals
If a free radical does not react with a spin trap, it will rapidly decay to more stable, non-radical products and become undetectable by spin trapping. For this reason DMPO must be present while the free radical still exists [20]. Therefore, stored samples of cells or tissues are of no use for spin trapping or IST assays.
In most cases in cells or whole animals, the higher the concentration of DMPO the better for IST assays, because both reduced glutathione (GSH) and L-ascorbate will react faster [27–28] with free radicals than DMPO will, or because they exist in a higher concentration than the spin trap (e.g. GSH) (See competing reactions in Fig. 1). Therefore, DMPO in cells or tissues will trap a relatively small fraction of the free radicals formed unless DMPO concentrations are high enough to outcompete other reactions of the free radicals, including, for example, peroxyl radical and dityrosine formation [21]. In vitro, DMPO concentrations as high as 100 mM are routinely used in incubation studies for 24 h with no changes in cell viability [29].
In this situation, the antioxidant nature of DMPO becomes apparent, and secondary and tertiary events including tissue damage and toxicity can be prevented [29]. For this reason a thorough study may require a range of non-cytotoxic DMPO incubation conditions (i.e., concentrations and incubation times). For exploratory work 2–100 mM DMPO is adequate. The final concentration of DMPO should be as high as possible, but not so high that it affects the biochemistry of the system under study [29].
In view of the necessary high concentrations of DMPO, it is fortunate that DMPO has low toxicity. At 50 mM, DMPO does not affect trypan blue exclusion or the clonogenicity of Chinese hamster ovary cells, but it does prevent colony formation by 9L tumor cells [30]. DMPO administered to Sprague Dawley rats at 2.32 g/kg body weight (average concentration of 20.5 mM) is nontoxic, so that necessarily high concentrations of DMPO can be used with little side effect [12].
In addition to all the usual controls, a typical IST assay in cells or whole animals should have an additional control, the no-DMPO control [31–32]. As with all polyclonal antibodies, non-specific binding can occur with the anti-DMPO antiserum. For instance, with the polyclonal rabbit anti-DMPO, non-specific binding to catalase in hepatocytes has been noticed [33]. This non-specific binding is independent of DMPO and therefore was obvious in the no-DMPO control. The greatest strength of IST, like spin trapping with ESR itself, is that false positives, although possible, are very rare. This absence of false positives is in contrast to other methods of detecting free radical-mediated oxidative modification of macromolecules. For example, the carbonyl assay, the measurement of 8-oxo-7,8-dihydro-2′-deoxyguanosine, and many other assays are characterized by high background signals of unknown origin. However, one should also consider artifactual sources of nitrone adducts produced by the chemistry of spin trapping [16, 18].
Any in vivo microenvironment that has the capability of forming a free radical certainly will have the ability to oxidize the radical adduct to its stable nitrone [20]. Although the oxidation of the radical adduct to the nitrone is a necessary step for detection with anti-DMPO, in principle the oxidation could occur ex vivo without compromising the assay. Unless either the radical or hydroxylamine adduct chemically fragments, as is known to occur for the superoxide and thiyl radical adducts, the nitrone adduct as the only stable product must accumulate until all of the radical and hydroxylamine adducts decompose one way or another [17].
As with any method, attention must be given to certain apparently small details if failure is to be averted. First, a horseradish peroxidase secondary antibody will be positive everywhere a protein with peroxidase activity is present, which usually occurs in tissues. This is true for all immunochemical detection using an horseradish peroxidase-labeled secondary antibody. When hemoproteins are known to be present, our laboratory uses an alkaline phosphatase-conjugated secondary antibody [31–32]. Moreover, DMPO and nearly all organic compounds are contaminated with trace quantities of H2O2 and reducing substances. All chemicals should be as pure as possible, because the assay with met-hoMb/H2O2 is as sensitive for H2O2 as Amplex Red. In the hoMb/H2O2 system, the DMPO-hoMb nitrone could easily be detected by ELISA with only 1 μM H2O2 [23]. Whether the assay detects a hoMb radical or H2O2 is only a matter of perspective.
5. IST study of protein radicals in purified proteins
Since the production of the anti-DMPO antibody [23], a number of protein radicals have been detected by DMPO-based IST [21, 32], including myoglobin [23, 34–36], hemoglobin [22, 35, 37–38], cytochrome c [39], neuroglobin [40], Cu, Zn-superoxide dismutase (SOD1) [41–43] and Mn-SOD [44], catalase [33], myeloperoxidase [45–46], eosinophil peroxidase [47], lactoperoxidase [48–49], microglobulin [50], albumin [43, 51], thyroid peroxidase (TPO)[25], α-lactalbumin [52] and Ras GTPase [53]. Oxidants used to generate these radicals included H2O2, peroxynitrite, Cu2+/H2O2, hypochlorous acid, tert-butylhydroperoxide, peroxymonocarbonate, and reactive species generated from ketoprofen plus UV-A, riboflavin plus UV-A irradiation and nano-TiO2 plus UV-A irradiation. Below, we briefly discuss a few examples of these studies of protein-centered radicals formed in models that use purified proteins treated with a bolus or slow generation of oxidants in the presence of DMPO, and highlight the medical significance of the biochemical findings, challenges, pitfalls and further considerations of the IST assay.
Myoglobin radical formation was observed in rat heart homogenates exposed to H2O2 by ESR-spin trapping [54] and IST [23]. The site of DMPO trapping on hoMb was determined to be Tyr103 by MS/MS and site-specific mutagenicity experiments [34, 55]. The LC-ELISA-MS-based strategy [36] is a powerful approach to identifying the sites of DMPO reaction with specific radicalized amino acid residues in the model hemoproteins sperm whale Mb, horse heart Mb and human oxyHb. Importantly, this study by Lardinois et [36] showed that peptides containing Tyr-DMPO adducts are better preserved under neutral or basic conditions. In fact, if a DMPO-protein nitrone adduct is not found, it does not mean that the protein was not radicalized, but that the reaction of DMPO with the radical site may have been slower than the reaction of the radical by other pathways (See competing reaction in Fig. 1).
Although a number of studies have identified sites of DMPO binding to proteins, these sites may or may not be the major sites of free radical damage or the cause of the observed effect on the change in function of the protein. Indeed, one of the major problems that potentially affect both traditional spin trapping and IST data is the rapid electron transfer between amino acid residues. Radical chemistry studies indicate that electrons can delocalize between Tyr, Trp, Cys, Met, Cys, and probably His residues, in proteins via electron or hydrogen atom transfer reactions. Then the spin trap will react with them at different rates, causing disequilibrium in the populations of radical sites in the protein. Therefore, a minor radical site in a protein can appear to be a major site of damage because of its rapid reaction with the spin trap or its greater accessibility. Thus the major radical site detected by DMPO-based IST in a protein may be determined more by the rate of addition of DMPO (and subsequent rate of oxidation of the nitrone), than the original concentration/abundance of the radical.
Bonini et al. [33] found that in vitro and in mouse hepatocytes the reaction of catalase with HOCl produced a protein radical, which is a primary event leading to catalase aggregation and loss of function. Catalase was immunoprecipitated with an anti-catalase antibody from hepatocytes treated with HOCl and DMPO, and then probed with an anti-DMPO antibody [33]. However, protein radicalization in cells exposed to a stressor and DMPO can be obtained when the anti-DMPO is first used to immunoprecipitate total adducted proteins, which can then be separated and identified [29]. A protein’s sensitivity to radicalization in a stressed cell depends on many factors, including its proximity to the source of oxidants, local concentration of oxidants, protein conformation, antioxidants in its microenvironment, etc. [56].
Ehrenshaft et al. [25] used IST to investigate protein radical formation in the reaction of TPO with H2O2, its normal substrate. IST has reduced the amount of protein needed for detection of radicals by 1,000- fold, from milligrams to micrograms [25]. We expect that the use of IST in TPO studies will provide a valuable tool for revealing the catalytic processes of this essential peroxidase. In order to translate these findings to the field of therapeutic interventions it would be necessary to adjust these experiments to a cell or in vivo model of thyroid deregulation by iodine deficiency or exposure to thyroid toxicants.
6. Protein radicals in models of diseases
Detection of protein radicals in vivo remains a major challenge in free radical research. Protein-DMPO radical adducts have been studied using IST in models of amyotrophic lateral sclerosis or Luc Gerig’s disease [57], idiosyncratic agranulocytosis [45–46, 58–60], host-parasite interaction [61–62], sepsis [63–65], LPS-induced lung injury [63], ischemia/reperfusion (I/R) [39, 66–70], asthma [43, 47], diabetes [71–72] and obesity [73–74]. These studies may provide a molecular mechanism of tissue dysfunction in toxicology and pathology. Some considerations in the trapping of radicals and detecting protein-DMPO in vivo have been discussed in section 4.
Before briefly discussing a few advances, challenges and pitfalls of IST two key points should be considered in the study protein radicals in vivo. Firstly, the fact that a particular protein can be detected as a DMPO nitrone adduct does not mean that it is important in pathology or damage. It may be exactly the opposite. The radicalized residue in a protein may have been detected because it did not undergo other reactions rapidly, and thus may be essentially an irrelevant species in pathology. It may be that radicals detected by IST are those that do not react rapidly with oxygen (e.g., Tyr and Trp residues) or do so reversibly (e.g., Cys). Thus in general, only radicals that do not undergo very rapid alternative reactions (See Fig. 1) appear to react competitively with DMPO, and hence give species that can be detected by spin trapping and IST. Secondly, the reaction of DMPO with reactive chemical species can affect the efficiency of the spin trap to react with protein radicals. Recently, we found that DMPO can prevent the activation of macrophages by interfering with ROS-mediated signaling pathways that lead to the activation of the master regulator of the inflammatory response nuclear factor (NF)-κB [75]. Most recently, Villamena’s team [17] has investigated the thermodynamics of the reactions of DMPO and other nitrones with a number of reactive nitrogen species, including •NO, •NO2, ONOOCO2− and ONOO2−, which are known to be generated in LPS-activated macrophages. This work indicated that these reactions of spin traps with reactive species should be carefully considered because they can diminish the local concentrations of the spin trap at sites of inflammation where protein radicalization more likely occurs.
Protein radicals have been determined in host-parasite interaction models. One of these studies from Radi’s team has shown peroxynitrite-mediated oxidative killing of Trypanosome cruzi trypomastigotes with formation of DMPO-nitrone adducts when phagocytized by J774.1 macrophages activated with interferon-γ and lipopolysaccharide (LPS) [62]. This study showed the trapping of a protein radical in a unique targeted compartment by loading a parasite with the spin trap, but did not consider protein radical formation in other compartments of the activated macrophage. See Section 7 and [29] for a discussion on the study of protein radicals in whole macrophages activated with LPS.
Recently Chatterjee and collaborators [64] reported the radicalization and trapping of carboxypeptidase B1 in the spleens of mice treated with a single bolus dose of LPS-a model of sepsis and DMPO. However, a careful assignment of a protein target of oxidation in cell or tissue homogenates should include an anti-DMPO Western blot of the total homogenate instead of immunoprecipitated anti-DMPO adducts with or without further detection of the specific protein [29, 76].
Dogan et al. [70] observed that DMPO prevented hepatic damage caused by reperfusion as assessed by reduction in the markers of hepatic damage, most likely by trapping protein and DNA radicals. Interestingly, their data showed that most nitrone adducts were localized in the cytosol as well as the mitochondria and the nucleus. The marked reduction in I/R induced thiobarbituric acid-reactive substances and 4-hydroxynonenal adducted proteins may indicate that DMPO can also trap lipid-centered radicals, or alternatively interfere with the chain of lipid peroxidation as previously suggested [77]. However, the development of a high-throughput immunoassay to determine DMPO-lipid nitrone adducts remains a challenge.
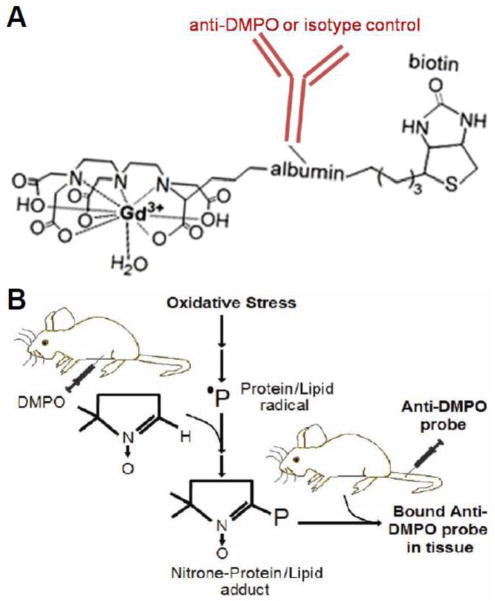
A recent study from Towner’s team [78] used a novel anti-DMPO probe (Fig. 3A) to study in vivo protein/lipid radicals in tissues of diabetic mice using a non-invasive, molecular magnetic resonance imaging (MRI) technique (Fig. 3B). The location of the anti-DMPO probe in excised tissues (lungs, liver and kidneys) was confirmed by confocal imaging via conjugation of streptavidin-Cy3, which targeted the probe biotin moiety. To our knowledge this is the first report showing protein radicals by DMPO-based IST in a mild in vivo model of chronic low-grade inflammation. Appropriate controls included non-diabetic mice administered the anti-DMPO probe and diabetic mice administered a non-specific IgG contrast agent of a similar molecular weight to the anti-DMPO probe [88]. However, the use of the anti-DMPO MRI molecular probe developed by Towner’s group because of its molecular size, the permeability throughout the intercellular space may limit its utility to detect protein/lipid radicals in hypoxic environments where blood perfusion is usually poor, but where inflammation and ROS can cause free radical-mediated damage. This group is now considering generating molecular MRI probes using nanoparticles and Fab antibody fragments, which may decrease the molecular size of the molecular imaging probes.
Figure 3. In vivo molecular MR imaging (mMRI) of DMPO-nitrone adducts.
A) Schematic representation of the anti-DMPO mMRI probe used to detect free radicals forming membrane-associated-DMPO nitrone adducts; B) scheme of the experimental design to form membrane-associated radicals and detection of these radicals using the probe shown in A with mMRI. See reference [78] for further details.
7. The future of immuno-spin trapping of protein radicals
Protein carbonyl formation is one acceptable marker of protein oxidation [79]. See also [80] in this special issue. Previously we found that trapping of protein radicals with DMPO prevented carbonyl formation [51]. A similar effect was observed in RAW264.7 macrophages activated with LPS. Ramirez’s team has developed a high-throughput cell-based anti-DMPO immunoassay that was used to quantify protein-DMPO nitrone adducts in LPS-activated RAW264.7 cells and helped to define the best conditions for further characterizing, identifying and localizing protein radicals formed in LPS-primed cells [29, 81]. This development may help advance the study of free radicals in high-throughput formats.
Radicalomics, the study of all protein radicals formed in time and space at a defined time or condition, is a very challenging area not only because free radicals can be formed in proteins [9], DNA [82], lipids [77] and even polysaccharides [83], but because several different free radicals can be formed within a single macromolecule and its constituents (amino acids, nucleotides, fatty acid and monosaccharides). More challenging yet is the fact that in a single constituent, the unpaired electron can delocalize between atoms and thus would determine the site where the DMPO will react and whether the radical adduct will be oxidized to the corresponding nitrone. These facts explain why a standard for nitrone adducts has not been developed yet.
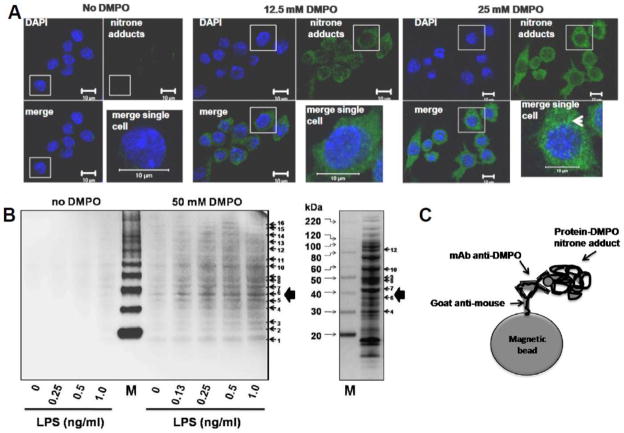
A broad search for identity and localization of specific radicalized proteins in stressed cells and the mechanism behind these findings is a challenge that Ramirez’s laboratory has taken up. DMPO was found to trap radicalized proteins to form protein-DMPO nitrone adducts, to decrease protein carbonyl formation, and to block LPS-induced cell death [29]. Microarray and isobaric tags for relative and absolute quantitation (iTRAQ) analyses showed that DMPO affects the transcriptome and proteome, respectively, in RAW 264.7 macrophages exposed to LPS (Ramirez laboratory, unpublished data). Moreover, confocal microscopy and cell fractionation data showed the subcellular localization of proteins labeled with DMPO (Fig. 4A). Cell fractionation data from macrophages treated with LPS and DMPO for 24 h clearly showed that DMPO nitrone adducts are mainly formed in the mitochondrial and microsomal fractions [32].
Figure 4. Localization and identification of protein-DMPO nitrone adducts in RAW 264.7 macrophages treated with LPS and DMPO.
A) Single-plane confocal images of nitrone adducts formed in cells treated with 1 ng/ml LPS and 12.5 or 50 mM DMPO for 24 h. Green indicates nitrone adducts and blue indicates nuclei. Insert is a high-power magnification of the image of a single and representative cell. The white arrowhead indicates the perinuclear localization of most nitrone adducts. B) Western blot analysis of protein-DMPO nitrone adducts in homogenates of cells treated with LPS and/or DMPO for 24 h. Right panel, coomassie blue staining of the homogenate of cells treated with 1 ng/ml LPS and 50 mM DMPO for 24 h, separated in a reducing gel and showing 7 representative bands that correspond to anti-DMPO-positive bands in the Western blot. C) Schematic representation of an anti-DMPO molecular “catcher”-protein-DMPO nitrone adduct complex used to pull-down proteins labeled with DMPO. M indicates Magic Mark Western XP molecular weight marker (Invitrogen). Modified from [29].
To identify radicalized and DMPO-tagged proteins, total homogenates of macrophages treated with LPS and DMPO were analyzed using an anti-DMPO Western blot and LC/MS-MS (Fig. 4B). Importantly, proteins identified in the anti-DMPO-positive bands might or might not be proteins labeled with DMPO [29]. For instance, the authors rationalized that some proteins not labeled with DMPO can be physically associated with proteins that are covalently bound to the spin trap and wrongly assigned as DMPO nitrone adducts. To simplify the analysis, we prepared an anti-DMPO molecular catcher (Fig. 4C) to pull down proteins or complexes of proteins in which at least one protein was adducted with DMPO [29]. This was the first report on radicalomics that warrants further corroboration and study of specific proteins labeled with DMPO during macrophage activation and their role in the fate of macrophages at sites of inflammation. In addition, the identified proteins need to be corroborated by knocking down specific proteins and mutating specific residues where DMPO adduction should be lost.
The identification of radicalized proteins during cell stress may allow them to be included in interactomes that will help define their role in the complex cell response to stressors. One of the major challenges in the characterization of free radical oxidation of specific proteins in cells or tissues from animals is the difficulty of isolating these DMPO-protein adducts and further identification of the specific residues where DMPO is bound. Cell and tissue fractionation experiments can help, but it is important to consider that DMPO adduction to some proteins and peptide fragments can be unstable under certain circumstances such as acid or basic pH [36]. In the damage and pathology discussed in sections 4 and 6, the significance of the identification of radical sites in a specific protein and the identity of the protein labeled with DMPO also applies to macrophage proteins labeled with DMPO.
8. Concluding Remarks
In this review article we have critically discussed recent advances, challenges and pitfalls of the investigation of protein-centered radicals using DMPO-based IST in models of purified proteins as well as in cells, tissues and whole animals. Indeed, IST has been successfully applied in mechanistic in vitro studies, and recently, macromolecule-centered radicals have been detected in models of drug-induced agranulocytosis, hepatotoxicity, cardiotoxicity, and I/R, as well as in animal models of neurological, immunological, and metabolic diseases. As in spin trapping, the fidelity of IST is determined by the reaction of a free radical with a spin trap, as well as the immune-specificity of the antibody, the technique used to visualize the antigen-antibody complex, and the techniques used to separate, localize and identify the site-specific reaction of the spin trap within the primary sequence of the protein. In addition, unlike other techniques used to determine protein oxidation, such as carbonyl and nitration assays, and because of the promiscuity of the free radical reactions, we have been unable to produce a standard to provide quantitative data. However, DMPO nitrone adducts can be presented as a signal relative to the amount of proteins or DNA in the sample or the relative intensity of images. Note that DMPO could trap a variety of macromolecular (and small) free radicals even though there are distinct reaction rate constants for each type of free radical. This relatively weak selectivity or specificity in trapping radicals should also be considered in assays of other types of macromolecular radicals.
Because the spin trap has to be present at high enough concentrations in the microenvironment where the radical is formed, the possible effects of the spin trap on gene expression, metabolism and cell physiology have to be considered. In trying to understand the role of protein radicalization in stressed cells or tissues, most published studies have been limited to immunoprecipitation of specific proteins from cell homogenates and further confirmation of DMPO adducted to a protein. However, because of the promiscuity of free radical reactions, mostly controlled by thermodynamic, kinetic and structural features, it is expected that more than one protein or amino acid residue can be radicalized. Consequently, to define the overall impact of protein radicalization on physiology, it is necessary to immunoprecipitate proteins labeled with DMPO and then confirm their identity, the site of reaction with DMPO, the location, and any change in function. Now the next goal should be to identify the specific proteins undergoing oxidation and to study the role of these modifications in pathogenesis and cell response to stress.
Finally, herein we have mentioned a number of examples of the use of IST to identify radicalized proteins in purified proteins, organelles, cells and tissues. We have also highlighted the important challenges regarding IST data interpretation: 1) the impact of the spin trap in physiology and gene expression; 2) the fidelity of spin trapping; 3) electron transfer and disequilibrium in the concentration of radical sites in the protein; 4) radical decay mechanisms and the significance of radicalized proteins in the cellular response to stress; 5) non-adducted proteins pulled down with DMPO-protein nitrone adducts; and 6) the need for appropriate controls. Only after meeting these challenges will we be able to assign a role to free radical modification of proteins in medicine.
Highlights.
DMPO-based immuno-spin trapping (IST) allows the study of protein radicals.
Protein radicals are usually formed and detected at sites of inflammation.
The fidelity of IST is determined by spin trapping and immunochemistry.
Electron transfer and decay kinetics in protein radicals should be considered.
DMPO effects on biology, gene expression and physiology should be evaluated.
Acknowledgments
DCR and SEGM acknowledge the Re-insertion Grant from the CONICET, Government of the Republic of Argentina (Res. D N° 1488). MCD-V and MDM are fellows from the CONICET. The project was supported in part by Award Number 5R00ES015415-04 to DCR from the National Institute of Environmental Health Sciences. SC is supported by grant 4R00ES019875-02 from NIEHS, NIH. KH’s research is supported by an MDA grant (MDA217526). RAT research is supported by the Oklahoma Medical Research Foundation. RPM’s research is supported by the Intramural Research Program of the NIEHS, NIH, DHHS. Authors acknowledge Dr. Ann Motten and Mary Mason for helping to edit this manuscript and to Dr Maria S. Gimenez and Sergio E. Alvarez (IMIBIO-SL-CONICET) for critical reading of this manuscript. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Environmental Health Sciences or the National Institutes of Health. Due to space limitations, we were unable to review all relevant work and apologize to those colleagues whose contributions were not discussed in this review article.
Abbreviations
- AG
aminoglutethimide
- BSA
bovine serum albumin
- DMPO
5,5-dimethyl-1-pyrroline N-oxide
- ESR
electron-spin resonance
- HPLC
high performance liquid chromatography
- hoMb
horse myglobin
- huHb/Mb
human hemoglobin/myoglobin
- I/R
ischemia/reperfusion
- IST
immuno-spin trapping
- LC
liquid chromatography
- LPS
lipopolysaccharide
- MRI
magnetic resonance imaging
- MS
mass spectrometry
- OA
octanoic acid
- SOD
superoxide dismutase
- TPO
thyroid peroxidase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mason RP. Assay of in situ radicals by electron spin resonance. Methods Enzymol. 1984;105:416–422. doi: 10.1016/s0076-6879(84)05058-8. [DOI] [PubMed] [Google Scholar]
- 2.Mason RP. In vivo spin trapping- from chemistry to toxicology. In: Rhodes CJ, editor. Toxicology of the Human Environment. The Critical Role of Free Radicals. Taylor and Francis; London: 2000. pp. 49–70. [Google Scholar]
- 3.Augusto O, Schreiber J, Mason RP. Direct ESR detection of a free radical intermediate during the peroxidase-catalyzed oxidation of the antimalarial drug primaquine. Biochem Pharmacol. 1988;37:2791–2797. doi: 10.1016/0006-2952(88)90042-1. [DOI] [PubMed] [Google Scholar]
- 4.Davies MJ. Detection and characterization of radicals using EPR methodology. BBA SPECIAL ISSUE: CURRENT METHODS TO STUDY REACTIVE OXYGEN SPECIES - PROS AND CONS. 2013 [Google Scholar]
- 5.Davies MJ, Slater TF. The use of electron-spin-resonance techniques to detect free-radical formation and tissue damage. Proc Nutr Soc. 1988;47:397–405. doi: 10.1079/pns19880058. [DOI] [PubMed] [Google Scholar]
- 6.Janzen EG. Spin trapping. Methods Enzymol. 1984;105:188–198. doi: 10.1016/s0076-6879(84)05025-4. [DOI] [PubMed] [Google Scholar]
- 7.Davies MJ, Hawkins CL. EPR spin trapping of protein radicals. Free Radic Biol Med. 2004;36:1072–1086. doi: 10.1016/j.freeradbiomed.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 8.Augusto O, Muntz Vaz S. EPR spin-trapping of protein radicals to investigate biological oxidative mechanisms. Amino Acids. 2007;32:535–542. doi: 10.1007/s00726-006-0429-4. [DOI] [PubMed] [Google Scholar]
- 9.Davies MJ, Gilbert BC, Haywood RM. Radical-induced damage to proteins: e.s.r. spin-trapping studies. Free Radic Res Commun. 1991;15:111–127. doi: 10.3109/10715769109049131. [DOI] [PubMed] [Google Scholar]
- 10.Janzen EG, West MS, Kotake Y, DuBose CM. Biological spin trapping methodology. III. Octanol-water partition coefficients of spin-trapping compounds. Journal of biochemical and biophysical methods. 1996;32:183–190. doi: 10.1016/0165-022x(96)00008-5. [DOI] [PubMed] [Google Scholar]
- 11.Anzai K, Aikawa T, Furukawa Y, Matsushima Y, Urano S, Ozawa T. ESR measurement of rapid penetration of DMPO and DEPMPO spin traps through lipid bilayer membranes. Arch Biochem Biophys. 2003;415:251–256. doi: 10.1016/s0003-9861(03)00260-1. [DOI] [PubMed] [Google Scholar]
- 12.Janzen EG, Poyer JL, Schaefer CF, Downs PE, DuBose CM. Biological spin trapping. II. Toxicity of nitrone spin traps: dose-ranging in the rat. Journal of biochemical and biophysical methods. 1995;30:239–247. doi: 10.1016/0165-022x(95)00012-1. [DOI] [PubMed] [Google Scholar]
- 13.Villamena FA, Das A, Nash KM. Potential implication of the chemical properties and bioactivity of nitrone spin traps for therapeutics. Future Med Chem. 2012;4:1171–1207. doi: 10.4155/fmc.12.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Floyd RA, Kopke RD, Choi CH, Foster SB, Doblas S, Towner RA. Nitrones as therapeutics. Free Radic Biol Med. 2008;45:1361–1374. doi: 10.1016/j.freeradbiomed.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janzen EG, Jandrisits LT, Shetty RV, Haire DL, Hilborn JW. Synthesis and purification of 5,5-dimethyl-1-pyrroline-N-oxide for biological applications. Chem Biol Interact. 1989;70:167–172. doi: 10.1016/0009-2797(89)90071-9. [DOI] [PubMed] [Google Scholar]
- 16.Ranguelova K, Mason RP. The fidelity of spin trapping with DMPO in biological systems. Magn Reson Chem. 2011;49:152–158. doi: 10.1002/mrc.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nash KM, Rockenbauer A, Villamena FA. Reactive nitrogen species reactivities with nitrones: theoretical and experimental studies. Chem Res Toxicol. 2012;25:1581–1597. doi: 10.1021/tx200526y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leinisch F, Ranguelova K, Derose EF, Jiang J, Mason RP. Evaluation of the Forrester-Hepburn Mechanism as an Artifact Source in ESR Spin-Trapping. Chem Res Toxicol. 2011;24:2217–2226. doi: 10.1021/tx2003323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hawkins CL, Morgan PE, Davies MJ. Quantification of protein modification by oxidants. Free Radical Biology and Medicine. 2009;46:965–988. doi: 10.1016/j.freeradbiomed.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 20.Mason RP. Using anti-5,5-dimethyl-1-pyrroline N-oxide (anti-DMPO) to detect protein radicals in time and space with immuno-spin trapping. Free Radic Biol Med. 2004;36:1214–1223. doi: 10.1016/j.freeradbiomed.2004.02.077. [DOI] [PubMed] [Google Scholar]
- 21.Gomez-Mejiba SE, Zhai Z, Akram H, Deterding LJ, Hensley K, Smith N, Towner RA, Tomer KB, Mason RP, Ramirez DC. Immuno-spin trapping of protein and DNA radicals: “tagging” free radicals to locate and understand the redox process. Free Radic Biol Med. 2009;46:853–865. doi: 10.1016/j.freeradbiomed.2008.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramirez DC, Chen YR, Corbett J, Mason RP. Detection of hemoglobin-tyrosyl-radical derived nitrone adducts by immuno-spin trapping. A first application to in vivo toxicology. Free Radical Biology and Medicine. 2002;33:434. [Google Scholar]
- 23.Detweiler CD, Deterding LJ, Tomer KB, Chignell CF, Germolec D, Mason RP. Immunological identification of the heart myoglobin radical formed by hydrogen peroxide. Free Radic Biol Med. 2002;33:364–369. doi: 10.1016/s0891-5849(02)00895-x. [DOI] [PubMed] [Google Scholar]
- 24.Schoneich C. Detection of posttranslational modifications of proteins. BBA SPECIAL ISSUE: CURRENT METHODS TO STUDY REACTIVE OXYGEN SPECIES - PROS AND CONS. 2013 [Google Scholar]
- 25.Ehrenshaft M, Mason RP. Protein radical formation on thyroid peroxidase during turnover as detected by immuno-spin trapping. Free Radic Biol Med. 2006;41:422–430. doi: 10.1016/j.freeradbiomed.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 26.Summers FA, Mason RP, Ehrenshaft M. Development of immunoblotting techniques for DNA radical detection. Free Radic Biol Med. 2012 doi: 10.1016/j.freeradbiomed.2012.10.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Domazou AS, Koppenol WH, Gebicki JM. Efficient repair of protein radicals by ascorbate. Free Radic Biol Med. 2009;46:1049–1057. doi: 10.1016/j.freeradbiomed.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 28.Gebicki JM, Nauser T, Domazou A, Steinmann D, Bounds PL, Koppenol WH. Reduction of protein radicals by GSH and ascorbate: potential biological significance. Amino Acids. 2010;39:1131–1137. doi: 10.1007/s00726-010-0610-7. [DOI] [PubMed] [Google Scholar]
- 29.Zhai Z, Gomez-Mejiba SE, Gimenez MS, Deterding LJ, Tomer KB, Mason RP, Ashby MT, Ramirez DC. Free radical-operated proteotoxic stress in macrophages primed with lipopolysaccharide. Free Radic Biol Med. 2012;53:172–181. doi: 10.1016/j.freeradbiomed.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khan N, Wilmot CM, Rosen GM, Demidenko E, Sun J, Joseph J, O’Hara J, Kalyanaraman B, Swartz HM. Spin traps: in vitro toxicity and stability of radical adducts. Free Radic Biol Med. 2003;34:1473–1481. doi: 10.1016/s0891-5849(03)00182-5. [DOI] [PubMed] [Google Scholar]
- 31.Ramirez DC, Gomez-Mejiba SE, Mason RP. Immuno-spin trapping analyses of DNA radicals. Nature protocols. 2007;2:512–522. doi: 10.1038/nprot.2007.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramirez DC, Mason RP. Immuno-spin trapping: detection of protein-centered radicals. Curr Protoc Toxicol. 2005;Chapter 17(Unit17 ):17. doi: 10.1002/0471140856.tx1707s24. [DOI] [PubMed] [Google Scholar]
- 33.Bonini MG, Siraki AG, Atanassov BS, Mason RP. Immunolocalization of hypochlorite-induced, catalase-bound free radical formation in mouse hepatocytes. Free Radic Biol Med. 2007;42:530–540. doi: 10.1016/j.freeradbiomed.2006.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Detweiler CD, Lardinois OM, Deterding LJ, de Montellano PR, Tomer KB, Mason RP. Identification of the myoglobin tyrosyl radical by immuno-spin trapping and its dimerization. Free Radic Biol Med. 2005;38:969–976. doi: 10.1016/j.freeradbiomed.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 35.Bhattacharjee S, Deterding LJ, Jiang J, Bonini MG, Tomer KB, Ramirez DC, Mason RP. Electron transfer between a tyrosyl radical and a cysteine residue in hemoproteins: spin trapping analysis. J Am Chem Soc. 2007;129:13493–13501. doi: 10.1021/ja073349w. [DOI] [PubMed] [Google Scholar]
- 36.Lardinois OM, Detweiler CD, Tomer KB, Mason RP, Deterding LJ. Identifying the site of spin trapping in proteins by a combination of liquid chromatography, ELISA, and off-line tandem mass spectrometry. Free Radic Biol Med. 2008;44:893–906. doi: 10.1016/j.freeradbiomed.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramirez DC, Chen YR, Mason RP. Immunochemical detection of hemoglobin-derived radicals formed by reaction with hydrogen peroxide: involvement of a protein-tyrosyl radical. Free Radic Biol Med. 2003;34:830–839. doi: 10.1016/s0891-5849(02)01437-5. [DOI] [PubMed] [Google Scholar]
- 38.Deterding LJ, Ramirez DC, Dubin JR, Mason RP, Tomer KB. Identification of free radicals on hemoglobin from its self-peroxidation using mass spectrometry and immuno-spin trapping: observation of a histidinyl radical. J Biol Chem. 2004;279:11600–11607. doi: 10.1074/jbc.M310704200. [DOI] [PubMed] [Google Scholar]
- 39.Chen Y-R, Chen C-L, Liu X, Li H, Zweier JL, Mason RP. Involvement of protein radical, protein aggregation, and effects on NO metabolism in the hypochlorite-mediated oxidation of mitochondrial cytochrome c. Free Radic Biol Med. 2004;37:1591–1603. doi: 10.1016/j.freeradbiomed.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 40.Lardinois OM, Tomer KB, Mason RP, Deterding LJ. Identification of protein radicals formed in the human neuroglobin-H2O2 reaction using immuno-spin trapping and mass spectrometry. Biochemistry. 2008;47:10440–10448. doi: 10.1021/bi800771k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramirez DC, Gomez Mejiba SE, Mason RP. Mechanism of hydrogen peroxide-induced Cu,Zn-superoxide dismutase-centered radical formation as explored by immuno-spin trapping: the role of copper- and carbonate radical anion-mediated oxidations. Free Radic Biol Med. 2005;38:201–214. doi: 10.1016/j.freeradbiomed.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 42.Ramirez DC, Gomez-Mejiba SE, Corbett JT, Deterding LJ, Tomer KB, Mason RP. Cu,Zn-superoxide dismutase-driven free radical modifications: copper- and carbonate radical anion-initiated protein radical chemistry. Biochem J. 2009;417:341–353. doi: 10.1042/BJ20070722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ranguelova K, Bonini MG, Mason RP. (Bi)sulfite Oxidation by Copper,Zinc-Superoxide Dismutase: Sulfite-Derived, Radical-Initiated Protein Radical Formation. Environmental Health Perspectives. 2010;118:970–975. doi: 10.1289/ehp.0901533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ansenberger-Fricano K, Ganini D, Mao M, Chatterjee S, Dallas S, Mason RP, Stadler K, Santos JH, Bonini MG. The peroxidase activity of mitochondrial superoxide dismutase. Free Radic Biol Med. 2013;54:116–124. doi: 10.1016/j.freeradbiomed.2012.08.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siraki AG, Bonini MG, Jiang J, Ehrenshaft M, Mason RP. Aminoglutethimide-induced protein free radical formation on myeloperoxidase: a potential mechanism of agranulocytosis. Chem Res Toxicol. 2007;20:1038–1045. doi: 10.1021/tx6003562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siraki AG, Deterding LJ, Bonini MG, Jiang J, Ehrenshaft M, Tomer KB, Mason RP. Procainamide, but not N-acetylprocainamide, induces protein free radical formation on myeloperoxidase: a potential mechanism of agranulocytosis. Chem Res Toxicol. 2008;21:1143–1153. doi: 10.1021/tx700415b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ranguelova K, Chatterjee S, Ehrenshaft M, Ramirez DC, Summers FA, Kadiiska M, Mason RP. Protein radical formation resulting from eosinophil peroxidase-catalyzed oxidation of sulfite. J Biol Chem. 2010 doi: 10.1074/jbc.M109.069054. Under review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo Q, Detweiler CD, Mason RP. Protein radical formation during lactoperoxidase-mediated oxidation of the suicide substrate glutathione: immunochemical detection of a lactoperoxidase radical-derived 5,5-dimethyl-1-pyrroline N-oxide nitrone adduct. J Biol Chem. 2004;279:13272–13283. doi: 10.1074/jbc.M310034200. [DOI] [PubMed] [Google Scholar]
- 49.Bonini MG, Siraki AG, Bhattacharjee S, Mason RP. Glutathione-induced radical formation on lactoperoxidase does not correlate with the enzyme’s peroxidase activity. Free Radic Biol Med. 2007;42:985–992. doi: 10.1016/j.freeradbiomed.2006.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo Q, Gao GH, Qian SY, Mason RP. Novel identification of a sulfur-centered, radical-derived 5,5-dimethyl-1-pyrroline N-oxide nitrone adduct formed from the oxidation of DTT by LC/ELISA, LC/electrospray ionization-MS, and LC/tandem MS. Chemical Research in Toxicology. 2004;17:1481–1490. doi: 10.1021/tx049837o. [DOI] [PubMed] [Google Scholar]
- 51.Ramirez DC, Gomez-Mejiba SE, Mason RP. Copper-catalyzed protein oxidation and its modulation by carbon dioxide: enhancement of protein radicals in cells. J Biol Chem. 2005;280:27402–27411. doi: 10.1074/jbc.M504241200. [DOI] [PubMed] [Google Scholar]
- 52.Dalsgaard TK, Triquigneaux M, Deterding L, Summers F, Ranguelova K, Mortensen G, Mason RP. Site-Specific Detection of Radicals on alpha-Lactalbumin after a Riboflavin-Sensitized Reaction, Detected by Immuno-spin Trapping, ESR, and MS. J Agric Food Chem. 2013;61:418–426. doi: 10.1021/jf303973b. [DOI] [PubMed] [Google Scholar]
- 53.Davis MF, Zhou L, Ehrenshaft M, Ranguelova K, Gunawardena HP, Chen X, Bonini MG, Mason RP, Campbell SL. Detection of Ras GTPase protein radicals through immuno-spin trapping. Free Radic Biol Med. 2012;53:1339–1345. doi: 10.1016/j.freeradbiomed.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Davies MJ. Detection of myoglobin-derived radicals on reaction of metmyoglobin with hydrogen peroxide and other peroxidic compounds. Free Radic Res Commun. 1990;10:361–370. doi: 10.3109/10715769009149905. [DOI] [PubMed] [Google Scholar]
- 55.Gunther MR, Tschirret-Guth RA, Witkowska HE, Fann YC, Barr DP, Ortiz De Montellano PR, Mason RP. Site-specific spin trapping of tyrosine radicals in the oxidation of metmyoglobin by hydrogen peroxide. Biochem J. 1998;330(Pt 3):1293–1299. doi: 10.1042/bj3301293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davies MJ. The oxidative environment and protein damage. Biochim Biophys Acta. 2005;1703:93–109. doi: 10.1016/j.bbapap.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 57.Cassina P, Cassina A, Pehar M, Castellanos R, Gandelman M, de Leon A, Robinson KM, Mason RP, Beckman JS, Barbeito L, Radi R. Mitochondrial dysfunction in SOD1G93A-bearing astrocytes promotes motor neuron degeneration: prevention by mitochondrial-targeted antioxidants. J Neurosci. 2008;28:4115–4122. doi: 10.1523/JNEUROSCI.5308-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Siraki AG, Jiang J, Mason RP. Investigating the mechanisms of aromatic amine-induced protein free radical formation by quantitative structure-activity relationships: implications for drug-induced agranulocytosis. Chem Res Toxicol. 2010;23:880–887. doi: 10.1021/tx900432d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ranguelova K, Rice AB, Khajo A, Triquigneaux M, Garantziotis S, Magliozzo RS, Mason RP. Formation of reactive sulfite-derived free radicals by the activation of human neutrophils: an ESR study. Free Radic Biol Med. 2012;52:1264–1271. doi: 10.1016/j.freeradbiomed.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ranguelova K, Rice AB, Lardinois OM, Triquigneaux M, Steinckwich N, Deterding LJ, Garantziotis S, Mason RP. Sulfite-mediated oxidation of myeloperoxidase to a free radical: Immuno-spin trapping detection in human neutrophils. Free Radic Biol Med. 2013;60C:98–106. doi: 10.1016/j.freeradbiomed.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ranguelova K, Suarez J, Magliozzo RS, Mason RP. Spin trapping investigation of peroxide- and isoniazid-induced radicals in Mycobacterium tuberculosis catalase-peroxidase. Biochemistry. 2008;47:11377–11385. doi: 10.1021/bi800952b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alvarez MN, Peluffo G, Piacenza L, Radi R. Intraphagosomal peroxynitrite as a macrophage-derived cytotoxin against internalized Trypanosoma cruzi: consequences for oxidative killing and role of microbial peroxiredoxins in infectivity. J Biol Chem. 2011;286:6627–6640. doi: 10.1074/jbc.M110.167247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gomez-Mejiba SE, Gimenez MS, Zhai Z, Ramirez DC. Trapping of protein-centered radicals with a nitrone spin trap prevents endotoxin-induced experimental acute respiratory distress syndrome mouse model. Free Radic Biol Med. 2010;49:S184. [Google Scholar]
- 64.Chatterjee S, Ehrenshaft M, Bhattacharjee S, Deterding LJ, Bonini MG, Corbett J, Kadiiska MB, Tomer KB, Mason RP. Immuno-spin trapping of a post-translational carboxypeptidase B1 radical formed by a dual role of xanthine oxidase and endothelial nitric oxide synthase in acute septic mice. Free Radic Biol Med. 2009;46:454–461. doi: 10.1016/j.freeradbiomed.2008.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chatterjee S, Lardinois O, Bonini MG, Bhattacharjee S, Stadler K, Corbett J, Deterding LJ, Tomer KB, Kadiiska M, Mason RP. Site-Specific Carboxypeptidase B1 Tyrosine Nitration and Pathophysiological Implications following Its Physical Association with Nitric Oxide Synthase-3 in Experimental Sepsis. Journal of Immunology. 2009;183:4055–4066. doi: 10.4049/jimmunol.0900593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen YR, Chen CL, Zhang L, Green-Church KB, Zweier JL. Superoxide generation from mitochondrial NADH dehydrogenase induces self-inactivation with specific protein radical formation. J Biol Chem. 2005;280:37339–37348. doi: 10.1074/jbc.M503936200. [DOI] [PubMed] [Google Scholar]
- 67.Zuo L, Chen YR, Reyes LA, Lee HL, Chen CL, Villamena FA, Zweier JL. The Radical Trap 5,5–Dimethyl-1-pyrroline N-oxide Exerts Dose-Dependent Protection Against Myocardial Ischemia-Reperfusion Injury Through Preservation of Mitochondrial Electron Transport. J Pharmacol Exp Ther. 2009;329:515–523. doi: 10.1124/jpet.108.143479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen Y-R, Chen C-L, Pfeiffer DR, Zweier JL. Mitochondrial complex II in the post-ischemic heart. Oxidative injury and the role of protein S-glutathionylation. J Biol Chem. 2007;45:32640–32654. doi: 10.1074/jbc.M702294200. [DOI] [PubMed] [Google Scholar]
- 69.Kang PT, Zhang L, Chen CL, Chen J, Green KB, Chen YR. Protein thiyl radical mediates S-glutathionylation of complex I. Free Radic Biol Med. 2012;53:962–973. doi: 10.1016/j.freeradbiomed.2012.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dogan S, Ozlem Elpek G, Kirimlioglu Konuk E, Demir N, Aslan M. Measurement of intracellular biomolecular oxidation in liver ischemia-reperfusion injury via immuno-spin trapping. Free Radic Biol Med. 2012;53:406–414. doi: 10.1016/j.freeradbiomed.2012.05.028. [DOI] [PubMed] [Google Scholar]
- 71.Stadler K, Bonini MG, Dallas S, Jiang J, Radi R, Mason RP, Kadiiska MB. Involvement of inducible nitric oxide synthase in hydroxyl radical-mediated lipid peroxidation in streptozotocin-induced diabetes. Free Radic Biol Med. 2008;45:866–874. doi: 10.1016/j.freeradbiomed.2008.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stadler K, Bonini MG, Dallas S, Duma D, Mason RP, Kadiiska MB. Direct evidence of iNOS-mediated in vivo free radical production and protein oxidation in acetone-induced ketosis. Am J Physiol Endocrinol Metab. 2008;295:E456–462. doi: 10.1152/ajpendo.00015.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Khoo NK, Cantu-Medellin N, Devlin JE, St Croix CM, Watkins SC, Fleming AM, Champion HC, Mason RP, Freeman BA, Kelley EE. Obesity-induced tissue free radical generation: An in vivo immuno-spin trapping study. Free Radic Biol Med. 2012;52:2312–2319. doi: 10.1016/j.freeradbiomed.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chatterjee S, Rana R, Corbett J, Kadiiska MB, Goldstein J, Mason RP. P2X7 receptor-NADPH oxidase axis mediates protein radical formation and Kupffer cell activation in carbon tetrachloride-mediated steatohepatitis in obese mice. Free Radic Biol Med. 2012;52:1666–1679. doi: 10.1016/j.freeradbiomed.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhai Z, Gomez-Mejiba SE, Zhu H, Lupu F, Ramirez DC. The spin trap 5,5-dimethyl-1-pyrroline N-oxide inhibits lipopolysaccharide-induced inflammatory response in RAW 264.7 cells. Life Sci. 2012;90:432–439. doi: 10.1016/j.lfs.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhai Z, Gomez-Mejiba SE, Ramirez DC. The Nitrone Spin Trap 5,5-Dimethyl-1-pyrroline N-oxide Affects Stress Response and Fate of Lipopolysaccharide-Primed RAW 264.7 Macrophage Cells. Inflammation. 2012;36:346–354. doi: 10.1007/s10753-012-9552-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Qian SY, Wang HP, Schafer FQ, Buettner GR. EPR detection of lipid-derived free radicals from PUFA, LDL, and cell oxidations. Free Radic Biol Med. 2000;29:568–579. doi: 10.1016/s0891-5849(00)00407-x. [DOI] [PubMed] [Google Scholar]
- 78.Towner RA, Smith N, Saunders D, Henderson M, Downum K, Lupu F, Silasi-Mansat R, Ramirez DC, Gomez-Mejiba SE, Bonini MG, Ehrenshaft M, Mason RP. In vivo imaging of immuno-spin trapped radicals with molecular magnetic resonance imaging in a diabetic mouse model. Diabetes. 2012;61:2405–2413. doi: 10.2337/db11-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Winterbourn CC, Buss IH. Protein carbonyl measurement by enzyme-linked immunosorbent assay. Methods Enzymol. 1999;300:106–111. doi: 10.1016/s0076-6879(99)00118-4. [DOI] [PubMed] [Google Scholar]
- 80.Dalle-Donne I. Protein carbonyls as oxidative biomarkers. BBA SPECIAL ISSUE: CURRENT METHODS TO STUDY REACTIVE OXYGEN SPECIES - PROS AND CONS. 2013 [Google Scholar]
- 81.Gomez-Mejiba SE, Zhai Z, Gimenez MS, Ashby MT, Chilakapati J, Kitchin K, Mason RP, Ramirez DC. Myeloperoxidase-induced genomic DNA-centered radicals. J Biol Chem. 2010;285:20062–20071. doi: 10.1074/jbc.M109.086579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ramirez DC, Gomez-Mejiba SE, Mason RP. Immuno-spin trapping of DNA radicals. Nat Methods. 2006;3:123–127. doi: 10.1038/nmeth852. [DOI] [PubMed] [Google Scholar]
- 83.Hawkins CL, Davies MJ. Detection of intermediates formed on reaction of hyaluronic acid and related materials with the hydroxyl radical. Biochem Soc Trans. 1995;23:248S. doi: 10.1042/bst023248s. [DOI] [PubMed] [Google Scholar]