Abstract
Transient receptor potential (TRP) channels are sensors for a wide range of cellular and environmental signals, but elucidating how these channels respond to physical and chemical stimuli has been hampered by a lack of detailed structural information. Here, we exploit advances in electron cryo-microscopy to determine the structure of a mammalian TRP channel, TRPV1, at 3.4Å resolution, breaking the side-chain resolution barrier for membrane proteins without crystallization. Like voltage-gated channels, TRPV1 exhibits four-fold symmetry around a central ion pathway formed by transmembrane helices S5–S6 and the intervening pore loop, which is flanked by S1–S4 voltage sensor-like domains. TRPV1 has a wide extracellular ‘mouth’ with short selectivity filter. The conserved ‘TRP domain’ interacts with the S4–S5 linker, consistent with its contribution to allosteric modulation. Subunit organization is facilitated by interactions among cytoplasmic domains, including N-terminal ankyrin repeats. These observations provide a structural blueprint for understanding unique aspects of TRP channel function.
TRP channels represent a large and diverse family of non-selective cation channels that respond to a wide range of chemical and physical stimuli1,2. Genetic and pharmacologic studies highlight the importance of TRP channels in numerous biological processes ranging from calcium adsorption to sensory transduction3, and thus elucidating how these channels respond to physiological stimuli or drugs is relevant to understanding disorders affecting every major organ system in the body. TRP channels are believed to resemble voltage-gated potassium (Kv) or sodium (Nav) channels in overall transmembrane topology and subunit organization1. However, aside from the fact that some TRP channels exhibit mild voltage sensitivity4, they otherwise share little in common with voltage-gated ion channels (VGICs) vis-à-vis pharmacological and biophysical properties. Whereas crystal structures have been determined for VGICs5–8, this has not been accomplished for any member of the TRP channel family. Thus, it is currently unknown whether, or to what extent, TRP and Kv or Nav channels share a common structural core, or how structural similarities and differences account for unique functionalities.
TRPV1 is the receptor for capsaicin, the pungent agent from chili peppers that elicits burning pain9. It is also the founding member of a subfamily of thermosensitive TRP channels that enable somatosensory neurons, or other cell types, to detect changes in ambient temperature. TRPV1 is activated by noxious heat and modulated by inflammatory agents, such as extracellular protons and bioactive lipids, which contribute to pain hypersensitivity9–13. TRPV1 is arguably the best-characterized member of the vertebrate TRP family; its widely validated role in pain physiology and the availability of well characterized pharmacological agents make it a ‘poster child’ for elucidating basic principles underlying TRP channel function and structure. Moreover, TRPV1 and other somatosensory TRP channels are considered important targets for analgesic drugs12,13, providing further impetus to determine a structure for any member of this extended protein family.
In recent years, single-particle electron cryomicroscopy (cryo-EM) has enabled three-dimensional (3D) reconstruction of large protein complexes to near atomic resolution14–17, but analysis of small membrane proteins, such as TRP channels18–20, remained at low resolution. Here, we exploit a newly developed direct electron detector and new image processing algorithms to correct motion-induced image blurring and improve signal and contrast of small particle cryo-EM images14,15. With these tools, we determine the structure of TRPV1 at 3.4Å resolution without crystallization. Thus, in addition to revealing a TRP channel structure, we showcase single-particle cryo-EM as a powerful and transformative advance in the structural analysis of membrane proteins.
General architecture of TRPV1
We first identified a rat TRPV1 deletion mutant with enhanced biochemical stability (Extended Data Figs. 1 and 2). When expressed in mammalian cells or oocytes this modified subunit responded to numerous TRPV1 stimuli, including capsaicin, resiniferatoxin, extracellular protons, spider toxins, and heat, and like the wild type channel showed relatively high permeability for calcium21,22. Thus, biophysical and structural information gleaned from this minimal TRPV1 construct should accurately reflect properties of the wild type channel.
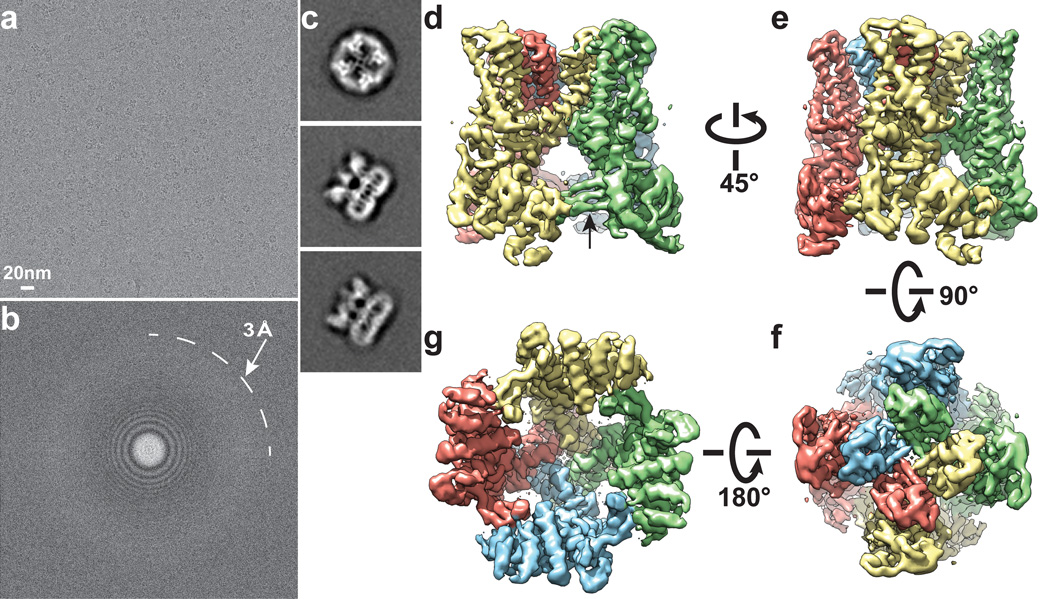
Figure 1. 3D reconstruction of TRPV1 determined by single-particle cryo-EM.
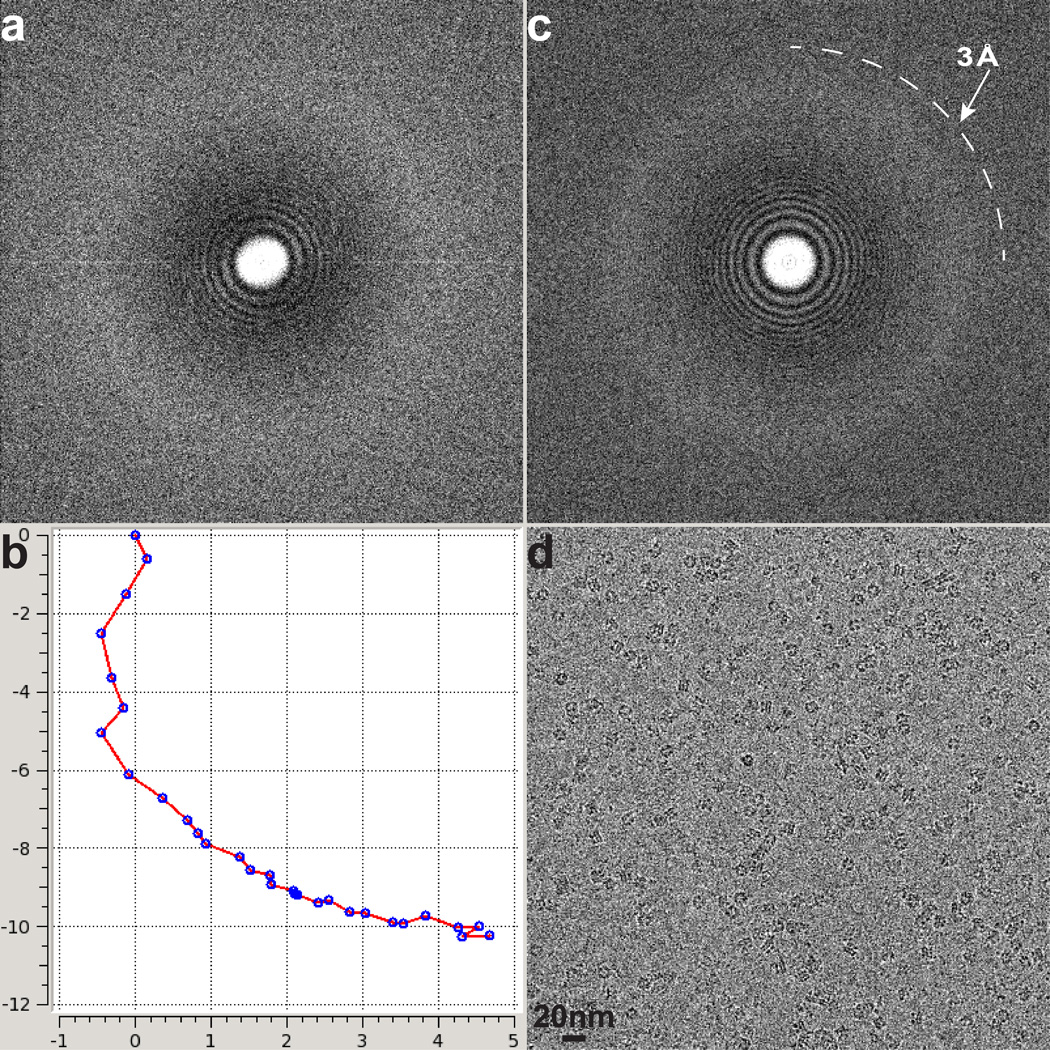
a, Representative electron micrograph of TRPV1 protein embedded in a thin layer of vitreous ice recorded at a defocus of 1.7µm. b, Fourier transform of micrograph shown in a, with Thon rings extending to nearly 3Å. c, Enlarged views of three representative 2D class averages show fine features of tetrameric channel complex. d–g, 3D density map of TRPV1 channel filtered to a resolution of 3.4Å (scaled to atomic structure) with each subunit color-coded. Four different views of the channel are shown, from side (d, e), top (f), and bottom (g). The arrow in panel d indicates β-sheet structure in the cytosolic domain of TRPV1.
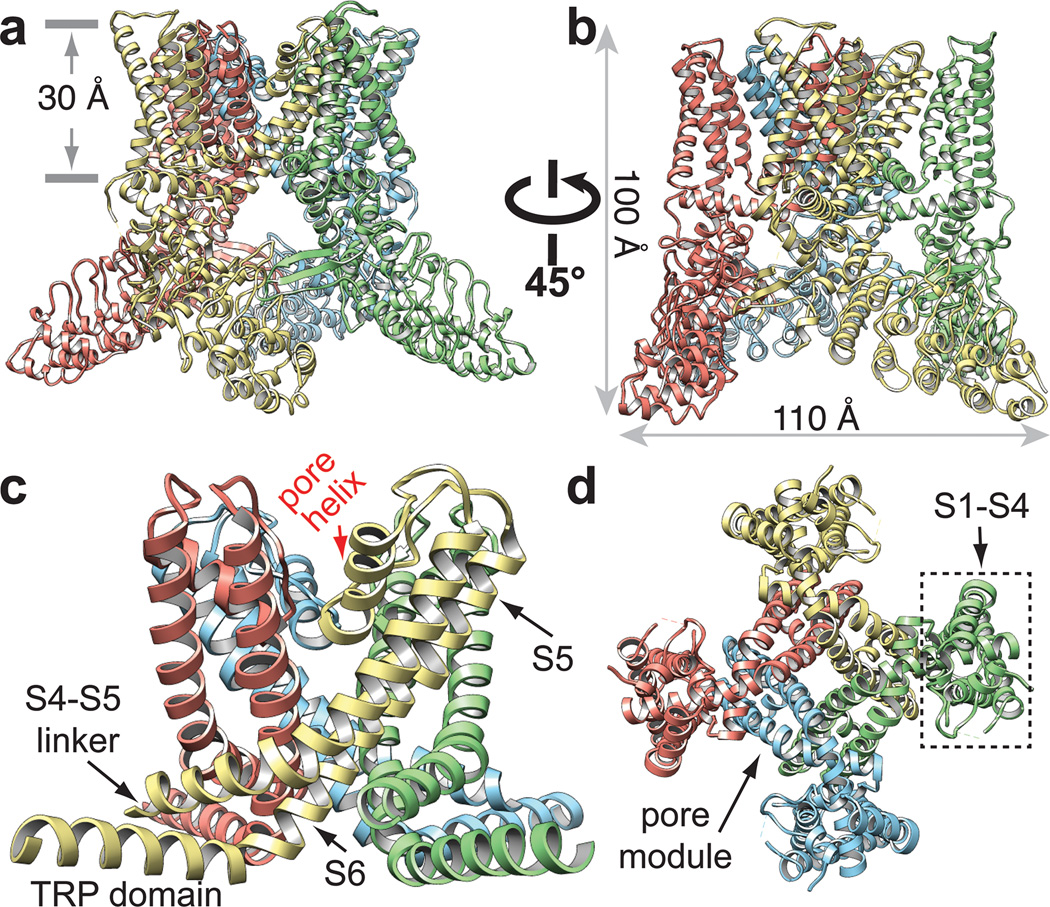
Figure 2. TRPV1 and VGICs share similar 4-fold symmetric architecture.
a–d, Ribbon diagram of TRPV1 atomic model with each of the four identical subunits color-coded, showing views from side (a and b). The dimensions of the channel and the length of the membrane-spanning helices are indicated. The dimensions of the TRPV1 tetramer are 100Å by 110Å by 110Å, as compared with 135Å by 95Å by 95Å for the rat Kv1.2 potassium channel7. c, Ribbon diagram focusing in on side view of S5-P-S6 pore with TRP domains. d, Bottom view focusing on transmembrane core, including S1–S4, S5-P-S6, and TRP domains. Note that S1–S4 domains flank and interact with S5-P-S6 pore modules from adjacent subunit, reminiscent of VGIC architecture.
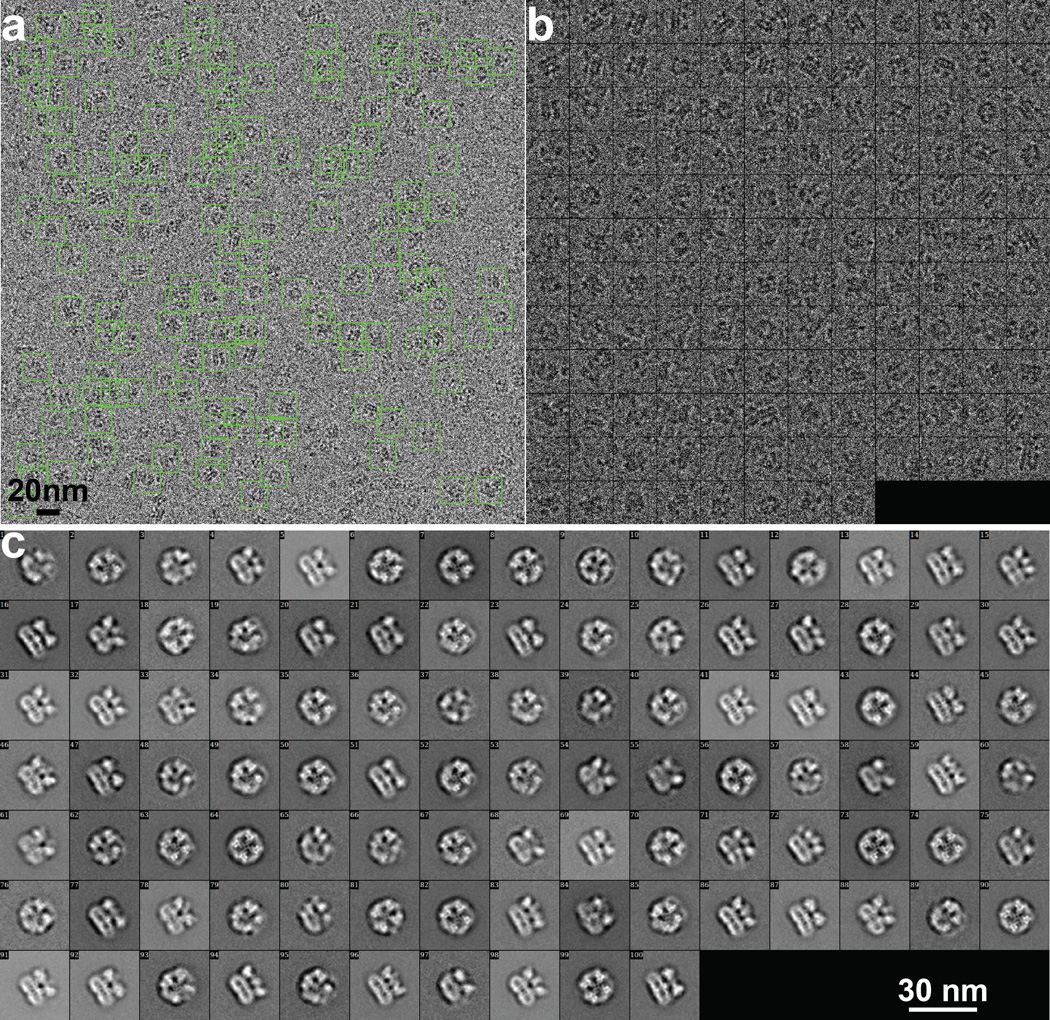
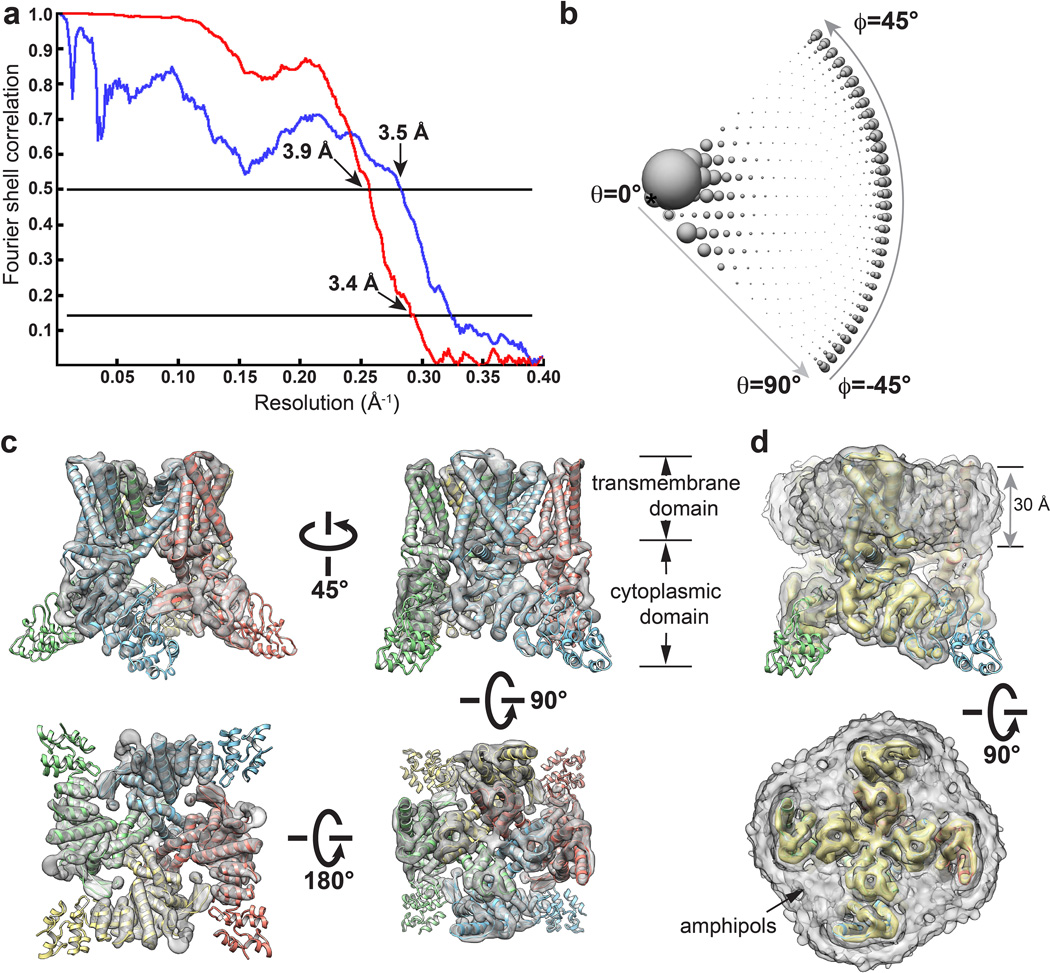
We initiated structural analysis using negative stain EM (Extended Data Fig. 3) followed by cryo-EM 3D reconstruction of images recorded at 200kV with a phosphor scintillator based CMOS camera (Extended Data Fig. 4 and 5). We then collected a cryo-EM dataset at 300kV using a direct detection camera following newly implemented procedures of dose fractionation and motion correction15. Thon rings were visible to ~3Å in Fourier power spectrum of almost every motion corrected image (Fig. 1a, b and Extended Data Fig. 6). Two-dimensional (2D) class averages of particle images (Fig. 1c and Extended Data Fig. 7) showed greater detail than those calculated from CMOS camera images (Extended Data Fig. 4e, f), demonstrating significant improvement of data quality. The final 3D reconstruction of TRPV1 has an overall resolution of 3.4Å, using gold standard Fourier shell correlation (FSC) = 0.143 criteria (Fig. 1d–g and Extended Data Fig. 8)23.
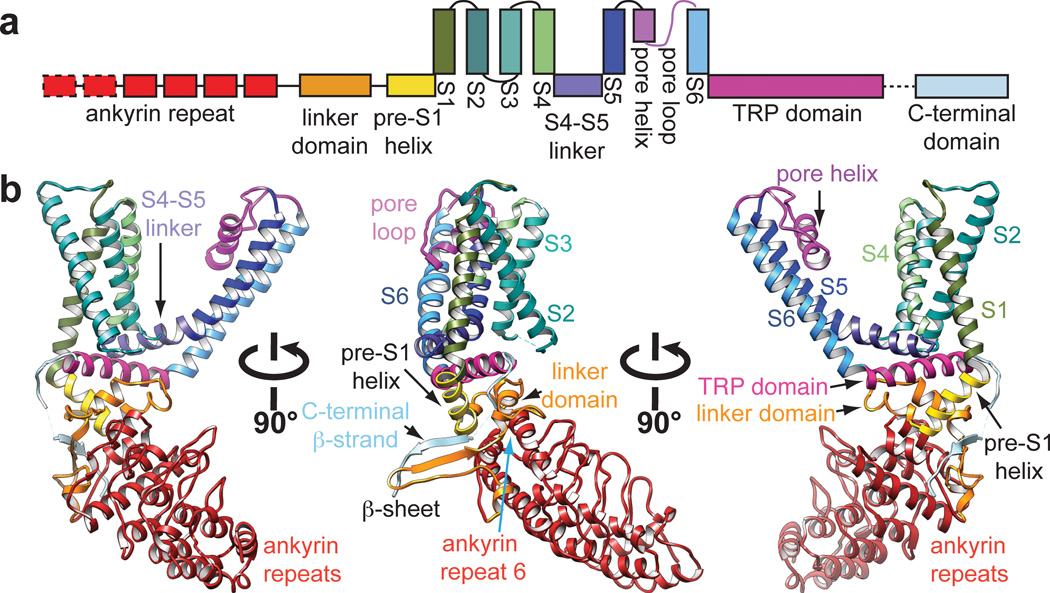
Figure 3. Structural details of a single TRPV1 subunit.
a, Linear diagram depicting major structural domains in a TRPV1 subunit, color coded to match ribbon diagrams below. Dashed boxes denote regions for which density was not observed (first two ankyrin repeats) or where specific residues could not be definitively assigned (C-terminal β-strand). b, Ribbon diagrams showing three different views of a TRPV1 monomer denoting specific domains.
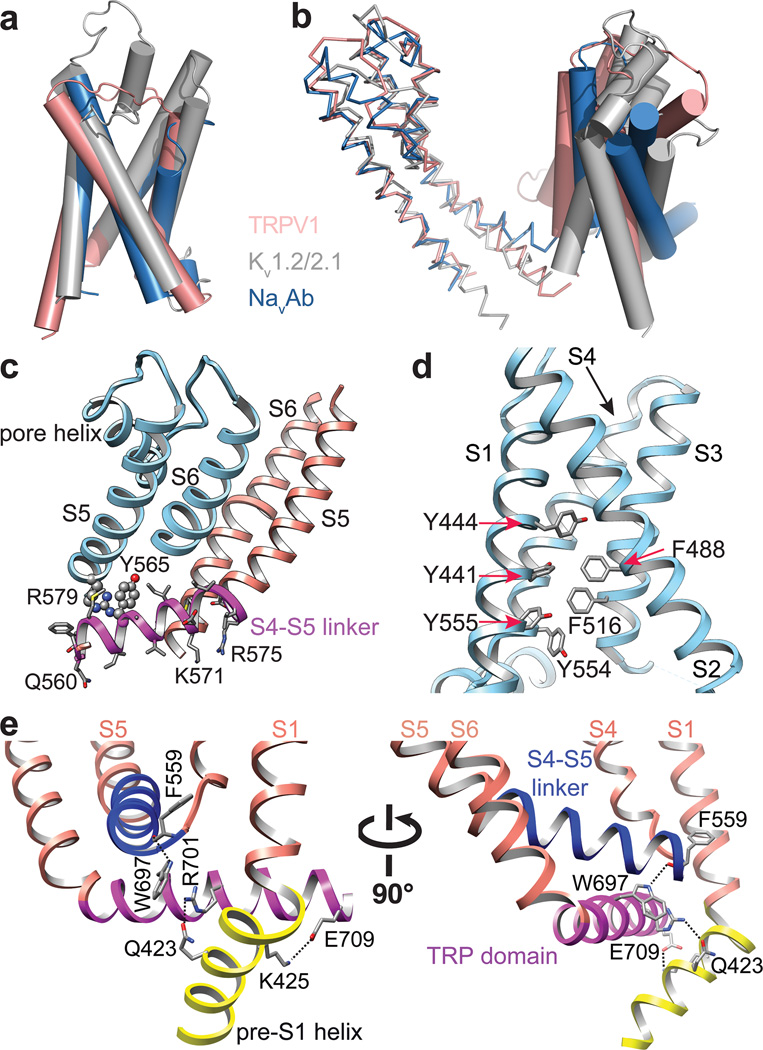
Figure 4. Unique structural features of TRPV1.
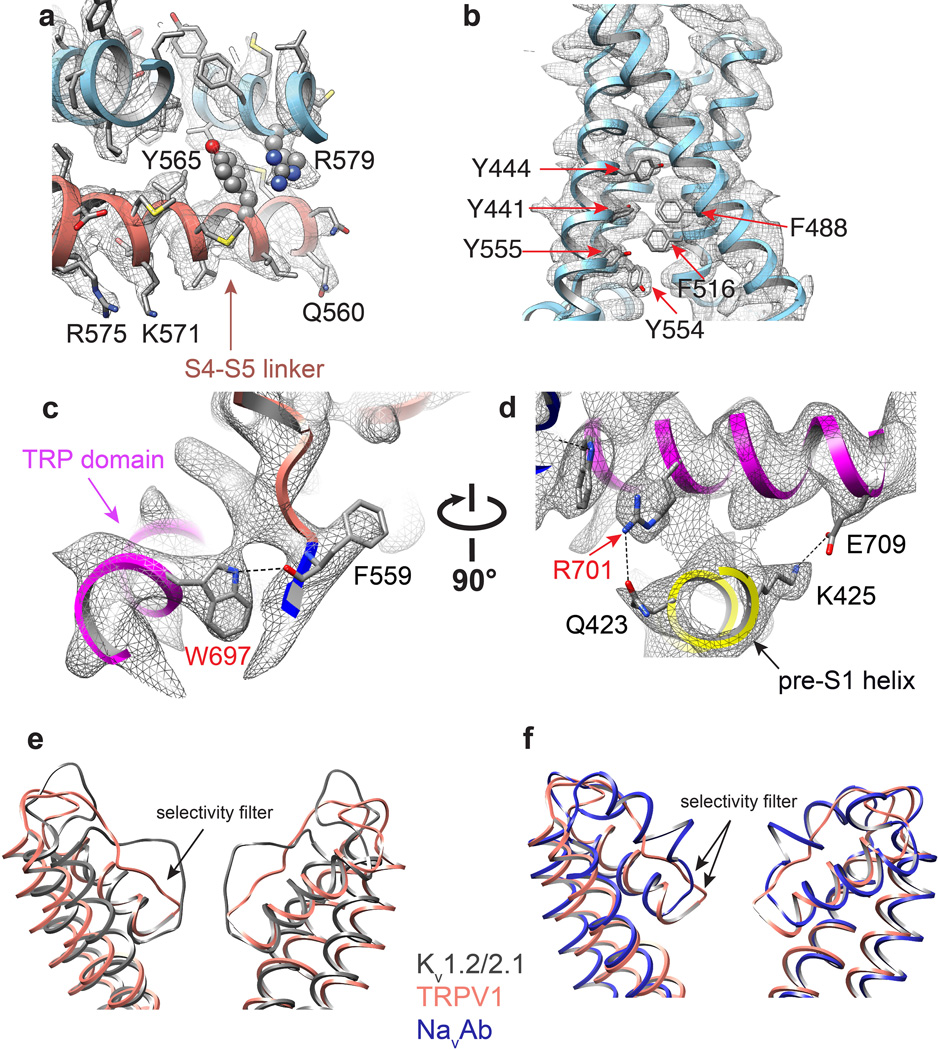
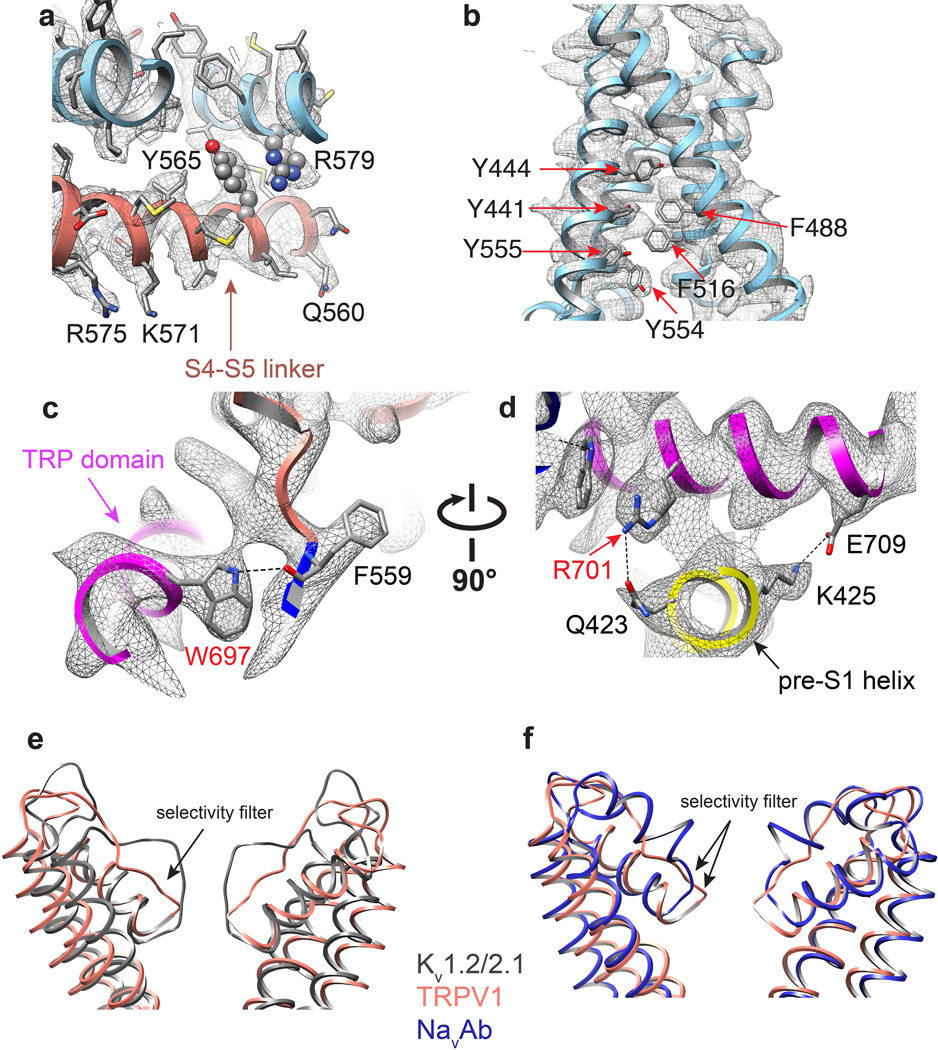
a, Alignment of S1–S4 transmembrane domains from TRPV1 (salmon), Kv1.2/2.1 chimera (PDB 2R9R; grey), and NavAb (PDB 3RVY; blue) show substantial overlap. b, When S5-P-S6 pore regions are aligned, the S1–S4 domains show differential relative orientations. c, The S4–S5 linker is an amphipathic α-helix whose charged surface faces the cytosol. Potential cation-π interactions between Y565 in the S4–S5 linker and R579 from S5 of the adjacent subunit are highlighted. Mutations of cognate residues in TRPV4 render the channel constitutively active and cause skeletal dysplasia. d, Aromatic side chains from S1, S3 and S4 helices create a hydrophobic interior in the S1–S4 domains, in contrast to the charged environment observed in VGICs. e, Two different views highlight interactions between TRP domain and S4–S5 linker and pre-S1 helix. Interactions (i.e. hydrogen bonds and salt bridge) are indicated by dashed lines.
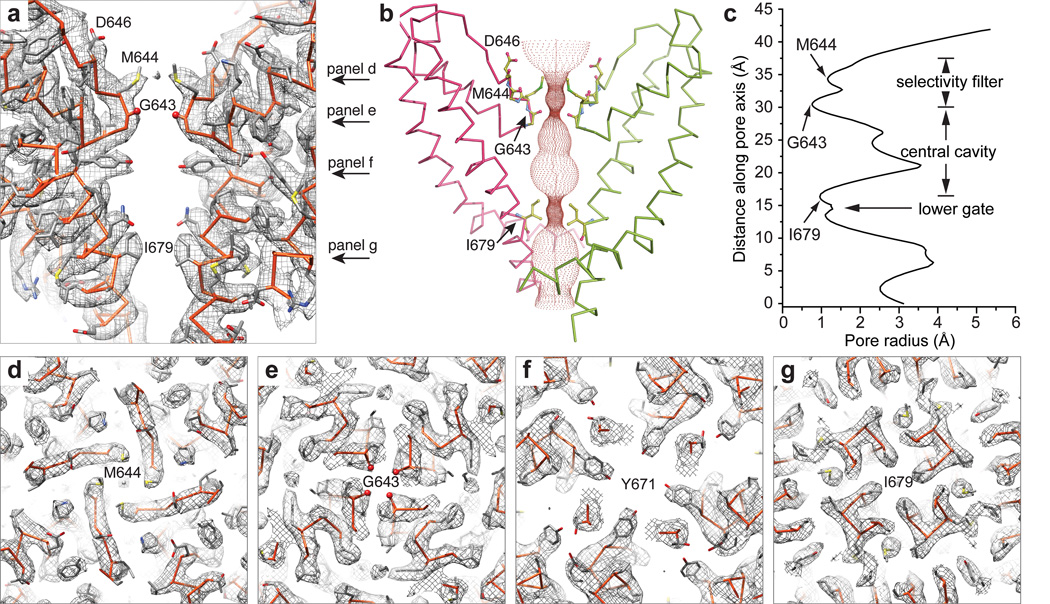
Figure 5. The ion permeation pathway of TRPV1.
a, Cryo-EM densities of the pore in longitudinal cross section are superimposed on an atomic model. Only two diagonally opposed subunits are shown for clarity. Several residues along the pore are labeled for orientation. Arrows denote positions of density maps for horizontal cross sections shown in panels d–g, as indicated. b, Solvent-accessible pathway along the pore mapped using the HOLE program. Residues located at the selective filter and lower gate are rendered as sticks. c, radius of the pore calculated with program HOLE. d–g, Cryo-EM densities of several residues along the pore are superimposed on the atomic model; all panels represent views along the four-fold axis, showing residues from each subunit of the homo-tetrameric channel.
Extended Data Figure 6. Motion correction improves the quality of images collected on Polora TF30 microscope using a K2 Summit direct electron detector.
a, Fourier transform of a representative cryo-EM image of TRPV1 embedded in a thin layer of vitreous ice over Quantifoil hole without supporting carbon film before motion correction. b, Path of motion of 30 individual subframes, determined as described in Methods. c, d, A nearly perfect Fourier transform (c) was restored after the EM image was corrected for motion (d).
Extended Data Figure 7. Picking and 2D classification of TRPV1 Cryo-EM particles collected on Polora TF30 microscope.
a, Representative cryo-EM image after motion correction. Green boxes indicate all particles that were selected by semi-automatic particle picking and 2D screening, as described in Methods. b, Gallery view of the particles shown in panel a. c, 2D class averages of cryo-EM particles show many fine features (also seen in enlarged views in Fig. 1c), and these features are not visible in the 2D class averages of cryo-EM particles from TF20 data (Supplementary Fig. 4e).
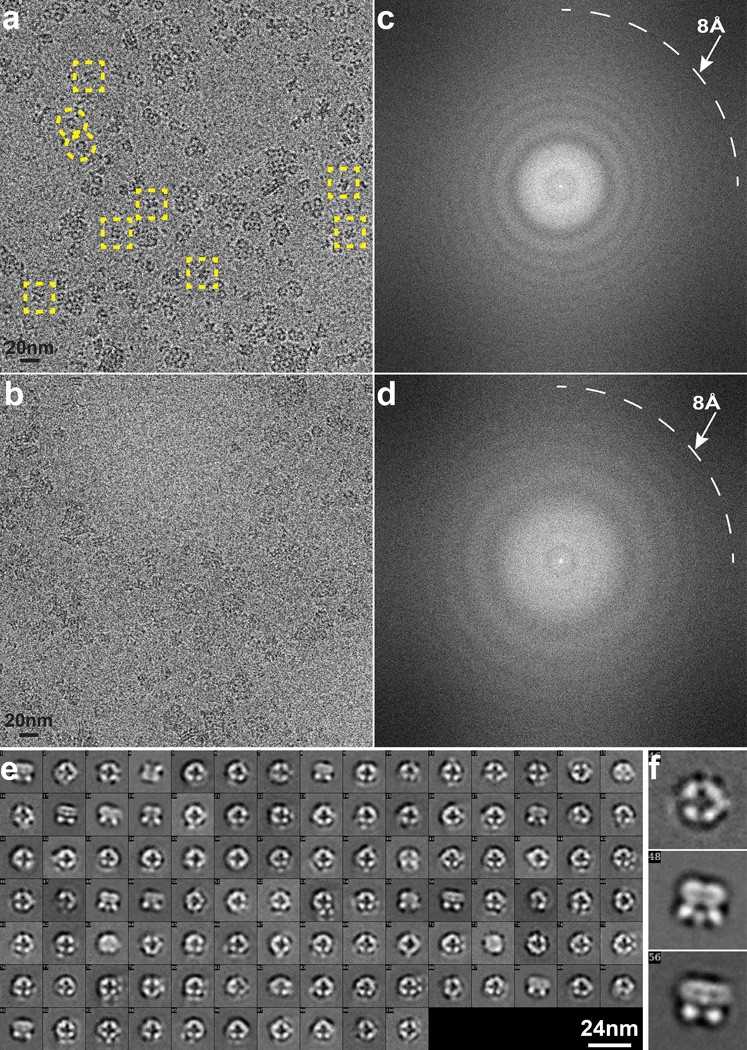
Extended Data Figure 4. Cryo-EM of TRPV1 using Tecnai TF20 microscope and TemF816 8K × 8K CMOS camera.
a–d, Representative images of frozen hydrated TRVP1 in amphipols taken at different defocus levels, 3.1 (a) and 1.5 µm (b) and their Fourier transforms (c, d). Thon rings extend to ~8Å. Dash-line squares or circles indicate representative particles showing two distinctive views. e, 2D class averages of TRPV1 particles. f, Enlarged view of three representative 2D class averages.
Extended Data Figure 8. 3D reconstruction of TRPV1 calculated from TF30 data.
a, Gold-standard FSC curve (red) of the final 3D reconstruction, marked with resolutions corresponding to FSC = 0.5 and 0.143. The FSC curve between the final map and that calculated from the atomic model is shown in blue. The relative low value of this FSC (blue) at low frequency range (> 10Å) is likely due to the presence of amphipol density in the experimental map. b, Euler angle distribution of all particles used for calculating the final 3D reconstruction. The sizes of balls represent the number of particles. The accuracy of rotation is 3.54°, as reported by RELION. c, Different views of the 3D reconstruction low-pass filtered at 6Å and amplified by a temperature factor of −100Å2, fitted with the atomic model of TRPV1. d, Two views of the 3D reconstruction displayed at two different isosurface levels (high in yellow and low in gray). At the low isosurface level, the belt-shaped density of amphipols is visible with a thickness of ~30Å.
Side chain densities of most residues within transmembrane helices S1–S6 were clearly resolved, as well as the TRP domain following S6 and most (~80%) residues in the N-terminal segment connecting the ankyrin repeat domain (ARD) to S1 (Extended Data Fig. 9). In other cytoplasmic regions, densities were less well resolved, but anti-parallel β-strands were well separated, indicating that cytoplasmic domains were resolved to ≤4.8Å (Fig. 1d, arrow). Densities for the first two ankyrin repeats were absent in our 3D density map (Fig. 1, Extended Data Figs. 5 and 8), even though the entire ARD was present in the expressed protein. Nonetheless, the crystal structure of isolated TRPV1 ankyrin repeats24 could be nicely docked to the density map as a rigid body without modification, as confirmed by bulky side-chain densities (Extended Data Fig. 9). Finally, we constructed an atomic model of TRPV1 in which residues L360 to A719 were built de novo, and L111 to E359 were docked from the ankyrin repeat crystal structure (Fig. 2). In the C-terminal domain, we could trace the main chain, but side chain densities were not sufficient for de novo model building.
Extended Data Figure 9. Cryo-EM densities of selected regions of TRPV1 at 3.4Å resolution.
a–d, Representative cryo-EM densities (gray mesh) are superimposed on atomic model (main chain in pink) for various TRPV1 domains, as indicated. e, f, Representative cryo-EM densities (gray mesh) are docked with crystal structure of TRPV1 ankyrin repeats (PDB ID: 2PNN). Accuracy of docking was supported by fitting of several bulky side chains. Map was low-pass filtered to 3.4Å and amplified by a temperature factor −100Å2.
Extended Data Figure 5. 3D reconstruction of TRPV1 calculated from TF20 data.
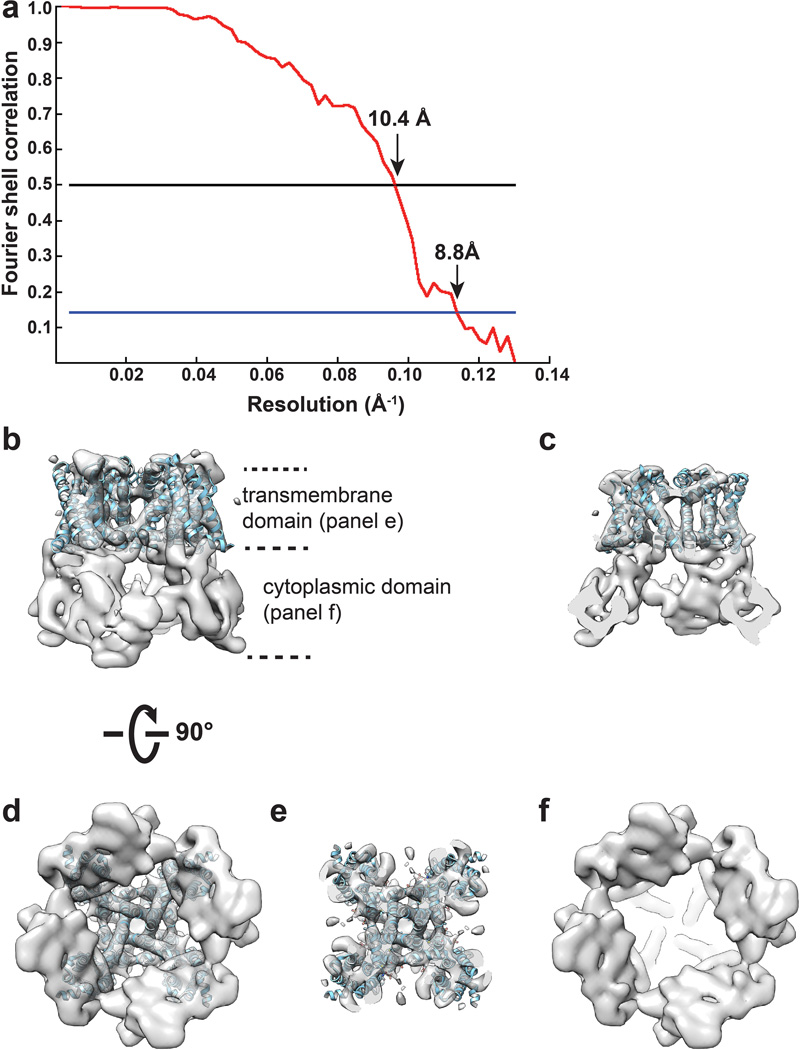
a, Gold-standard FSC curve for the 3D reconstruction, marked with resolutions corresponding to FSC = 0.5 and 0.143. b, Side view of the 3D reconstruction low-pass filtered at 9Å and amplified by a temperature factor −1500Å2, showing transmembrane (top) and cytoplasmic (bottom) domains. The transmembrane domain roughly fitted by the atomic model of NavAb (PDB ID: 3RVY). c, Longitudinal cross section view focused on central transmembrane helices. d, Bottom-up view of the 3D reconstruction shows overall structure. e, f, Bottom-up cross section views showing the arrangement of transmembrane (e) and cytoplasmic (f) domains.
Extended Data Figure 2. Sequence alignment of TRPV1 to other TRPV family members.
The rat TRPV1 construct used for this study consists of residues 110 to 764 (indicated by red arrows), excluding the highly divergent region (604–626, highlighted by cyan box). Secondary structure elements are indicated above the sequence. The starting points of six ankyrin repeats are based on a crystal structure of ARD of TRPV1 (PDB ID: 2PNN). Several critical residues discussed in the text are labeled in blue, and conserved glycine and proline residues at the turn of a β-sheet (highlighted in Figure 6) are indicated with red stars.
Our TRPV1 structure demonstrates that, despite the rather low (<20%) sequence similarity between TRPs and VGICs, these two channel families share a similar tetrameric architecture in which subunits are arranged in four-fold symmetry around a central ion permeation path. Each subunit consists of six transmembrane α-helices (S1–S6) that span the lipid bilayer (Fig. 2 and Extended Data Fig. 8), plus a re-entrant loop with a pore helix located between S5 and S6 that together assume an ‘inverted teepee’ arrangement resembling that of VGICs (Fig. 2c).
Domains unique to TRP channels are immediately evident. First, we see four of six known ankyrin repeats at the N-terminus, arranged as previously described24, followed by a linker (P360 - V415) that is conserved among TRPV subtypes and which connects the ARD to the pre-S1 helix (Fig. 3). This linker forms a tightly packed domain containing β-strand and α-helical elements, and is sandwiched between the sixth ankyrin repeat on one side and the pre-S1 helix and TRP domain on the other. Interestingly, an anti-parallel β-sheet from the linker region and a β-strand from the C-terminus make contact with two ankyrin repeats from an adjacent subunit (Figs. 1d, 2a, and 3b), an interaction that likely plays a role in channel assembly, reminiscent of the T1 domain in Kv channels. Another unique feature is the extended and kinked interfacial helix following S6, which corresponds to the signature ‘TRP domain’ found in many TRP channels2. This domain is strategically located to interact with both the S4–S5 linker and pre-S1 helix (E416 - R428), likely providing a physical substrate through which stimuli allosterically affect pore conformation (Fig. 3).
The transmembrane core
In VGICs, the transmembrane core is organized into two distinct clusters that include the central ion-conducting pore formed by tetrameric assembly of S5-P-S6 domains, and surrounding S1–S4 voltage sensing domains25,26. Each S1–S4 domain associates with the S5-P-S6 region of an adjacent subunit, forming a pinwheel-like structure through this ‘domain swap’ organization. Remarkably, we find that TRPV1 adopts this same overall configuration (Fig. 2d), despite minimal sequence and functional similarity to VGICs1,4. When pore regions (S5-P-S6) of three channels (TRPV1, Kv1.2/2.1 chimera and NavAb) are aligned, the relative positions of S1–S4 domains differ, even though the domains themselves show substantial overlap (Fig. 4a, b), perhaps reflecting intrinsic structural differences or distinct physiological states (e.g. open, closed, or desensitized). Within each TRPV1 subunit, the S1–S4 and S5-P-S6 units are connected by a helical S4–S5 linker that runs parallel to the membrane, again reminiscent of VGICs (Figs. 3 and 4c, Extended Data Fig. 10a).
Extended Data Figure 10. Details of domain interactions and outer pore configurations.
a–d, Cryo-EM densities (gray mesh) of highlighted regions of TRPV1, as indicated, at 3.4Å resolution are superimposed onto atomic model. Map was low-pass filtered to 3.4Å and amplified by a temperature factor −100Å2. e, Superimposition of TRPV1 (salmon) with Kv1.2/2.1 chimera (PDB ID: 2R9R; grey). f, Superimposition of TRPV1 (salmon) with NavAb (PDB ID: 3RVY; blue). In each case, substantial structural differences are observed in the outer pore region. Structural alignments are based on the pore domain (S5-P-S6).
In VGICs, S3 and S4 helices engage in extensive side chain interactions, forming a charge transfer center that facilitates movement of S4 in response to changes in membrane potential5,7. Interestingly, the equivalent region of TRPV1 lacks charged side chains and is, instead, replete with aromatic residues (Fig. 4d and Extended Data Fig. 10b). The importance of this aromatic cluster is underscored by studies demonstrating that replacement of these amino acids (Y441, Y444, Y554, and Y555) with small non-aromatic residues produces non-functional channels27,28. Such hydrophobic packing might impart rigidity to the S1–S4 domain, allowing it to serve as a relatively stationary anchor upon which the S4–S5 linker moves to facilitate TRPV1 gating. Indeed, as discussed in the accompanying study29, the entire S1–S4 domain remains static during channel activation, but provides an external surface for the binding of lipophilic ligands, such as capsaicin and resiniferatoxin. Thus, unique biochemical interactions within this structurally conserved S1–S4 domain likely reflect differential sensitivities of VGICs and TRPs to specific physiologic stimuli (i.e. voltage versus lipid metabolites).
The TRP domain and S4–S5 linker
The TRP domain, a 23–25 amino acid-long region located just after S6, is found in many TRP family members and has been proposed to engage in subunit assembly or allosteric modulation of channel gating1,2,30. We find that the TRP domain assumes a α-helical structure that runs parallel to the inner leaflet of the membrane by virtue of a sharp bend after S6 (Fig. 2c). This α-helix encompasses the first two-thirds of the TRP domain, after which the structure transitions to a random coil (Fig. 3b). An invariant tryptophan (W697) near the middle of the TRP domain forms a hydrogen bond with the main chain carbonyl oxygen of F559 at the beginning of the S4–S5 linker (Extended Data Fig. 10c). Interestingly, gain-of-function mutations at this equivalent tryptophan in TRPV3 underlie a congenital disorder, Olmsted syndrome, in humans31. Moreover, charged side chains within the TRP domain are located on the side of the helix facing the cytoplasm, where they interact with the pre-S1 helix via hydrogen bonding and salt bridging (Fig. 4e and Extended Data Fig. 10c, d). Taken together, these observations are consistent with a role for the TRP helix as a point of structural integration that facilitates allosteric coupling between channel domains.
In VGICs, the S4–S5 linker plays a critical role in coupling the movement of the S1–S4 voltage sensor to gating of the pore5–8,32. Gain-of-function mutations within the S4–S5 linker or S5 helix of TRPV4 enhance basal open probability and result in skeletal dysplasia syndromes33. Interestingly, a cation-π interaction can be seen between residues at equivalent positions in TRPV1, namely Y565 in the S4–S5 linker of one subunit and R579 in S5 of a neighboring subunit (Fig. 4c and Extended Data Fig. 10a). Thus, in addition to covalent intra-subunit interaction between the S4–S5 linker and S5-P-S6 pore region, functional coupling may also be achieved through weak trans-subunit interactions.
The pore and ion permeation pathway
The outer pore region of TRPV1 is wide open compared to Kv channels, with a broad funnel-like structure that likely enhances accessibility to both small and large pharmacophores (Fig. 5a, b). Indeed, as described in the accompanying study29, the uppermost regions of the pore loop proximal to S5 and S6 form a binding site for spider toxins. Further down the central canal, we see a short selectivity filter (643GMGD646) in which backbone carbonyls or side chains point into the central pathway (Fig. 5a–e). TRPV1 and related subtypes are highly calcium permeable9,34 and molecular modeling studies predict a selectivity filter diameter of ~6Å35,36. In our structure, we see a restriction point of 4.6Å between diagonally opposed carbonyl oxygens at G643 (Fig. 5a–c, e), suggesting that the selectivity filter is in a non-conducting state, reminiscent of inactivated bacterial NavRh channels8. Indeed, as we show in the accompanying study29, the selectivity filter diameter increases substantially when TRPV1 is in its activated state. The overall arrangement that we see for TRPV1 – namely, a wide outer pore with a short selectivity filter - more closely resembles that of bacterial Nav channels than Kv channels (Extended Data Fig. 10e, f)5–8. However, TRPV1 lacks hydrogen bonding within and between adjacent pore helices, in contrast to Nav channels, where such elaborate interactions are believed to impart rigidity to the selectivity filter7. This more flexible architecture, together with a short selectivity filter, may underlie the phenomenon of pore dilation seen in several TRP channels, including TRPV1, whereby prolonged or repetitive activation renders the channel permeable to large organic cations21,37. Interestingly, when compared with TRPV1-4, TRPV5/6 subtypes bear distinct sequences at the selective filter (i.e., T-V/I-I-D versus G-M/L-G-D/E), as well as the pore helix, suggesting a different architecture of the outer pore domain (Extended Data Fig. 2). This divergence may underlie the fact that TRPV5/6 exhibit more pronounced selectivity for calcium over monovalent cations (PCa2+/PNa+ > 100) compared to other TRPV subtypes (PCa2+/PNa+ ≈ 3–10) (ref34).
Continuing down the pore, we see a constriction site in which I679 in S6 helices from each subunit come together to form a hydrophobic seal measuring 5.3Å between side chains, thus constituting the most constricted point in the lower gate (Fig. 5a–c, g). Modeling studies, together with substituted cysteine accessibility analysis, have previously implicated I679, Y671, and L681 as candidates for this position35,38. Our structure indicates that side chains of Y671 are located ~10Å above the narrowest point formed by I679 (Fig. 5a, f), while the side chain of L681 points away from the ion conduction pathway, making Y671 and L681 unlikely major contributors to the lower gate. Of note, TRPV subtypes exhibit high sequence similarity throughout the S6 helix, and all contain an isoleucine at the position equivalent to I679 in TRPV1 (Extended Data Fig. 2), suggesting a similar architecture in the lower gate. Whereas the narrowest constriction site in bacterial KcsA and NavAb channels is located at the apex of the inverted teepee7,39, the I679 restriction point is higher in the TRPV1 permeation pathway, sitting ~10Å above the S6 bundle crossing. The structure shown here apparently represents TRPV1 in the closed state, a conclusion that is validated by comparison with structures representing agonist bound states of the channel described in the accompanying study29.
Ankyrin repeats form an assembly domain
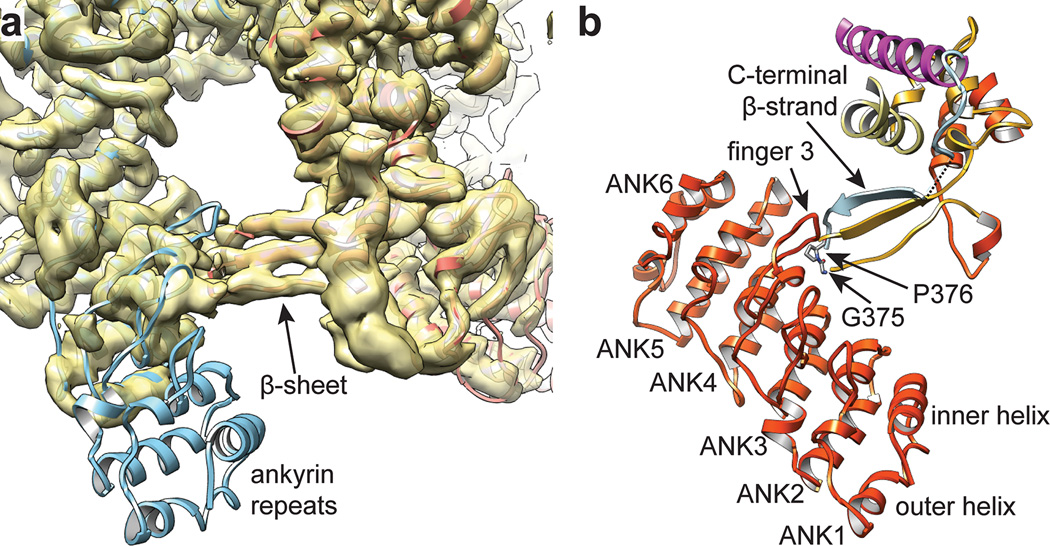
A characteristic feature of many TRP channels is the presence of ankyrin repeats within the cytoplasmic N-terminus40. In our structure, residues in the atypically long finger 3 and inner helices of ankyrin repeats 3 and 4 from one subunit interact with a three-stranded anti-parallel β-sheet formed by the ARD-S1 linker region (K368-D383) and C-terminus of an adjacent subunit, packing the cytosolic part of the channel together (Fig. 6). Moreover, the β-sheet structure tethers cytoplasmic N- and C-terminal domains together within the same subunit, reminiscent of GIRK channels, where N- and C-termini interact through a short parallel β-sheet41,42. Interestingly, two residues (G375 and P376) that produce the sharp turn connecting the N-terminal β-strands are invariant among all TRPV subtypes (Extended Data Fig. 2), supporting the proposed functional importance of the β-sheet in subunit structure and assembly.
Figure 6. Cytosolic interactions mediated by ankyrin repeat domains.
a, Cryo-EM density is well fitted by crystal structure of TRPV1 ankyrin repeats 3–6 (PDB code: 2PNN) as a rigid body. Ankyrin repeats 1 and 2 are not observed, presumably due to high flexibility. b, Fingers 3 and inner helices from ankyrin repeat 3 and 4 on the concave surface of the ARD interact with β–strands from the linker and C-terminus of an adjacent subunit.
In the TRPV1 ARD crystal structure, ATP is bound within the concave surface formed by ankyrin repeats 1–3 and has been proposed to both stabilize the ARD fold and regulate Ca2+-calmodulin-dependent desensitization24. Moreover, a number of mutations that render TRPV1 or other TRPV channels constitutively active lie in the vicinity of the proposed ATP binding site22,43, further indicating that the ARD is an important locus for channel modulation. Our structural data suggest that modulation may also involve perturbation of subunit-subunit interactions. At the same time, the ARD surface forms an extensive cytoplasmic skirt, much of which remains available for interaction with other, as yet unidentified cellular factors. Whether and how these regions contribute to channel regulation may be revealed by structures of TRPV channels in complex with physiological modulators that target this domain.
Concluding remarks
TRP channels represent one of the last major ion channel families to yield to structure determination at the atomic level. Obtaining crystals for membrane proteins, particularly of mammalian origin, is generally challenging, perhaps especially so for TRP channels, which respond to diverse stimuli (chemical and physical) and are therefore believed to be conformationally dynamic. If so, then this represents an additional obstacle to coaxing these proteins into forming well-ordered crystal lattices required for X-ray or electron crystallographic analysis. We have circumvented this problem by taking advantage of recent technological breakthroughs in single-particle cryo-EM to gain high-resolution structural information of purified TRP channels under conditions that do not require absolute conformational homogeneity (supported by the fact that only a subset of the dataset was used to generate the final 3D reconstruction), or large amounts of material. We expect that this approach will be particularly powerful for analyzing heterogeneous populations of channels with the goal of identifying and characterizing different conformational states en route to gating, or in complex with agonists, drugs, or other modulators. Indeed, we provide a demonstration of this in the accompanying study29.
Methods Summary
A minimal functional rat TRPV1 construct was cloned into a modified Bacmam vector (Invitrogen) containing an N-terminal fusion cassette (Kozac-MBP-Tev protease site) for purification on amylose resin. TRPV1 protein was expressed in HEK293S GnTI− cells grown in suspension at 37°C. Cells were harvested 48 hours post transduction for preparation of crude membrane and subsequent protein purification, as described10. Negative stain EM and cryo-EM were carried out following established protocols15,44. At 3.4Å resolution, the cryo-EM map was of sufficient quality for de novo atomic model building. A poly-alanine model was first built in Coot and amino acid assignment subsequently achieved based mainly on the clearly defined side chains densities of bulky residues such as Phe, Tyr, and Trp, as well as some Arg and Lys residues.
Online Methods
Protein expression and purification
A Bacmam vector was generated from pFastBac1 (Invitrogen) to direct protein expression in mammalian cells after baculovirus transduction45. In brief, the polyhedron promoter (PPH) in pFastBac1 was replaced with a mammalian cell active promoter (PCMV), immediately followed by an N-terminal fusion cassette (Kozac-MBP-Tev protease site) for affinity purification with amylose resin (New England Biolabs). A minimal functional rat TRPV1 construct, composed of amino acids 110 to 603 and 627 to 764 was cloned into this modified Bacmam vector, and recombinant baculoviruses obtained following the manufacturer’s protocol (Bac-to-Bac expression system, Invitrogen). For protein expression, HEK293S GnTI− cells46, grown in suspension at 37°C in an orbital shaker, were transduced when cell density reached ~ 2×106/ml. Sodium butyrate was added to the culture 24 hour post transduction at a final concentration of 10 mM to boost protein expression, and cells harvested 48 hour post transduction for preparation of crude membrane and subsequent protein purification, as described10 with slight modification. TRPV1 channel was eluted from amylose resin with buffer composed of 150 mM NaCl, 2 mM TCEP, 10% Glycerol, 20 mM HEPES, 0.5 mM DDM, 0.1 mg/ml soybean lipids, and 20 mM maltose, then incubated with TEV protease for 4 hours at 4°C. The cleaved protein sample was then mixed with amphipols at 1:3 (w/w) with gentle agitation for another 4 hours. Detergent was removed with Bio-Beads SM-2 (4°C overnight, 15 mg per 1 ml channel/detergent/amphipols mixture). Bio-beads were then removed over a disposable polyprep column, and eluent cleared by centrifugation before further separation on a Superdex 200 column in buffer composed of 150 mM NaCl, 20 mM HEPES, 2 mM TCEP, pH 7.4. The peak corresponding to tetrameric TRPV1 channels was collected for analysis by cryo-EM.
Cell imaging and electrophysiology
Forty-eight hours post transduction, HEK293S GnTI− cells were loaded with Fura-2 AM in physiologic Ringer’s buffer (140 mM NaCl, 5 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 10 mM HEPES, pH 7.4) for ratiometric calcium imaging. Currents were recorded at room temperature in the whole-cell patch-clamp configuration. The intracellular solution contained 150 mM NaCl, 10 mM HEPES, 10 mM EGTA, pH 7.4. DkTx and capsaicin were freshly diluted into this buffer and applied via an in-line perfusion system (Automatic Scientific). To obtain proton-evoked current, buffer consisting of 150 mM NaCl, 10 mM sodium acetate, 10 mM EGTA, pH 5.0 was perfused onto TRPV1-expressing cells. Purified TRPV1 protein was also reconstituted into soybean lipid liposomes, and activation by capsaicin or heat examined by patch-clamp analysis, as described51. To estimate ion permeability ratios, the intracellular solution contained: 150 mM NaCl, 10 mM HEPES, 10 mM EDTA, pH 7.4, and extracellular solutions contained: (A) 150 mM KCl, 10 mM HEPES, 10 mM EDTA, pH 7.4; (B) 150 mM CsCl, 10 mM HEPES, 10 mM EDTA, pH 7.4; (C) 110 mM CaCl2, 10 mM HEPES, pH 7.4. Resiniferatoxin, capsaicin, and DkTx were diluted in appropriate solution immediately before use, and applied to the TRPV1-expressing cell using an in-line perfusion system.
EM data acquisition
Detergent solubilized TRPV1 particles were monodispersed as assessed by negative stain EM (Supplementary Fig 3a, b), enabling us to obtain a 3D reconstruction by random conical tilt (Supplementary Fig. 3c, d), which served as an initial model for cryo-EM 3D refinement. Grids for negative stain EM were prepared following the established protocol44. Specifically, 2.5µl of purified TRPV1 was applied to glow-discharged EM grids covered by a thin layer of continuous carbon film and was stained with 0.75% (w/v) uranyl formate. Negatively stained EM grids were imaged on a Tecnai T12 microscope (FEI Company, Hillsboro, OR) operated at 120 kV. Images were recorded at a nominal magnification of 67 kX using a 4K × 4K CCD camera (UltraScan 4000, Gatan, Inc., Pleasanton, CA), corresponding to a pixel size of 1.73Å/pixel on the specimen. Tilt pair images for random conical tilt 3D reconstruction were manually recorded at 50° and 0°.
For cryo-EM, detergent was replaced by amphipols47,48 in which purified TRPV1 remained stable and monodispersed (Supplementary Fig 3e–g). 2µl of purified TRPV1 sample at a concentration of ~0.3mg/ml was applied to a glow-discharged Quantifoil holey carbon grid (1.2µm hole size, 400 mesh), blotted inside a Vitrobot Mark III (FEI Company) using 6s blotting time with 90% humidity, and then plunge-frozen in liquid ethane cooled by liquid nitrogen. Cryo-EM images were collected at liquid nitrogen temperature on a Tecnai TF20 electron microscope (FEI) operated at 200 kV using a CT3500 side entry holder (Gatan Inc.), following the low-dose procedure. On the TF20 microscope, images were recorded at a nominal magnification of 80kX using a phosphor scintillator based TemF816 8K × 8K CMOS camera (TVIPS GmbH, Gauting, Germany), corresponding to a pixel size of 0.9Å/pixel on the specimen. Images were recorded with a defocus in the range from 1.5 to 3.5µm (Extended Data Fig. 4). We determined a 3D reconstruction with C4 symmetry to an overall resolution of 8.8Å, using gold-standard Fourier Shell Correction (FSC) = 0.143 criteria23 (Extended Data Fig. 5a). In this density map, transmembrane α-helices are clearly resolved. The shape of the cytoplasmic domain is clearly defined, but secondary structural features are not well resolved (Extended Data Figs 5b–f), likely indicating greater flexibility of this region.
Another dataset of frozen hydrated TRPV1 particles were collected on a TF30 Polara electron microscope (FEI Company) operated at 300 kV. It is equipped with a K2 Summit direct electron detector camera (Gatan Inc.). Images were recorded using super-resolution counting mode following an established protocol15. Specifically, images from TF30 were recorded at a nominal magnification of 31kX, corresponding to a calibrated super resolution pixel size of 0.6Å/pixel on the specimen. The dose rate on the camera was set to be ~8 counts (corresponding to ~9.9 electron) per physical pixel per second. The total exposure time was 6 seconds, leading to a total accumulated dose of 41e−/Å2 on the specimen. Each image was fractionated into 30 subframes, each with an accumulation time of 0.2 s/frame. All dose-fractionated cryo-EM images were recorded using a semi-automated acquisition program UCSFImage4 (written by Xueming Li). Images were recorded with a defocus in a range from 1.5 to 3.0µm.
Image processing
SamViewer, an interactive image analysis program written in wxpython, was used for all 2D image display and particle picking. Negative stain-EM images were 2 × 2 binned for manual particle picking. Defocus was determined using CTFFIND and CTFTILT49. Individual particles were cut out and normalized to have a mean of 0 and a standard deviation of 1. For 2D classification, particles were first corrected for contrast transfer function (CTF) by flipping the phase using “ctfapply” (written by Xueming Li), and subjected to 10 cycles of correspondence analysis, k-means classification and multi-reference alignment (MRA), using SPIDER operations “CA S”, “CL KM” and “AP SH”55. For random conical tilt (RCT) 3D reconstruction, SamViewer was used for picking particles from the tilt-pair images, as well as determination of the tilting axes and angles. After 2D classification of untilted particles, RCT 3D reconstructions of each 2D class were calculated using FREALIGN50.
Low-dose images of frozen hydrated TRVP1 collect on TF20 were binned 2 × 2, resulting with a pixel size of 1.9Å, for image processing. For particle picking and 2D classification, images were 2 × 2 binned further to a pixel size of 3.8Å. Dose-fractionated super-resolution image stacks of frozen hydrated TRPV1 images collected using K2 Summit camera were first binned 2 × 2 resulting with a pixel size of 1.2Å for motion correction and further image processing. After motion correction15, a sum of all subframes in each image stack was used for further processing. Particle picking and 2D classification used 6 × 6 binned images (3.6Å/pixel), and 3D classification used 4 × 4 binned images (2.4Å/pixel). Final 3D reconstruction was calculated from 2×2 binned images (1.2Å/pixel). Image binning was calculated using Fourier cropping. First, ~2,000 particles were picked interactively and classified into ~10 2D classes using the same classification procedure described above. Then, for each micrograph, the entire image was cut into a set of overlapping small images with a window of 64 × 64 pixels, all windowed images were subject to MRA against the 2D class averages generated from manually selected particles (implemented in a python script, “samautopick.py”). All particles were then displayed in SamViewer, in the order of their cross-correlation values from MRA, and a threshold was interactively set to remove the particles with cross-correlation values below the threshold. A small number of particles with blurry appearance or with obvious wrong shape and size were also removed interactively. We picked 70,585 particles from 300 cryo-EM images collected on the TF20, and 97,166 particles from 946 image stacks collected on the TF30. The selected particles were further screened by a reference-free 2D classification. A total of 45,625 particles from the TF20 data and 88,915 particles from the TF30 data were kept for determinations of 3D reconstructions.
All 3D classification and refinement were carried out in RELION51. The RCT 3D reconstruction from negatively stained TRPV1 was low-pass filtered to 60Å, and used as the starting model for 3D classification of the TF20 data. After 25 iterations of 3D classification with 6 classes, particles in the classes that produced similar 3D reconstructions were combined for 3D auto-refinement. In this step, the 3D reconstructions from the 3D classification were low-pass filtered to 60Å and used as the starting models. 10,357 particles were used to calculate the final map (Extended Data Fig. 5). The resolution was estimated by the gold-standard FSC = 0.143 criteria, after applying a soft spherical mask on the two reconstructions refined from the half-data sets independently23. The resolution of the 3D reconstruction from TF20 data was estimated as 8.8Å. This 3D reconstruction was again low-passed filtered to 60Å, and used as the starting model for 3D classification of the TF30 Polara data. 3D classification and refinement were carried out using the same procedure as for the TF20 data. The 3D reconstruction calculated from selected 35,645 particles each averaged from 30 subframes of motion-corrected image stack reached a resolution of 3.6Å. Continued refinement using the same particles but averaged from subframe 3–16 improved the resolution further to 3.4Å. This was because the first 2 frames contain the most severe motion and the later frames accumulate more radiation damage15. The accumulated dose of the first 16 frames is 21e−/Å2 on specimen. Final gold-standard FSC curve was calculated using a soft spherical mask (with a 5-pixel fall-off) on the two independent reconstructions (Extended Data Fig. 8). The accuracies of rotation and translation reported by RELION were 3.54° and 1.358Å. RELION post-processing with auto-mask and auto-bfactor determined the resolution of the final map as 3.28Å and bfactor as -101.228. For model building and visualization, amplitude of the final 3D density map of 3.4Å resolution was amplified either by a temperature factor, −100Å2 (Fig. 5) or by frequency dependent scaling factor determined by comparing the experimental 3D density map with the ideal density map calculated from the atomic model (Fig. 1d–f, 6a)15. All maps shown in the figures were low-pass filtered to the stated resolutions. The map deposited to the EMDB database is the raw map without amplitude sharpening, masking or filtering. UCSF Chimera52 was used to visualize and segment the cryo-EM maps, and the rigid body fitting of atomic model was done using the fit-in-map function from Chimera.
Model building in Coot
At 3.4Å resolution, the cryo-EM map was of sufficient quality for de novo atomic model building, except for the ankyrin repeat domain, where the density is not as well resolved as other regions of the channel. Taking advantage of the defined geometry of α-helices and clear bumps for Cα atoms, a poly-alanine model was first built in Coot53,54. Amino acid assignment was subsequently achieved based mainly on the clearly defined side chains densities of bulky residues such as Phe, Tyr, and Trp, as well as some Arg and Lys residues. The ankyrin repeat domain structure (PDB ID: 2PNN) was docked into the density as a rigid body without further modification. The final model includes almost all residues, except for those located at S2–S3 loop one (503–507) and a region C-terminal to the TRP domain (720–751). Although a C-terminal region of the channel (registered as 752–762 in the model) clearly assumes a β-strand secondary structure, amino acids could not be assigned since side chain density is not well resolved at this segment. The final model exhibits good geometry as indicated by the Ramachandran plot (preferred region, 95.73%; allowed region, 3.92%; outliers, 0.34%). Of note, the six transmembrane helices, TRP domain, and pre-S1 helix are devoid of any such outliers. One outlier is located at S3–S4 loop and the other resides at docked ankyrin repeats. Pore radii were calculated using the HOLE program55.
Extended Data Figure 1. A minimal TRPV1 channel that is functional and biochemically stable.
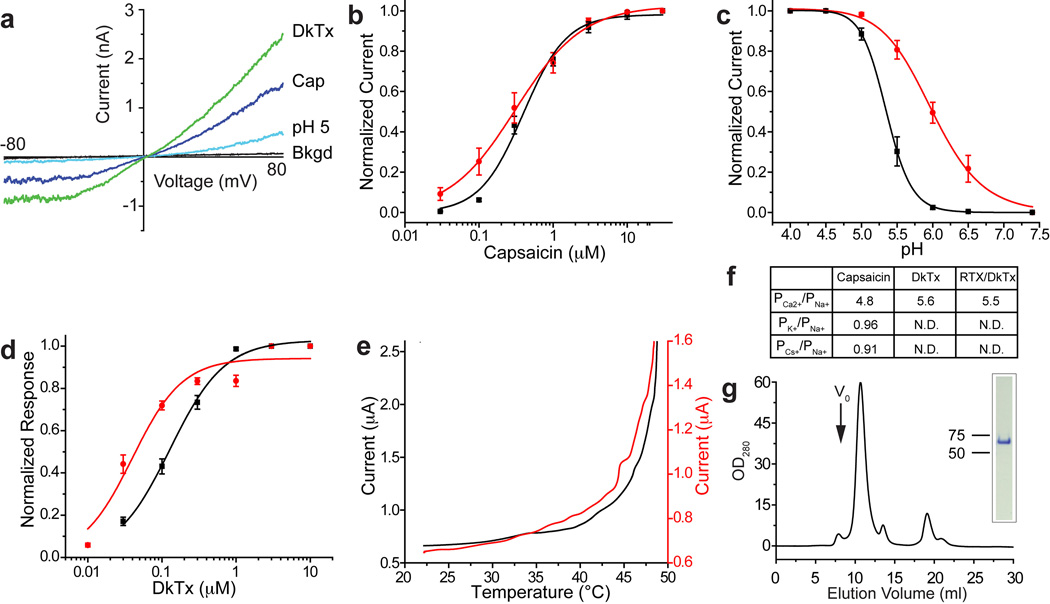
a, Mammalian (HEK293) cells expressing a minimal construct (with an N-terminal GFP tag) responded to various TRPV1 agonists, including capsaicin (Cap; 0.5 µM), extracellular protons (pH 5.0), and double-knot spider toxin (DkTx; 2 µM). Electrophysiological responses were measured in whole-cell patch-clamp configuration. b–c, Dose responsive curves for capsaicin (b) or protons (c) were determined for minimal (black) or full length (red) TRPV1, both of which contained an N-terminal GFP fusion. Values were normalized to maximal currents evoked by 30 µM capsaicin (b) or pH 4.0 (c) (n=6 independent whole cell recordings). d, DkTx dose response curves for minimal (black) or full length (red) TRPV1 as in (b) and (c) determined by calcium imaging. Values were normalized to maximal capsaicin- (10 µM) evoked response in transfected HEK293 cells (n>30 per point). e, Thermal response profiles for minimal (black) or full length (red) TRPV1 expressing oocytes reveal similar heat sensitivity. f, Ion permeability ratios of agonist-evoked currents from minimal TRPV1 were estimated from reversal potential shifts in whole-cell patch-clamp recordings of transfected HEK293 cells, revealing no significant differences from full length channel. g, Gel filtration profile (Superdex-200) of detergent solubilized TRPV1 after purification on amylose affinity resin and proteolytic removal of MBP tag. The major species elutes as a symmetrical peak after the void volume (V0). Inset shows that peak material migrates as a single, homogeneous band on SDS-PAGE (4–12% gradient gel; Coomassie stain).
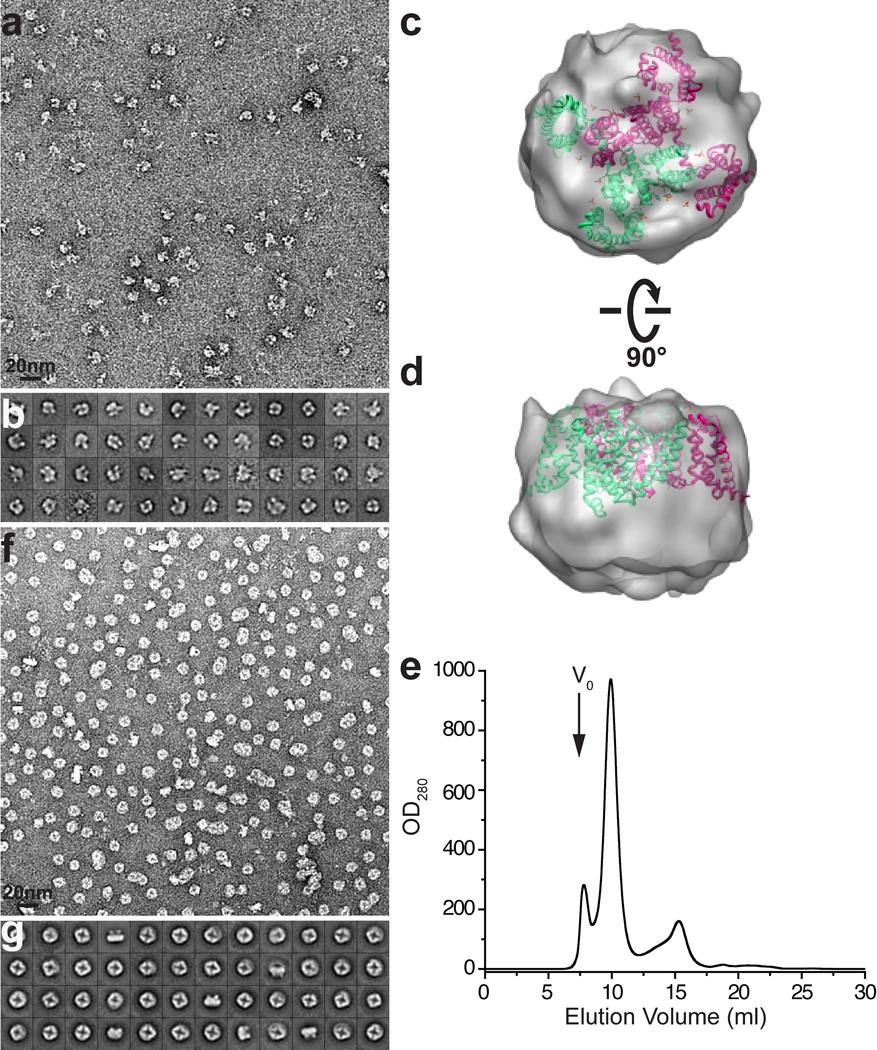
Extended Data Figure 3. Negative stain EM of TRPV1.
a, Representative negative stain image of purified minimal TRPV1 protein in detergent (DDM) after proteolytic removal of MBP tag. b, 2D class averages of negatively stained particles in DDM. c, d, Two views of a random conical tilt (RCT) reconstruction from negatively stained TRPV1 in DDM. The RCT reconstruction was low pass filtered at 30Å, and fitted with the structure of NavAb (PDB ID: 3RVY) to indicate the size and general shape. e, Gel filtration profile (Superdex-200) of purified minimal TRPV1 protein after exchange from DDM into amphipols. The major species elutes as a symmetrical peak after the void volume (V0). f, Representative negative stain image of purified minimal TRPV1 protein without MBP tag in amphipols. g, 2D class averages of negative stain particles in amphipols.
Acknowledgements
We thank X. Li for assistance with data acquisition using TF30 Polara and K2 Summit camera, S. Zhou and D. King for help with protein microsequencing, and J.P Armache, C. Bohlen, J. Cordero-Morales, and J. Osteen for discussion and critical reading of the manuscript. This work was supported by grants from the National Institute of Health (R01GM098672 and S10RR026814 to Y.C. and R01NS065071 and R01NS047723 to D. J.), the National Science Foundation (DBI-0960271 to D. Agard and Y.C.), and the UCSF Program for Breakthrough Biomedical Research (Y.C.). E.C. was a fellow of the Damon Runyon Cancer Research Foundation.
Footnotes
Author contributions
All authors designed experiments. E.C. expressed, purified all protein samples used in this work and performed all functional studies. M.L carried out all cryo-EM experiments, including data acquisition and processing. E.C. built atomic model based on the cryo-EM maps. All authors analyzed data and wrote the manuscript.
3D cryoEM density map of TRPV1 complexes without low pass filter and amplitude modification have been deposited in the Electron Microscopy Data Bank under the accession number: EMD-5778 (TRPV1). Particle images related to this entry are available for download at http://www.ebi.ac.uk/~ardan/aspera/em-aspera-demo.html with id number 10005. The coordinates of atomic model of TRPV1 have been deposited in the Protein Data Bank under the accession number: 3J5P.
The authors declare no competing financial interests.
References
- 1.Ramsey IS, Delling M, Clapham DE. An introduction to TRP channels. Annu Rev Physiol. 2006;68:619–647. doi: 10.1146/annurev.physiol.68.040204.100431. [DOI] [PubMed] [Google Scholar]
- 2.Venkatachalam K, Montell C. TRP channels. Annu Rev Biochem. 2007;76:387–417. doi: 10.1146/annurev.biochem.75.103004.142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nilius B, Owsianik G. Transient receptor potential channelopathies. Pflugers Arch. 2010;460:437–450. doi: 10.1007/s00424-010-0788-2. [DOI] [PubMed] [Google Scholar]
- 4.Nilius B, et al. Gating of TRP channels: a voltage connection? J Physiol. 2005;567:35–44. doi: 10.1113/jphysiol.2005.088377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 6.Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 7.Payandeh J, Scheuer T, Zheng N, Catterall WA. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475:353–358. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X, et al. Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature. 2012;486:130–134. doi: 10.1038/nature11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caterina MJ, et al. The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature. 1997;389:816–824. doi: 10.1038/39807. [DOI] [PubMed] [Google Scholar]
- 10.Cao E, Cordero-Morales JF, Liu B, Qin F, Julius D. TRPV1 channels are intrinsically heat sensitive and negatively regulated by phosphoinositide lipids. Neuron. 2013;77:667–679. doi: 10.1016/j.neuron.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yao J, Liu B, Qin F. Kinetic and energetic analysis of thermally activated TRPV1 channels. Biophys J. 2010;99:1743–1753. doi: 10.1016/j.bpj.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brederson JD, Kym PR, Szallasi A. Targeting TRP channels for pain relief. Eur J Pharmacol. 2013 doi: 10.1016/j.ejphar.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 13.Julius D. TRP channels and pain. Annu Rev Cell Dev Biol. 2013;29:355–384. doi: 10.1146/annurev-cellbio-101011-155833. [DOI] [PubMed] [Google Scholar]
- 14.Bai XC, Fernandez IS, McMullan G, Scheres SH. Ribosome structures to near-atomic resolution from thirty thousand cryo-EM particles. eLife. 2013;2:e00461. doi: 10.7554/eLife.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nature methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu X, Jin L, Zhou ZH. 3.88 A structure of cytoplasmic polyhedrosis virus by cryo-electron microscopy. Nature. 2008;453:415–419. doi: 10.1038/nature06893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X, Jin L, Fang Q, Hui WH, Zhou ZH. 3.3 A cryo-EM structure of a nonenveloped virus reveals a priming mechanism for cell entry. Cell. 2010;141:472–482. doi: 10.1016/j.cell.2010.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mio K, et al. The TRPC3 channel has a large internal chamber surrounded by signal sensing antennas. Journal of molecular biology. 2007;367:373–383. doi: 10.1016/j.jmb.2006.12.043. [DOI] [PubMed] [Google Scholar]
- 19.Moiseenkova-Bell VY, Stanciu LA, Serysheva II, Tobe BJ, Wensel TG. Structure of TRPV1 channel revealed by electron cryomicroscopy. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:7451–7455. doi: 10.1073/pnas.0711835105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shigematsu H, Sokabe T, Danev R, Tominaga M, Nagayama K. A 3.5-nm structure of rat TRPV4 cation channel revealed by Zernike phase-contrast cryoelectron microscopy. J Biol Chem. 2010;285:11210–11218. doi: 10.1074/jbc.M109.090712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chung MK, Guler AD, Caterina MJ. TRPV1 shows dynamic ionic selectivity during agonist stimulation. Nature neuroscience. 2008;11:555–564. doi: 10.1038/nn.2102. [DOI] [PubMed] [Google Scholar]
- 22.Myers BR, Bohlen CJ, Julius D. A yeast genetic screen reveals a critical role for the pore helix domain in TRP channel gating. Neuron. 2008;58:362–373. doi: 10.1016/j.neuron.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nature methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lishko PV, Procko E, Jin X, Phelps CB, Gaudet R. The ankyrin repeats of TRPV1 bind multiple ligands and modulate channel sensitivity. Neuron. 2007;54:905–918. doi: 10.1016/j.neuron.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 25.Catterall WA. Ion channel voltage sensors: structure, function, and pathophysiology. Neuron. 2010;67:915–928. doi: 10.1016/j.neuron.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swartz KJ. Sensing voltage across lipid membranes. Nature. 2008;456:891–897. doi: 10.1038/nature07620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boukalova S, Marsakova L, Teisinger J, Vlachova V. Conserved residues within the putative S4–S5 region serve distinct functions among thermosensitive vanilloid transient receptor potential (TRPV) channels. J Biol Chem. 2010;285:41455–41462. doi: 10.1074/jbc.M110.145466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boukalova S, Teisinger J, Vlachova V. Protons stabilize the closed conformation of gain-of-function mutants of the TRPV1 channel. Biochim Biophys Acta. 2013;1833:520–528. doi: 10.1016/j.bbamcr.2012.11.017. [DOI] [PubMed] [Google Scholar]
- 29.Cao E, Liao M, Cheng Y, Julius D. TRPV1 structures in distinct conformations reveal mechanisms of activation. Nature. doi: 10.1038/nature12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Latorre R, Zaelzer C, Brauchi S. Structure-functional intimacies of transient receptor potential channels. Q Rev Biophys. 2009;42:201–246. doi: 10.1017/S0033583509990072. [DOI] [PubMed] [Google Scholar]
- 31.Lin Z, et al. Exome sequencing reveals mutations in TRPV3 as a cause of Olmsted syndrome. Am J Hum Genet. 2012;90:558–564. doi: 10.1016/j.ajhg.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Long SB, Campbell EB, Mackinnon R. Voltage sensor of Kv1.2: structural basis of electromechanical coupling. Science. 2005;309:903–908. doi: 10.1126/science.1116270. [DOI] [PubMed] [Google Scholar]
- 33.Loukin S, Su Z, Kung C. Increased basal activity is a key determinant in the severity of human skeletal dysplasia caused by TRPV4 mutations. PLoS One. 2011;6:e19533. doi: 10.1371/journal.pone.0019533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Owsianik G, Talavera K, Voets T, Nilius B. Permeation and selectivity of TRP channels. Annual review of physiology. 2006;68:685–717. doi: 10.1146/annurev.physiol.68.040204.101406. [DOI] [PubMed] [Google Scholar]
- 35.Susankova K, Ettrich R, Vyklicky L, Teisinger J, Vlachova V. Contribution of the putative inner-pore region to the gating of the transient receptor potential vanilloid subtype 1 channel (TRPV1) J Neurosci. 2007;27:7578–7585. doi: 10.1523/JNEUROSCI.1956-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voets T, Janssens A, Droogmans G, Nilius B. Outer pore architecture of a Ca2+-selective TRP channel. J Biol Chem. 2004;279:15223–15230. doi: 10.1074/jbc.M312076200. [DOI] [PubMed] [Google Scholar]
- 37.Binshtok AM, Bean BP, Woolf CJ. Inhibition of nociceptors by TRPV1-mediated entry of impermeant sodium channel blockers. Nature. 2007;449:607–610. doi: 10.1038/nature06191. [DOI] [PubMed] [Google Scholar]
- 38.Salazar H, et al. Structural determinants of gating in the TRPV1 channel. Nat Struct Mol Biol. 2009;16:704–710. doi: 10.1038/nsmb.1633. [DOI] [PubMed] [Google Scholar]
- 39.Doyle DA, et al. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 40.Gaudet R. A primer on ankyrin repeat function in TRP channels and beyond. Mol Biosyst. 2008;4:372–379. doi: 10.1039/b801481g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Inanobe A, Matsuura T, Nakagawa A, Kurachi Y. Structural diversity in the cytoplasmic region of G protein-gated inward rectifier K+ channels. Channels (Austin) 2007;1:39–45. [PubMed] [Google Scholar]
- 42.Nishida M, MacKinnon R. Structural basis of inward rectification: cytoplasmic pore of the G protein-gated inward rectifier GIRK1 at 1.8 A resolution. Cell. 2002;111:957–965. doi: 10.1016/s0092-8674(02)01227-8. [DOI] [PubMed] [Google Scholar]
- 43.Inada H, Procko E, Sotomayor M, Gaudet R. Structural and biochemical consequences of disease-causing mutations in the ankyrin repeat domain of the human TRPV4 channel. Biochemistry. 2012;51:6195–6206. doi: 10.1021/bi300279b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Booth DS, Avila-Sakar A, Cheng Y. Visualizing proteins and macromolecular complexes by negative stain EM: from grid preparation to image acquisition. Journal of visualized experiments : JoVE. 2011 doi: 10.3791/3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 45.Dukkipati A, Park HH, Waghray D, Fischer S, Garcia KC. BacMam system for high-level expression of recombinant soluble and membrane glycoproteins for structural studies. Protein expression and purification. 2008;62:160–170. doi: 10.1016/j.pep.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reeves PJ, Callewaert N, Contreras R, Khorana HG. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Althoff T, Mills DJ, Popot JL, Kuhlbrandt W. Arrangement of electron transport chain components in bovine mitochondrial supercomplex I1III2IV1. The EMBO journal. 2011;30:4652–4664. doi: 10.1038/emboj.2011.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Popot JL, et al. Amphipols from A to Z. Annual review of biophysics. 2011;40:379–408. doi: 10.1146/annurev-biophys-042910-155219. [DOI] [PubMed] [Google Scholar]
- 49.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. Journal of structural biology. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 50.Grigorieff N. FREALIGN: high-resolution refinement of single particle structures. Journal of structural biology. 2007;157:117–125. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 51.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. Journal of structural biology. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 53.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 54.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MS. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph. 1996;14:354–360. 376. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]