Abstract
Oligonucleotides can be used to direct the alteration of single nucleotides in chromosomal genes in yeast. Rad51 protein appears to play a central role in catalyzing the reaction, most likely through its DNA pairing function. Here, we re-engineer the RAD51 gene in order to produce proteins bearing altered levels of known activities. Overexpression of wild-type ScRAD51 elevates the correction of an integrated, mutant hygromycin resistance gene ∼3-fold. Overexpression of an altered RAD51 gene, which encodes a protein that has a higher affinity for ScRad54, enhances the targeting frequency nearly 100-fold. Another mutation which increases the affinity of Rad51 for DNA was also found to increase gene repair when overexpressed in the cell. Other mutations in the Rad51 protein, such as one that reduces interaction with Rad52, has little or no effect on the frequency of gene repair. These data provide the first evidence that the Rad51 protein can be modified so as to increase the frequency of gene repair in yeast.
INTRODUCTION
The Rad51 protein is required for conferring resistance to ionizing radiation and for regulating mitotic recombination, as well as the induction of meiotic recombination in Saccharomyces cerevisiae (Sc) (1–4). Rad51 has sequence and functional similarity to the Escherichia coli RecA protein and, thus, can catalyze a variety of ATP-dependent DNA pairing reactions (5,6). The mechanism of Rad51-promoted DNA pairing remains to be fully elucidated, but the binding of single-stranded DNA and Rad51’s interactions with Rad54 and Rad52 appear to be critically important steps (3,7,8). Rad51 has also been shown to increase the frequency of a process known as gene repair or targeted nucleotide exchange in which modified single-stranded oligonucleotides hybridize to a complementary DNA sequence and direct the alteration of single base targets (9–15). The use of oligonucleotides for site-specific mutagenesis in yeast originates from the work of Sherman and colleagues (16–18), which has recently been extended and expanded by Liu et al. (9,12,19,20) and others (13,20). Appropriately, overexpression of RAD51 and RAD54 has been shown to enhance gene repair on an integrated copy of a hygromycin-eGFP fusion gene in the yeast strain LSY678 [LSY678(Int)Hyg)] (9); these observations confirmed biochemical/cell-free extract results that unveiled the importance of members of the RAD52 epistasis groups, particularly RAD51, RAD52 and RAD54, in this reaction. Furthermore, when the reaction was carried out in cells deficient in RAD51 or RAD54, the level of gene repair was reduced by at least 70% (21).
While the repair of genetic mutations directed by an oligonucleotide within the context of the chromosome is the logical and ultimate form of gene therapy, such approaches have suffered from unpredictable and variable correction frequencies (see 15 for review). The recent observations by Drury et al. (13) strongly suggest that the assimilation of the single-stranded vector into the complementary target site is likely the rate-limiting step of the reaction, which could impact significantly on the frequency. Thus, a detailed examination of the proteins that regulate this step is warranted. As described above, Rad51 is the most logical candidate since this recombinase catalyzes DNA pairing in the gene repair process; previous results have defined its importance (9). But, other activities such as helix destabilization may also be important in the pathway as structural disruptions may enable chromatin remodeling which in turn increases the accessibility of the target site in genomic DNA to the single-stranded oligonucleotide vector.
From a practical and perhaps ‘applications’ standpoint, the introduction of multiple plasmids containing multiple expression constructs into cells seems unrealistic, and recapitulating balanced effective expression levels is clearly quite challenging. To circumvent such complexities, we have engineered the yeast RAD51 gene creating altered Rad51 proteins that elevate or reduce levels of particular activities. The yeast protein contains certain domains which may regulate its interactions with DNA and other proteins (22), providing a reservoir of sites within domains that can be mutated and tested. Our focus on Rad51 is dictated by previous observations (23), as is our other selection of certain areas within the protein as sites for alteration. For these experiments, we created four individual amino acid changes in Rad51 that lead to an altered function of the protein (22). In this paper, we report that re-engineered Rad51 containing either increased single-strand DNA binding activity or increased interaction with the Rad54 protein enhanced the frequency of gene repair significantly.
MATERIALS AND METHODS
Plasmids and oligonucleotides
The integrative targeting plasmid, pAUR101Hyg(rep)eGFP, was constructed by inserting a fusion gene containing the mutant hygromycin gene and the eGFP gene into pAUR101 (Panvera). The mutation is in the hygromycin coding sequence at nucleotide 138 (TAT→TAG), resulting in a stop codon. Plasmid pYNRAD51 (9) served as the template to create mutations in the RAD51 gene using a QuickChange site directed mutagenesis kit (Stratagene, Cedar Creek, TX), and all variants were subsequently verified by DNA sequence analyses. The single-stranded oligonucleotide, Hyg3S/74NT (9,19) (74 nt long with three phosphorothioate linkage modification at each end designed to target the non-transcribed strand of hygromycin gene), was synthesized by IDT (Coralville, IA) and purified by reverse phase HPLC (24). Plasmids pYNRAD54, pYNSGS1, pYN132 and pYNU132 are described by Liu et al. (9)
Yeast transformation and targeting
Yeast transformation and targeting were carried out as described previously (9). Briefly, LSY678 (Mata leu2-3, 112 trp1-1 ura3-1 his3-11, 15 ade2-1 can1-100) was grown to a density of 2 × 107 cells/ml in YPD medium and transformed with the integrative targeting plasmid pAUR101-Hyg(rep)eGFP. The transformed cell line, LSY678(Int), thus created, was selected on YPD-aureobasidin A plates and integration confirmed by colony PCR and Southern blot analyses. A second transformation with pYN132, pYNRAD51 plasmids and/or pYNU132 plasmids was carried out and each vector maintained in these cells by continual growth on SC-trp plates and/or SC-trp-ura plates. LSY678(Int), bearing one type of pYN132 and pYNRAD51 plasmid, was cultured in -trp selective media until a cell density of 2 × 107/ml was reached, then electroporated using BioRad Gene Pulser apparatus (Richmond, CA) (0.2 cm cuvette, 1.5 kV, 25 µF, 200 Ω, 1 pulse, 5 ms per pulse length) with 10 µg of Hyg3S/74NT. Recovery took place in 3 ml of YPD media supplemented with 1 M sorbitol for 16 h prior to the cells (200 µl) being spread on YPD-Hygromycin (300 µg/ml) plates or (200 µl of a 105 dilution) spread on YPD-aureobasidin A (0.5 µg/ml) plates. Cells were grown for 3 days at 30°C and resistant colonies counted. Independent experiments were carried out in triplicate; the mean and standard deviations of hygr and aureor colonies, as well as their statistical significance, were analyzed by the Analysis of Variance (ANOVA) procedure in the SAS system. The significance was gated when P < 0.05.
MMS sensitivity assay
LSY678 cells or LSY402(Δrad51) bearing plasmid pYN132 or various mutant pYNRAD51 plasmids were cultured in -trp selective media to a density of ∼2 × 107 cells/ml. Cells were diluted to a density of 0.6 × 107 cells/ml (equivalent to OD600 = 0.2) and from this cells were further diluted 10-fold; 10 µl of cells was taken from each dilution and spread on YPD plates containing either 0 or 0.025% (v/v) of MMS, then cultured at 30°C for 3 days and observed for cell growth. Plates were photographed using an Alpha Imager™ 2200 (Alpha Imager, San Leandro, CA).
RT–PCR and western blot analyses
LSY678(Int) cells or LSY402(LSY678 rad51::LEU2) bearing pYNRAD51 mutant plasmids or the pYN132 plasmid were cultured to a density of 2 × 107 cell/ml in SC-trp selective medium, and RNA was isolated from 1–5 × 107 cells using RNeasy mini-kit (Qiagen, Valencia, CA). The first cDNA strand was synthesized and amplified using a Superscript II transcription kit (Invitrogen, Carlsbad, CA) with inside primers of Rad51F (5′ gag gtg gag ctt aag atg gca atg cag atg cag ctt gaa gca aat gca g) and Rad51B (5′ gtg gag gtg tct aga tca gtc ttt ggc atc tcc cac tcc atc tgc). The PCR products (1.2 kb in size) were electrophoresed through 1% of agarose gel using a 1 kb ladder (BioRad, Richmond, CA) as a standard, stained with EtBr and photographed under UV light.
For western blot analyses, LSY402(Δrad51) and LSY678(Int) cells bearing pYNRAD51 mutant plasmids or pYN132 were cultured to a density of ∼2 × 107 cells/ml in SC-trp selective medium. The cells were harvested and washed with 10 ml of extraction buffer (200 mM Tris pH 8.0, 150 mM ammonium sulfate, 1 mM EDTA, 10% glycerol, 2 mM DTT) containing a cocktail of protease inhibitors and resuspended in 300 µl of extraction buffer plus 200 µl of glass beads. Cells were vortexed for 5 min and centrifuged for 30 min. The supernatant was collected and protein concentration was measured. Protein samples (10 µg) were separated on an 8% SDS–PAGE gel and electroblotted to a nitrocellulose membrane (BioRad, Richmond, CA). The membrane was incubated with the rabbit antibody raised against ScRad51 (a kind gift of Dr Patrick Sung, University of Texas) in a 1:2500 dilution. A second antibody (Santa Cruz Inc., Santa Cruz, CA) conjugated with horse radish peroxidase (HRP) in a dilution of 1:5000 (mouse anti rabbit antibody) was used to visualize the primary reaction. Expression of ScRad51 protein (43 kDa) was visualized using enhanced chemiluminescence ECL™ Reagents (Amersham, Piscataway, NJ) and hyperfilm (Kodak), and quantitated with a Typhoon 8600 Variable Mode Imager (Molecular Dynamics Inc., Sunnyvale, CA). An internal control was set up by utilizing a goat β-actin antibody (Santa Cruz Biotech Inc., Santa Cruz, CA) in a 1:500 dilution and visualized by anti-goat IgG-HRP (Sigma, St Louis, MO) in a dilution of 1:500 in the same ECL™ developing system.
eGFP visualization using confocal microscopy
LSY678(Int) yeast cells containing pYNRAD51(K342E) were targeted with 10 µg of Hyg3S/74NT and plated on YPD-hygromycin plates for 3 days. The hygromycin-resistant colonies were picked and cultured 2 h before loading into a two-chamber slide (Nunc). Green fluorescence indicating the expression of the fusion gene was visualized with a Zeiss LSM510 NLO confocal microscope using the 488 nm excitation line of the Argon laser and a 500–550 nm band pass filter for detecting green fluorescence emission.
DNA sequence analyses
Hygromycin-resistant colonies were picked from YPD plates containing hygromycin and cultured in YPD for 16 h. Yeast cells were grown to a density of 3 × 107 cells/ml and harvested. The cells were resuspended in 200 µl of lysis buffer (2% Triton X-100, 1% SDS, 0.1 M NaCl, 1 mM EDTA, 1 mM Tris–HCl, pH 8.0), 300 µl of glass beads and 200 µl of phenol/chloroform (1:1), vortexed for 5 min, an additional 200 µl of TE was added and the samples were centrifuged for 5 min more. The aqueous supernatant was collected to which 2-fold volume of 100% ethanol was added and spun immediately. The DNA pellet was rinsed with 70% ethanol and re-dissolved in TE. PCR was performed with primers of pAUR123F (5′ tct gca caa tat ttc aag c) and Hyg1560R (5′ aaa tca cgc cat gta gtg) (9). The 500 bp PCR products were sequenced directly using an ABI prism kit as specified by the manufacturer on an automated ABI 310 capillary sequencer (Applied Biosystems, Foster City, CA).
RESULTS AND DISCUSSION
Based on many of the observations described above, we created a series of mutations that are known to affect specific functions of Rad51, initially to define the domains critical for gene repair activity and then to use these functions to elevate the frequency of gene repair even further. One mutation in Rad51 (K342E) increases its interaction with Rad54 (22), a functional pairing that has been shown to be important for chromosomal repair (9). The second Rad51 mutation (I345T), which increases the DNA binding activity of Rad51 (25), was chosen for study because previous data have shown that the binding of the modified, single-stranded oligonucleotide is a prerequisite for efficient recombinase-mediated gene repair (13). Finally, two mutations that reduce Rad51’s interaction with Rad52 (Y388H and G393D) were also tested since Rad52 has exhibited antagonist behavior within the gene repair reaction (9,21). Thus, our overall strategy was to construct mutant RAD51 genes, place them in a low copy expression vector and evaluate their influence in LSY678(Int), which contains two integrated copies of a mutant hygromycin-resistance-eGFP fusion gene (clarified by Southern blot). For simplicity, this strain is referred to as LSY678(Int) throughout this manuscript.
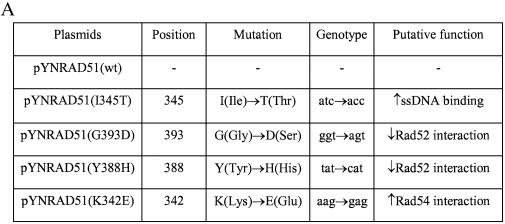
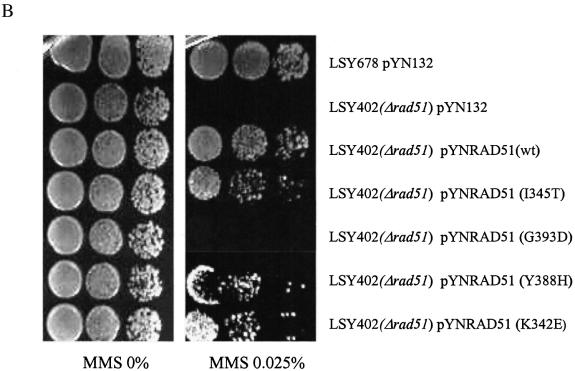
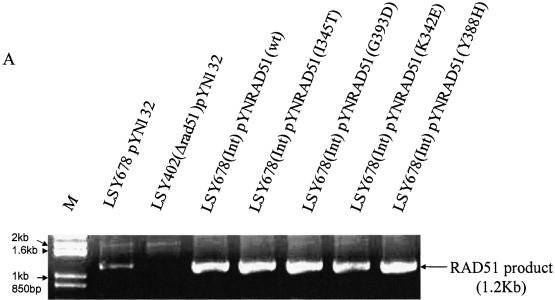
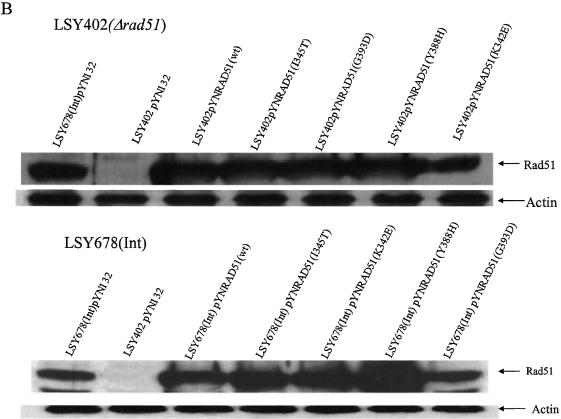
The RAD51 alleles used in this study are presented in Figure 1A along with a description of the Rad51 protein functions that the mutations are known to alter (22,25). The genes were cloned into the vector pYN132, a CEN-ARS plasmid in which expression is regulated by the constitutive promoter, TPI (9). We used several standard assays to evaluate the expression of the mutant RAD51 alleles: first, rescue of MMS sensitivity in LSY402(Δrad51); second, RT–PCR analyses of the overexpressed RAD51 genes; and third, western blotting designed to measure Rad51 levels in two different genetic backgrounds. As shown in Figure 1B, MMS resistance is restored to LSY402(Δrad51) by the expression of wild-type Rad51, but not by Rad51(G393D), a mutation that reduces interaction with Rad52. A modest low level of MMS resistance is observed when Rad51(Y388H), Rad51(I345T) and Rad51(K342E) are used to complement the Δrad51 strain. Complementation with the empty pYN132 vector does not restore the MMS-resistant phenotype. RT–PCR analyses confirm that the mutant strain, LSY402(Δrad51), produces no detectable RAD51 transcript, but that overexpression of each pYNRAD51 plasmid produces a fragment of correct length generated from primers designed within the RAD51 gene (Fig. 2A). Note that the relative level of expression from each plasmid exceeds that of the endogenous RAD51 gene in LSY678(Int), as expected. Western blot analyses indicate that each plasmid is actively producing protein when placed in the LSY402(Δrad51) strain, which provides a clear measure of expression vector activity. From this, we can conclude that expression of the various Rad51 proteins is also observed in LSY678(Int) (Fig. 2B), the host strain for the gene repair reaction.
Figure 1.
Altered RAD51 genes and analyses of gene or protein expression. (A) RAD51 mutant plasmids. Plasmid pYNRAD51(K342E) contains a mutation in RAD51 cDNA that will result in the substitution of K(aag, Lys) with E(gag, Glu) at amino acid position 342. This substitution increases its interaction with Rad54; the I345T substitution increases the DNA binding activity of Rad51; the Y388H substitution and the G393D substitution each decrease the interaction of Rad51 with Rad52. pYN132 is an empty vector as indicated in the Materials and Methods. (B) The MMS sensitivity assay. Yeast cells carrying the indicated plasmids were cultured and serial dilutions were made on YPD plates containing 0 or 0.025% of MMS to assess survival.
Figure 2.
The expression of RAD51 allele from various pYNRAD51 plasmids. (A) RT–PCR analysis of total RNA extracts from LSY678(Int) strain harboring various pYNRAD51 mutant plasmids. A 1.2 kb RAD51 product was amplified using primers of Rad51F and Rad51B and electrophoresed through 1% agarose gel. (B) Western blot analysis of whole-cell extracts from LSY402(Δrad51) and LSY678(Int) containing the various pYNRAD51 expression plasmids. Protein extracts from LSY402(Δrad51) or LSY678(Int), alone or containing the different pYNRAD51 expression plasmids, were evaluated by blotting with rabbit α Rad51 antibody and visualized by an anti-rabbit antibody conjugated with HRP in an enhanced chemiluminescence ECL™ system. Protein extracts from LSY402(Δrad51) with empty vector (pYN132) were used as a negative control. An internal control was set up by utilizing a goat β-actin antibody and visualized by anti-goat IgG-HRP antibody in the same ECL™ system.
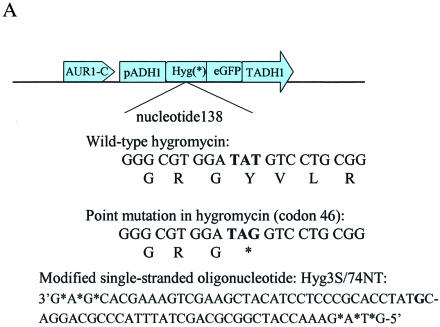
LSY678(Int) contains a stop codon (TAG) at nucleotide position 138 of the hygromycin gene (9), which prevents the translation of a hygromycin-resistant-eGFP fusion protein. This targeting system is depicted in Figure 3A with the mutant codon highlighted in bold. The targeting vector Hyg3S/74NT, a modified, 74 base, single-stranded oligonucleotide containing three phosphorothioate linkages at each terminus designed to direct the repair of the mutation is also presented. This oligomer will hybridize to the non-transcribed (NT) strand of the mutant fusion gene, except at one site where a GG mismatch is created. The repair of this mismatch through nucleotide exchange restores hygromycin resistance and eGFP expression by converting a TAG codon to a TAC codon. The possibility that these phenotypes arise as a result of contamination is diminished because the directed alteration does not revert the codon to a wild type, TAT, but rather to TAC. The integrated cell line also contains a functional aureobasidin resistance gene, allowing us to monitor the total number of cells surviving electroporation, the process used to introduce the oligonucleotide into the yeast cell. Dividing the number of hygromycin-resistant colonies (growing on hyg-agar plates) by the number of aureobasidin-resistant colonies (growing on aureo-agar plates) produces a value known as the correction efficiency index (CE). This value provides an internal control to offset differences among experiments that may arise during yeast electroporation. For each data point, five independent experiments were carried out in triplicate, averaged, and the hygr colony numbers, the aureor colony numbers, as well as the CE values are presented with the appropriate standard deviations.
Figure 3.
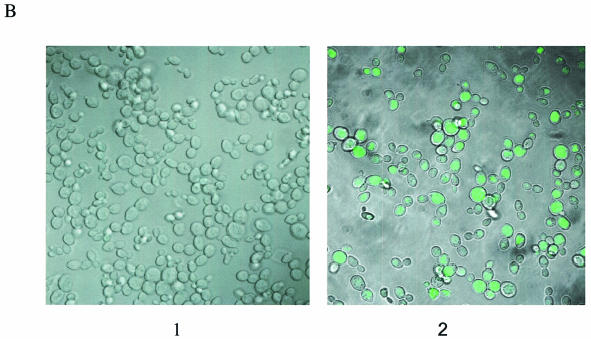
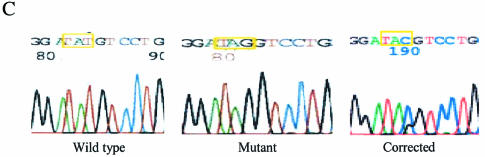
(A) Gene targeting system for chromosomal nucleotide repair. Plasmid pAUR101(Int)Hygs-eGFP, which carries a Hygs-eGFP mutant cassette, was integrated into LSY678. The wild-type hygromycin gene contains TAT at codon 46, while the mutant gene contains a TAG, and the correction creates a TAC codon, restoring hygromycin resistance and eGFP expression. The single-stranded DNA oligonucleotide vector Hyg3S/74NT—74 nt long and containing three phosphorothioate linkages at each end—is designed to target the non-transcribed strand of the fusion gene, repairing codon 46. The targeting nucleotide in oligonucleotide Hyg3S/74NT was highlighted. (B) Evidence of eGFP expression from the Hyg-eGFP fusion gene in LSY678(Int) bearing pYNRAD51(K342E). Yeast cells observed by Zeiss LSM510 confocal microscope (1) before and (2) after the cells were targeted with Hyg3S/74NT. (C) DNA sequence confirmation of fusion gene correction. Direct sequence from PCR amplification showed that the TAG mutation in the integrated Hygs-eGFP cassette was corrected. Wild-type (TAT), mutant (TAG) and corrected (TAC) sequences are provided. The corrected colony sequence comes from an experiment in which pYNRAD51(K342E) was overexpressed in LSY678(Int).
To initialize the reaction, cells were grown to cell density of 2 × 107 cells/ml, at which time they were electroporated with 10 µg of Hyg3S/74NT. After a 16 h recovery period, the cells were spread on plates laden with hygromycin or aureobasidin. The average colony count and correction efficiencies, with standard deviations, are depicted in Table 1. Addition of pYNRAD51(wt) increases the CE ∼3-fold, confirming previous observations (9), while the expression of Rad51(I345T) or Rad51(K342E) enhance the targeting frequency to a greater extent. In particular, the Rad51(K342E) construct increases gene repair nearly 100-fold. Overexpression of Rad51(Y388H) does not enhance the gene repair frequency, to a statistically significant level. Plasmids pYNRAD51(I345T) and pYNRAD51(K342E) produce a statistically significant increase with a P-value of <0.05, compared to the empty vector pYN132 value (see Materials and Methods).
Table 1. Targeted nucleotide repair is elevated by over-expression of certain RAD51 mutants.
| Strains | Hygr a | Aurr (×105) | CE (×10–5)b | Fold |
|---|---|---|---|---|
| LSY678(Int) pYN132 | 2170 ± 295 | 167 ± 18 | 13.95 ± 2.09 | 1 |
| LSY678(Int) pYNRAD51(wt) | 7666 ± 898 | 164 ± 23 | 47.49 ± 8.74 | 3.4 |
| LSY678(Int) pYNRAD51(1345T) | 17 232 ± 2224 | 117 ± 35 | 166.80 ± 44.58c | 11.9 |
| LSY678(Int) pYNRAD51(K342E) | 76 640 ± 6339 | 67 ± 24 | 1354.67 ± 210.30c | 97.1 |
| LSY678(Int) pYNRAD51(Y388H) | 4018 ± 601 | 138 ± 45 | 32.93 ± 9.99 | 2.3 |
| LSY678(Int) pYNRAD51(G393D) | 1393 ± 99 | 111 ± 10 | 12.40 ± 1.12 | 0.9 |
aAverage hygromycin-resistant colony counts from five experiments are presented.
bCorrection efficiency calculated as hygromycin-resistant colonies per aureobasidin-resistant colonies. Statistical analysis was done with AVOVA by SAS Program.
cCompared to that of LSY678(Int) pYN132, P < 0.05 by ANOVA test.
Since the chromosomal target is a fusion gene, hygromycin-resistant cells generated from a correction event should also express eGFP. Hygr cells from a reaction in which Rad51(K342E) was overexpressed were visualized by confocal microscopy and found, in fact, to express eGFP (Fig. 3B), demonstrating that the fusion protein is entirely active. To confirm that the designated base was altered, DNA sequencing was carried out; as shown in Figure 3C, a specific nucleotide alteration (repair) has been made—TAG to TAC—using DNA isolated from a hygr colony. We routinely sequence 20 colonies from each electroporation, and each colony tested in this series was found to contain a mixed peak of G and C, indicating that a subset of two integrated fusion genes is corrected.
Our long-term goal is to enhance the frequency of targeted nucleotide exchange in eukaryotic cells; first, by identifying genes that can influence the frequency, and, second, by introducing them into the targeted cell. Our earlier studies (9,19,21) focused on RAD51, RAD54 and RAD52, with a particular emphasis on RAD51, and it was determined that two activities—the interaction of Rad51 with DNA and the Rad51–Rad54 interaction—were central to the gene repair process. We now report that enhancing these properties by modulating the appropriate Rad51 domains can, in fact, lead to a significant elevation of the gene repair frequency.
Using a tentative 3D model of the Rad51 protein, Krejci et al. (22) identified a possible binding domain for Rad54 on the outer face of the DNA/Rad51 filament. This mutation, K342E, reverses the charge of the wild-type residues and, as a consequence, may result in an increase in its interaction with Rad54. Such a change would logically enhance the capacity of the Rad51/Rad54 complex to stimulate ATP-hydrolysis and catalyze strand separation more readily (26). The K342E mutation in Rad51 corresponds to a position in the RecA protein proposed to be involved in DNA binding and conformational changes that follow ATP-hydrolysis (27,28). Thus, the mutation might affect a cascade of folding events that ultimately leads to a change in the binding of Rad54.
Rad54 has been implicated as a chromatin remodeling protein that helps to destabilize nucleosomal structure (29), providing a more accessible target by reducing steric hindrance. Recent data from Alexeev et al. (30) indicate that Rad54 remodels chromatin, stimulated by the Rad51-ssDNA filament. Mutations in the DNA binding domains of Rad51 that lead to a higher DNA binding affinity would require less protein per filament (25) and would, therefore, be more effective in initiating any homologous pairing activity, a reaction we found to be required for the initialization of gene repair (13). These data support our observation that the Rad51(I345T) mutant, which increases Rad51-ssDNA association, is more active in the gene repair reaction. Under these conditions, the Rad51-ssDNA filament might be more productive in stimulating or solidifying the interaction with Rad54. The Rad51 mutants (Y388H and G393D) that decrease association with Rad52 were also tested because earlier observations suggested that the Rad52 protein was suppressing the gene repair reaction (21). Our results using Rad51(Y388H) partially support this notion since its overexpression elevates the frequency of gene repair to some extent, but overexpression of Rad51(G393D) was found not to influence the gene repair reaction in the assays reported herein. It may be that the G393D mutation is outside the edge of the binding domain, near the COOH terminus of RAD51 and, thus, the ‘normal’ (suppressive?) interaction between Rad51 and Rad52 is left intact. But, the effect on gene repair activity induced by these two mutants is minimal and judged not to be statistically significant. Interestingly, these two mutant genes exhibit differences in their capacities to rescue MMS sensitivity (Fig. 1B). We have also tested a ‘double mutant’ bearing K342E and I345T in the MMS sensitivity functional assay and the gene repair assay. This mutant was unable to rescue the Δrad51 MMS sensitivity and showed no enhancement of gene repair. Thus, the mutations appear to inactivate several protein functions.
We have tested the effect of expression of the various altered RAD51 genes on gene repair in a cell line that is not deficient in RAD51. Thus, the enhancement we observe occurs within a genetic background composed of the normal complement of DNA repair genes, including RAD51. Rad51(I345T) and Rad51(K342E) are, in essence, gain-of-function mutants (22,25) for gene repair and appear to exert a dominant effect. As shown in Figure 2B, the level of Rad51 expressed from each of the vectors is only slightly higher than the wild-type levels of Rad51 produced in LSY678. Thus, it is likely that cells bearing these vectors contain nearly stoichiometric levels of the mutant Rad51 proteins. Since this pathway of repair relies on multiple proteins acting in concert, it will be important to continue to design genetic systems that test the function of these proteins further.
Based on our previous data (9,12) and the data presented herein, we have proposed a tentative model to account for the enhancement of gene repair directed by Rad51(K342E) (31). The Rad51–DNA filament is first bound by Rad54 in preparation for the strand invasion of a chromosome. When Rad51(K342E) protein is present in the reaction, we predict that either more Rad54 will bind to the nucleofilament or Rad54 will bind with a higher affinity (or be retained more efficiently), enabling the complex to maintain or extend an active conformation. The activated filament then invades the chromosomal target and aligns in homologous register. The oligonucleotide is designed, however, to create a single base mismatch with the target site, a structural distortion that is subsequently recognized and acted upon by DNA repair pathways (15).
This model makes several predictions, one of which is that the enhancement of gene repair seen with Rad51(K342E) should be mimicked by co-expression of RAD51 and RAD54 in LSY678(Int). To test this prediction, several types of plasmids in different combinations were electroporated into the wild-type strain and maintained under selection. Plasmids pYN132 and pYNU132 are empty control vectors. The gene repair reaction was initiated by introduction of Hyg3S/74NT into LSY678(Int), with hygromycin-resistant colonies quantified as described above. As shown in Table 2, overexpression of RAD51 and RAD54 together resulted in a significant enhancement of gene repair overexpression of either RAD51 or RAD54 alone, whereas other combinations, such as RAD51 and SGS1, did not. SGS1 is a helicase that is required for replication and transcription in yeast (32) and may also suppress genome instability (33). Overexpression of RAD51 results in a 4- to 5-fold increase, whereas dual expression of RAD51 and RAD54 led to a 40-fold increase. Again, expression of both of these genes surpasses the level of endogenous Rad51. These results indicate that elevated levels of Rad51 and Rad54 can improve the frequency of gene repair, lending support to the tentative model presented previously (31). These data are also consistent with observations that indicate that Rad54 requires the presence of Rad51, and vice versa, to catalyze D-loop formation on a chromatin template (34). In addition, Rad54 requires the presence of Rad51 to act as a chromatin remodeler. Since the gene repair reaction is dependent on gaining access to the chromosomal target, any genetic modulation that enhances the Rad51-Rad54 association should elevate the frequency. These data also provide the first evidence that specific re-engineering of the central protein in the gene repair process, Rad51, improves the frequency of gene repair, reaching a rate of ∼1.0% in some cases. These frequencies rival those achieved using the elegant ‘delitto perfetto’ technique described recently by Resnick and colleagues (35).
Table 2. Targeted nucleotide repair is elevated by over-expression of RAD51 and RAD54.
| Strainsa | Hygr | Aurr (×105) | CE (×10–5)b | Fold |
|---|---|---|---|---|
| LSY678 (Int) pYN132+pYNU132 | 1249 ± 55 | 111 ± 12 | 12.39 ± 1.10 | 1 |
| LSY678 (Int) pYNRAD51+pYNU132 | 7134 ± 445 | 133 ± 11 | 53.60 ± 5.29 | 4.3 |
| LSY678 (Int) pYNRAD54+pYNU132 | 3372 ± 409 | 128 ± 35 | 26.18 ± 8.68 | 2.1 |
| LSY678 (Int) pYNRAD51+pYNURAD54 | 44 008 ± 5064 | 87 ± 18 | 488.01 ± 191.38c | 39.4 |
| LSY678 (Int) pYNRAD51+pYNUSGS1 | 7121 ± 459 | 98 ± 16 | 74.36 ± 9.83 | 6.0 |
| LSY678 (Int) pYN132+pYNUSGS1 | 1498 ± 109 | 141 ± 21 | 10.94 ± 1.38 | 0.9 |
aLSY678(Int) with a combination of pYN132 and pYNU132 expression vectors were targeted with Hyg3S/74NT and plated on Hyg-agar and Aur-agar plates as described in Materials and Methods.
bHygromycin- or aureobasidin- resistant colonies, as well as the correction efficiency from five experiments, are presented as mean and standard deviation. Statistical analysis was done by ANOVA test at SAS program.
cCompared to that of LSY678(Int) pYN132+pYNU132, P < 0.05 by ANOVA test.
It is widely recognized that a major stumbling block for the application of this technique to gene therapy, especially for the repair of genetic mutations, is the low frequencies observed in certain cell types [see Andersen et al. (36) for a review]. Factors that enhance and regulate the process are likely to be required to stabilize or elevate the frequency of repair. This work presents the possibility that engineering recombination or repair proteins to function more efficiently in the gene repair process may provide an exciting solution to these therapeutic problems. We do not mean to suggest that we have exhausted the list of potential Rad51 mutants, but rather have based our choice on functional activities suspected to be important for gene repair. We hope to extend this list on a regular basis as the other players in this reaction reveal themselves.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Dr Patrick Sung for the supply of Rad51 antibodies and Dr Lorraine Symington for the mutant yeast strains. The advice of our colleagues, members of the Kmiec laboratory and Dr Anja van Brabant, is appreciated. This work was supported, in part, by a grant from NaPro BioTherapeutics and NIH (RO1 CA89325).
REFERENCES
- 1.Aboussekhra A., Chanet,R., Adjiri,A. and Fabre,F. (1992) Semidominant suppressors of Srs2 helicase mutations of Saccharomyces cerevisiae map in the RAD51 gene, whose sequence predicts a protein with similarities to procaryotic RecA proteins. Mol. Cell Biol., 12, 3224–3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basile G., Aker,M. and Mortimer,R.K. (1992) Nucleotide sequence and transcriptional regulation of the yeast recombinational repair gene RAD51. Mol. Cell Biol., 12, 3235–3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shinohara A., Ogawa,H. and Ogawa,T. (1992) Rad51 protein involved in repair and recombination in S.cerevisiae is a RecA-like protein. Cell, 69, 457–470. [DOI] [PubMed] [Google Scholar]
- 4.Shinohara A., Gasior,S., Ogawa,T., Kleckner,N. and Bishop,D.K. (1997) Saccharomyces cerevisiae recA homologues RAD51 and DMC1 have both distinct and overlapping roles in meiotic recombination. Genes Cells, 2, 615–629. [DOI] [PubMed] [Google Scholar]
- 5.Sugiyama T., Zaitseva,E.M. and Kowalczykowski,S.C. (1997) A single-stranded DNA-binding protein is needed for efficient presynaptic complex formation by the Saccharomyces cerevisiae Rad51 protein. J. Biol. Chem., 272, 7940–7945. [DOI] [PubMed] [Google Scholar]
- 6.Sung P. and Robberson,D.L. (1995) DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell, 82, 453–461. [DOI] [PubMed] [Google Scholar]
- 7.Donovan J.W., Milne,G.T. and Weaver,D.T. (1994) Homotypic and heterotypic protein associations control Rad51 function in double-strand break repair. Genes Dev., 8, 2552–2562. [DOI] [PubMed] [Google Scholar]
- 8.Jiang H., Xie,Y., Houston,P., Stemke-Hale,K., Mortensen,U.H., Rothstein,R. and Kodadek,T. (1996) Direct association between the yeast Rad51 and Rad54 recombination proteins. J. Biol. Chem., 271, 33181–33186. [DOI] [PubMed] [Google Scholar]
- 9.Liu L., Cheng,S., van Brabant,A.J. and Kmiec,E.B. (2002) Rad51p and Rad54p, but not Rad52p, elevate gene repair in Saccharomyces cerevisiae directed by modified single-stranded oligonucleotide vectors. Nucleic Acids Res., 31, 2742–2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cole-Strauss A., Gamper,H., Holloman,W.K., Munoz,M., Cheng,N. and Kmiec,E.B. (1999) Targeted gene repair directed by the chimeric RNA/DNA oligonucleotide in a mammalian cell-free extract. Nucleic Acids Res., 27, 1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gamper H.B., Hou,Y.M. and Kmiec,E.B. (2000) Evidence for a four-strand exchange catalyzed by the RecA protein. Biochemistry, 39, 15272–15281. [DOI] [PubMed] [Google Scholar]
- 12.Liu L., Rice,M.C., Drury,M., Cheng,S., Gamper,H. and Kmiec,E.B. (2002) Strand bias in targeted gene repair is influenced by transcriptional activity. Mol. Cell Biol., 22, 3852–3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drury M.D. and Kmiec,E.B. (2003) DNA pairing is an important step in the process of targeted nucleotide exchange. Nucleic Acids Res., 31, 899–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brachman E.E. and Kmiec,E.B. (2003) Targeted gene repair of cyc1 mutations in Saccharomyces cerevisiae directed by modified single-stranded DNA oligonucleotides. Genetics, 163, 527–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brachman E.E. and Kmiec,E.B. (2002) The ‘biased’ evolution of targeted gene repair. Curr. Opin. Mol. Ther., 4, 171–176. [PubMed] [Google Scholar]
- 16.Yamamoto T., Moerschell,R.P., Wakem,L.P., Komar-Panicucci,S. and Sherman,F. (1992) Strand-specificity in the transformation of yeast with synthetic oligonucleotides. Genetics, 131, 811–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamamoto T., Moerschell,R.P., Wakem,L.P., Ferguson,D. and Sherman,F. (1992) Parameters affecting the frequencies of transformation and co-transformation with synthetic oligonucleotides in yeast. Yeast, 8, 935–948. [DOI] [PubMed] [Google Scholar]
- 18.Moerschell R.P., Tsunasawa,S. and Sherman,F. (1988) Transformation of yeast with synthetic oligonucleotides. Proc. Natl Acad. Sci. USA, 85, 524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu L., Rice,M.C. and Kmiec,E.B. (2001) In vivo gene repair of point and frameshift mutations directed by chimeric RNA/DNA oligonucleotides and modified single-stranded oligonucleotides. Nucleic Acids Res., 29, 4238–4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parekh-Olmedo H., Drury,M. and Kmiec,E.B. (2002) Targeted nucleotide exchange in Saccharomyces cerevisiae directed by short oligonucleotides containing locked nucleic acids. Chem. Biol., 9, 1073–1084. [DOI] [PubMed] [Google Scholar]
- 21.Rice M.C., Bruner,M., Czymmek,K. and Kmiec,E.B. (2001) In vitro and in vivo nucleotide exchange directed by chimeric RNA/DNA oligonucleotides in Saccharomyces cerevisae. Mol. Microbiol., 40, 857–868. [DOI] [PubMed] [Google Scholar]
- 22.Krejci L., Damborsky,J., Thomsen,B., Duno,M. and Bendixen,C. (2001) Molecular dissection of interactions between Rad51 and members of the recombination-repair group. Mol. Cell Biol., 21, 966–976. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Liu L., Cheng,S., van Brabant,A.J. and Kmiec,E.B. (2002) Rad51p and Rad54p, but not Rad52p, elevate gene repair in Saccharomyces cerevisiae directed by modified single-stranded oligonucleotide vectors. Nucleic Acids Res., 30, 2742–2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gamper H.B., Parekh,H., Rice,M.C., Bruner,M., Youkey,H. and Kmiec,E.B. (2000) The DNA strand of chimeric RNA/DNA oligonucleotides can direct gene repair/conversion activity in mammalian and plant cell-free extracts. Nucleic Acids Res., 28, 4332–4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fortin G.S. and Symington,L.S. (2002) Mutations in yeast Rad51 that partially bypass the requirement for Rad55 and Rad57 in DNA repair by increasing the stability of Rad51-DNA complexes. EMBO J., 21, 3160–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mazin A.V., Bornarth,C.J., Solinger,J.A., Heyer,W.D. and Kowalczykowski,S.C. (2000) Rad54 protein is targeted to pairing loci by the Rad51 nucleoprotein filament. Mol. Cell, 6, 583–592. [DOI] [PubMed] [Google Scholar]
- 27.Story R.M., Weber,I.T. and Steitz,T.A. (1992) The structure of the E.coli recA protein monomer and polymer. Nature, 355, 318–325. [DOI] [PubMed] [Google Scholar]
- 28.Roca A.I. and Cox,M.M. (1997) RecA protein: structure, function, and role in recombinational DNA repair. Prog. Nucleic Acid Res. Mol. Biol., 56, 129–223. [DOI] [PubMed] [Google Scholar]
- 29.Fyodorov D.V. and Kadonaga,J.T. (2001) The many faces of chromatin remodeling: SWItching beyond transcription. Cell, 106, 523–525. [DOI] [PubMed] [Google Scholar]
- 30.Alexeev A., Mazin,A. and Kowalczykowski,S.C. (2003) Rad54 protein possesses chromatin-remodeling activity stimulated by the Rad51-ssDNA nucleoprotein filament. Nat. Struct. Biol., 10, 182–186. [DOI] [PubMed] [Google Scholar]
- 31.Liu L., Parekh-Olmedo,H. and Kmiec,E.B. (2003) The development and regulation of gene repair. Nat. Rev. Genet., 4, 679–689. [DOI] [PubMed] [Google Scholar]
- 32.Lee S.K., Johnson,R.E., Yu,S.L., Prakash,L. and Prakash,S. (1999) Requirement of yeast SGS1 and SRS2 genes for replication and transcription. Science, 286, 2339–2342. [DOI] [PubMed] [Google Scholar]
- 33.Myung K., Datta,A., Chen,C. and Kolodner,R.D. (2001) SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nature Genet., 27, 113–116. [DOI] [PubMed] [Google Scholar]
- 34.Alexiadis V. and Kadonaga,J.T. (2002) Strand pairing by Rad54 and Rad51 is enhanced by chromatin. Genes Dev., 16, 2767–2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Storici F., Lewis,L.K. and Resnick,M.A. (2001) In vivo site-directed mutagenesis using oligonucleotides. Nat. Biotechnol., 19, 773–776. [DOI] [PubMed] [Google Scholar]
- 36.Andersen M.S., Sorensen,C.B., Bolund,L. and Jensen,T.G. (2002) Mechanisms underlying targeted gene correction using chimeric RNA/DNA and single-stranded DNA oligonucleotides. J. Mol. Med., 80, 770–781. [DOI] [PubMed] [Google Scholar]