Abstract
The RNA-dependent RNA polymerase (RdRP) qde-1 is an essential component of post-transcriptional gene silencing (PTGS), termed ‘quelling’ in the fungus Neurospora crassa. Here we show that the overexpression of QDE-1 results in a dramatic increase in the efficiency of quelling, with a concomitant net increase in the quantity of al-1 siRNAs. Moreover, in overexpressed strains there is a significant reduction in the number of transgenes required to induce quelling, and an increase in the phenotypic stability despite progressive loss of tandemly repeated transgenes, which normally determines reversion of a silenced phenotype to wild type. These data suggest that the activation and maintenance of silencing in Neurospora appear to rely both on the cellular amount of QDE-1 and the amount of transgenic copies producing RNA molecules that act as a substrate for the RdRP, implicating QDE-1 as a rate-limiting factor in PTGS.
INTRODUCTION
Post-transcriptional gene silencing (PTGS) mechanisms are highly conserved: quelling in fungi, co-suppression in plants and RNA interference (RNAi) in animals all occur due to the presence of foreign nucleic acid sequences such as transgenes, transposons and viral RNAs or double-stranded RNA (dsRNA) (1). The silencing mechanism is based on the cleavage of dsRNA by dicer (2,3), producing small interfering 21–25 nucleotide (nt) RNAs (siRNA) which, in conjunction with the RNA-induced silencing complex (RISC) (4), degrade all homologous mRNAs. While dsRNA has been shown to trigger PTGS directly (5), it is still unclear how dsRNA originates in transgene-induced silencing. Current models (6,7) propose that aberrant single-stranded transgenic transcripts (abRNA) are converted to dsRNA by a cellular RNA-dependent RNA polymerase (RdRP). The identification of the quelling-defective gene qde-1 in Neurospora was the first experimental evidence of the involvement of an RdRP in PTGS (8). QDE-1 RdRP activity in vitro was shown to catalyse de novo or primer-independent RNA polymerization on a single-stranded RNA (ssRNA) template (9). As well as initiating the transgene-induced silencing mechanism, RdRP may also be responsible for the amplification and maintenance of the silencing signal by synthesis of secondary dsRNA trigger molecules, which in turn would be processed into secondary siRNAs.
In general, the introduction or the direct expression of dsRNA in a eukaryotic cell is sufficient to initiate silencing. When transgenes are introduced in plants and fungi, however, this sequence-specific degradation does not always occur. As the mere presence of transgenic DNA, although necessary, is not sufficient to activate silencing, it has been proposed that only particular transgenic loci are able to work as silencing inducers. In plants for instance, highly expressed transgenes are better inducers than poorly expressed ones (10), suggesting that transgenic mRNAs may only trigger silencing when they exceed a given threshold. Moreover, a correlation between the presence of tandemly arranged transgenic loci and the occurrence of silencing has been observed in both plants and fungi (11,12). It has therefore been suggested that tandem transgenic repeats are good inducers of silencing, even without a high rate of transcription, because they produce RNAs that are somehow ‘aberrant’, which are specifically used as substrates for RdRPs. However, the presence of a tandem repeat per se is not sufficient to elicit a silencing response (13,14). Furthermore, it is still unclear whether the correlation between tandem repeats or highly expressed transgenes and the activation of silencing reflects special features of such transgenes, or suggests the existence of a threshold for either transgenic RNAs or transgenic copy number above which cells activate the silencing machinery. A simple model would be that transgenes or duplicated DNA are able to produce a silencing trigger (i.e. an RNA substrate for RdRP) per se, and only when such a trigger reaches a given threshold is silencing efficiently elicited.
In order to gain insight into the initiation of transgene-induced gene silencing, we decided to overexpress the RdRP qde-1 in Neurospora crassa. This was based on the hypothesis that augmenting the level of QDE-1 may increase conversion of transgenic RNA into dsRNA, leading to the activation of silencing in all transgenic strains, including those that supposedly express the RNA trigger below a given threshold. In this paper we present data showing that the overexpression of QDE-1 leads to a dramatic increase in quelling efficiency (i.e. percentage of transgenic strains showing silencing). Moreover, in overexpressed strains there is a significant reduction in the number of transgenes required to induce quelling, and a concomitant increase in phenotypic stability. Thus, the activation and maintenance of silencing in Neurospora appears to rely on the relative concentration of both QDE-1 and its transgenic RNA substrates.
MATERIALS AND METHODS
Neurospora crassa strains
The N.crassa wild-type strain 74-OR23A (FGSC No. 987) was obtained from the Fungal Genetics Stock Center, University of Kansas (KA, USA). The qde-1-overexpressing strain OQ1 was created by transforming a wild type with plasmid pMXY2:qde-1 and purified by isolation of microconidia to obtain a homokaryon.
Neurospora crassa media and growth conditions
Strains were grown in Vogel’s minimal medium for Neurospora (NMM) (15) plus benomyl (1 µg/ml) or hygromycin (0.2 mg/ml in slants and liquid media, or 0.3 mg/ml in solid media), as required. Neurospora strains transformed with al-1 were grown for 48 h in liquid medium at 28°C in the dark, with constant shaking at 150 r.p.m. [for growth in induced conditions, liquid medium contained 0.5% sucrose, 1× Vogel’s and 0.6% quinic acid (QA)]. Expression of al-1 was induced by constant saturating light for 20 min at 10 W/m2.
Plasmids
The qde-1 overexpression plasmid pMXY2:qde-1 was created by insertion of the qde-1 gene into the SmaI site immediately downstream of the inducible QA dehydrogenase (qa-2) promoter in the pMXY2 vector that contains benomyl resistance as a selectable marker (16). Plasmid pCSN44 carrying the hygromycin resistance gene (17) was used in co-transformation with the pX16 plasmid carrying the al-1 sequence (18) to induce silencing.
Transformation
Preparation of N.crassa spheroplasts and transformation with recombinant plasmids was performed as described by Vollmer and Yanofsky (19). Transformants were selected by growth on plates containing selectable markers, as required, and subsequently transferred to slants.
Southern analysis
Genomic N.crassa DNA was prepared as described in (20). Five micrograms of chromosomal DNA was digested with SmaI/HindIII and fractionated by electrophoresis on a 1% agarose gel. DNA was transferred onto Gene Screen Plus (NEN) filters by capillary blotting. Filters were hybridized according to the manufacturer’s instructions at 65°C with a labelled 1.3-kb XbaI–ClaI fragment of the al-1 gene, which is able to detect both endogenous and transgenic al-1 sequences. Probes were 32P-labelled using a Random Primed DNA Labelling Kit (Roche) according to the manufacturer’s instructions.
Northern analysis
Total RNA was extracted from frozen mycelia. Five micrograms of RNA, quantified by spectrophotometric analysis, were run on a 1% agarose formaldehyde gel and blotted onto HybondN membranes (Amersham Corp). Membranes were hybridized with a 500-bp fragment of the qde-1 gene amplified by PCR from wild-type DNA using Amplitaq DNA polymerase (Perkin Elmer) using forward primer 5′-GCTGGACACTTGATTGAG-3′ and reverse primer 5′-GTCATTGCGGTCACGAAC-3′, and labelled as described above. The al-1 probe was same as that used for the Southern analysis. Quantitative analysis was carried out by electronic autoradiography (Packard Instant Imager).
Purification by microconidia
Purification was carried out as described previously (21).
Small RNA purification and hybridization
These were performed as described previously (4). Hybridization was carried out with a labelled XbaI–ClaI fragment of the al-1 gene.
RESULTS
Overexpression of qde-1 increases silencing efficiency
In N.crassa, the albino genes involved in the carotenoid biosynthetic pathway are used as a visual reporter system, since quelling of albino-1 (al-1) encoding phytoene dehydrogenase confers an albino phenotype. The N.crassa strain OQ1 (carrying an inducible qde-1 transgene) (Fig. 1) and a wild-type strain were co-transformed with plasmids pX16 (17), to trigger silencing of al-1, and pCSN44, conferring hygromycin resistance, as a marker of co-transformation (see Materials and Methods). Transformants grown in conditions of induced and non-induced qde-1 overexpression may be divided into those that are ‘non-silenced’ (NS) or ‘silenced’ (S), irrespective of qde-1 overexpression, and those that are ‘inducibly silenced’ (IS), exhibiting a silenced phenotype only when qde-1 is overexpressed (Fig. 2). The percentage of transformed wild-type colonies with an albino phenotype due to silencing of al-1 was 22%, while for OQ1 transformants the level of silencing increased to 92% in induced conditions, compared with 21% observed in non-induced conditions (Table 1). In order to verify that the high level of silencing was a general phenomenon, and not specific to al-1, OQ1 was transformed with albino-2 (al-2) encoding phytoene synthase, which is also involved in carotenogenesis. Similar levels of silencing of al-2, as indicated by an albino phenotype, were observed (Table 1).
Figure 1.

Northern analysis of wild-type strains transformed with the qde-1 overexpression cassette. Overexpression of qde-1, induced by the addition of quinic acid, was revealed in transformants 3 and 6. Both transformants showed the same silencing efficiency (data not shown). Subsequent experiments were carried out with transformant 6 as the qde-1-overexpressing strain OQ1. The shadow seen in the non-induced lanes reflects a low level of QDE-1 expression. N, non-induced; I, induced.
Figure 2.
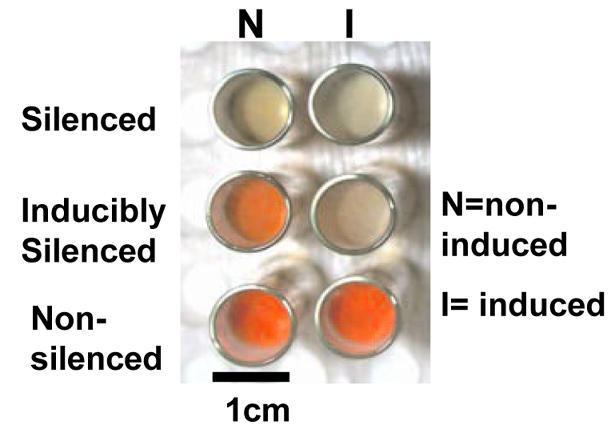
Effect of qde-1 overexpression on the phenotype of al-1 transformants. Phenotype of a non-silenced (orange) and a silenced (albino) strain, irrespective of qde-1 overexpression, and an inducibly silenced strain where silencing (albino phenotype) was a result of qde-1 overexpression induced by quinic acid.
Table 1. Effect of qde-1 overexpression on the efficiency of silencing in al-1 and al-2 transformants in OQ1 and wild-type (WT) strains.
| Gene | Non-silenced (%) | Silencing in non-induced conditions (%) | Silencing in induced conditions (%) | Frequency of silencing excluding NS transformants with no al-1 transgene (%) | Total numbers analysed (n) |
|---|---|---|---|---|---|
| OQ1 strain | |||||
| al-1 | 22 | 21 | 78 | 92 | 1400 |
| al-2 | 23 | 23 | 73 | 90 | 780 |
| Wild-type strain | |||||
| al-1 | 78 | 22 | 22 | NA | 500 |
| al-2 | 81 | 19 | 20 | NA | 220 |
NS, non-silenced; NA, not available.
Colonies were either non-silenced (orange phenotype) or silenced (albino phenotype) irrespective of qde-1 overexpression, or inducibly silenced (albino phenotype) due to qde-1 overexpression induced by quinic acid. The frequency of silencing considers only those transformants that contain the al-1 transgene and was calculated by analysing non-silenced transformants for copy number. Two-thirds (66%) of the NS strains (eight out of 12 strains analysed) contained no copies of al-1 and are therefore ‘false negatives’, putting the actual level of silencing in conditions of qde-1 overexpression at ∼92%.
The results above indicate that in some transgenic strains (S strains), the silencing signal is sufficient to induce substantial mRNA degradation, whereas in other transformants (IS) silencing only occurs when qde-1 is overexpressed. Consistent with this hypothesis, we found that al-1 siRNA levels are increased in induced conditions, both in constitutively silenced (OQ1-S1) and inducibly silenced (OQ1-IS60) strains (Fig. 3). Characteristically, both constitutively and inducibly silenced strains show an accumulation of siRNAs in induced and non-induced conditions, but in the OQ1-IS60 strain the siRNA level, which is lower than in the OQ1-S1 strain, does not appear to be sufficient for mRNA degradation.
Figure 3.
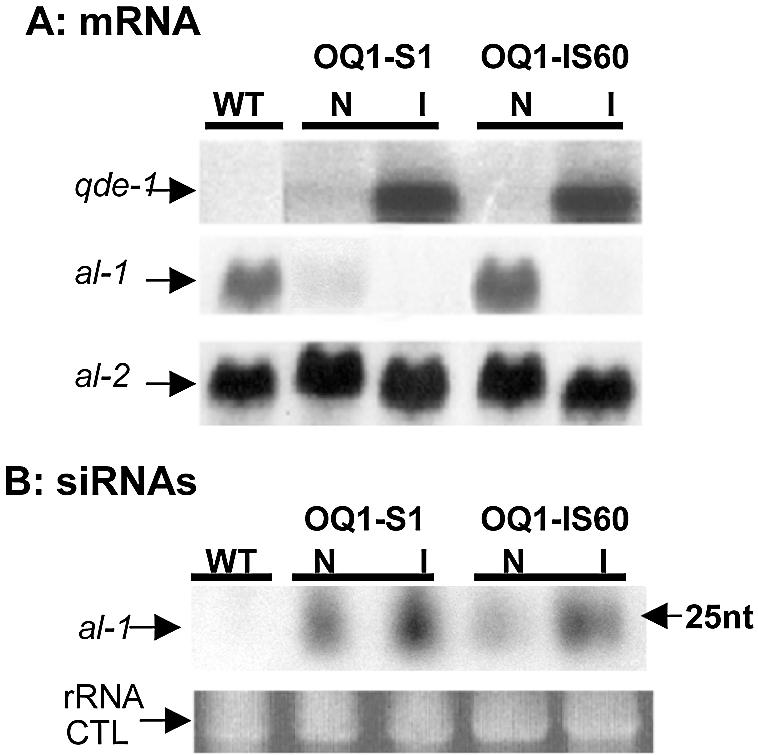
Northern analysis of mRNA and siRNA. (A) Northern analysis of qde-1 and al-1 mRNA in a wild-type (WT) control and two overexpressed strains in both non-induced (N) and induced (I) conditions. al-1 mRNA was silenced in OQ1-S1, whereas for OQ1-IS60 it was only silenced when qde-1 was overexpressed. Expression of the al-2 gene was assessed for normalization purposes. (B) Northern analysis of al-1 siRNAs in the same strains as in (A). As expected, no al-1 siRNAs were detected in the wild type, whereas in OQ1-S1 and OQ1-IS60 an increase of al-1 siRNA was detected under induced conditions. The ribosomal RNA is shown as a control for equal loading.
The increase in silencing frequency and the isolation of silencing-inducible transgenic strains strongly suggests that the overexpression of qde-1 acts on a (pre-existing) silencing signal (present in almost all transgenic strains) by increasing the amount of dsRNA synthesized, consequently augmenting the level of siRNAs. It could be argued, however, that the increased silencing frequency is affected by uncontrollable parameters during transformation, altering the number of transgenic copies integrated, the site(s) of integration and/or organization. To overcome this objection we carried out a forced heterokaryon experiment where a non-silenced wild-type transformant containing the al-1 transgene was forced with the qde-1 overexpresssing strain (no copies of transgenic al-1). The resulting heterokaryon showed a silenced phenotype when QDE-1 was overexpressed by quinic acid. Instead, when the non-silenced wild-type al-1 transformant was forced with a wild-type strain, no silencing was seen. These data suggest that QDE-1 is a ‘limiting factor’, since the level of transgenic RNA remained the same in both heterokaryons. In addition, these data support the notion that in a wild-type background a given transgenic locus is able to produce a silencing signal, but that this signal is not always sufficient to induce silencing.
Overexpression of qde-1 reduced the number of copies required for silencing
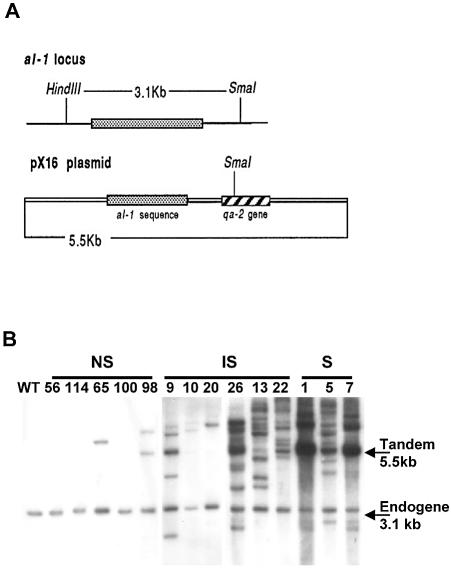
Given that a non-silenced (NS) wild-type al-1 with a few copies can be silenced by creating a heterokaryon with overexpressed qde-1, we asked if the number of transgenic copies required to elicit a silencing response is reduced when OQ1 is overexpressed. We therefore carried out a Southern analysis of al-1 copy number in 52 OQ1 al-1 transformants, including a wild-type strain as a control for the endogenous copy (giving a 3.1-kb band) (Fig. 4A and B). Tandem arrays of the al-1 transgene revealed by a 5.5-kb band were present in S strains (OQ1-S1, -5 and -7), confirming previous data demonstrating that a high transgene copy number is required for efficient silencing. In comparison, the IS strains (OQ1-IS9, -10, -20, -26, -13 and -22) harboured fewer copies of the al-1 transgene, even as few as one or two copies (OQ1-IS10 and -20), indicating that overexpressing qde-1 compensates for the lower al-1 copy number, i.e. the number of transgenes required to trigger silencing is reduced when QDE-1 is not limiting. Interestingly, 66% of the NS strains (8 out of 12 strains) contained no copies of al-1, indicating that a large portion of the 22% of NS OQ1 al-1 transformants may have been transformed with pCSN44 only (conferring hygromycin resistance) and are therefore ‘false negatives’, suggesting that the actual level of silencing in conditions of qde-1 overexpression is ∼92% (see Table 1). As quelling is known to progressively revert phenotype back to wild type, which is associated with a loss of tandemly repeated copies over a prolonged culture time (12), and since we have IS strains containing one or two copies, we may expect the stability of the phenotype in overexpressed strains to be increased. We therefore compared the level of reversion between six OQ1 and five wild-type al-1 transformants. All transformants were purified by isolation of microconidia to ensure that they were homokaryotic (21). Table 2 shows that wild-type al-1 strains have a reversion rate of up to 28% in al-1 transformants. Only one of the OQ1 strains (OQ1-IS14) showed reversion of a single colony out of 5700 descendents. The other five strains showed no reversion. To determine whether these strains still continued to lose copies of the transgene, we analysed 50 single colonies of OQ1-S1, OQ1-IS60 and WT-S3. Whilst OQ1-S1 and OQ1-IS60 showed no phenotypic reversion (Fig. 5A), Southern analysis revealed a loss of the al-1 tandem array at a rate similar to that of wild-type strains, i.e. ∼30% (Fig. 5B). Quantification of copy number using electronic autoradiography revealed that reversion of silencing occurred when less than six transgenic al-1 copies remained in tandem, whereas in OQ1 strains, silencing was maintained even in the presence of only two copies. This confirms that there is a threshold number of copies required to elicit a silencing response, and that this number is reduced when qde-1 is overexpressed.
Figure 4.
NS, non-silenced; IS, inducibility silenced; S, constitutely silenced. (A) Southern analysis of al-1 copy number in transformed wild-type and OQ1 strains. Schematic representation of the al-1 endogenous locus and of plasmid pX16 carrying the al-1 transgene. (B) Southern analysis of al-1 copy number in wild-type and OQ1 strains transformed with pX16. Eleven out of a total data set of 52 are shown. Lane 1: wild type (control); lanes 2–6: non-silenced OQ1 strains; lanes 7–12: inducibly- silenced OQ1 strains; lanes 13–15: silenced OQ1 strains. The 3.1-kb band corresponds to the endogenous al-1 gene and the 5.5-kb band corresponds to ectopic al-1 transgenes. The more intense 5.5-kb signal in silenced OQ1 strains denotes tandem arrays of transgenic al-1 sequences, while diverse bands represent randomly integrated transgene copies. Non-silenced OQ1 transformants harbour only one, two or no al-1 transgenes.
Table 2. Phenotypic reversion of al-1 transformants of OQ1 and wild-type strains.
| Transformant | Number analysed | No. of reversions (%) |
|---|---|---|
| 1. OQ1-S7 | 764 | 0 (0) |
| 2. OQ1-S1 | 1925 | 0 (0) |
| 3. OQ1-IS13 | 3160 | 0 (0) |
| 4. OQ1-IS14 | 5700 | 1 (0.017) |
| 5. OQ1-IS60 | 3570 | 0 (0) |
| 6. OQ1-IS72 | 3934 | 0 (0) |
| 7. WT-S1 | 1600 | 1 (0.06) |
| 8. WT-S2 | 3500 | 100 (2.86) |
| 9. WT-S3 | 1880 | 540 (28) |
| 10. WT-S12 | 1580 | 0 (0) |
| 11. WT-S207 | 1100 | 70 (6.3) |
Figure 5.
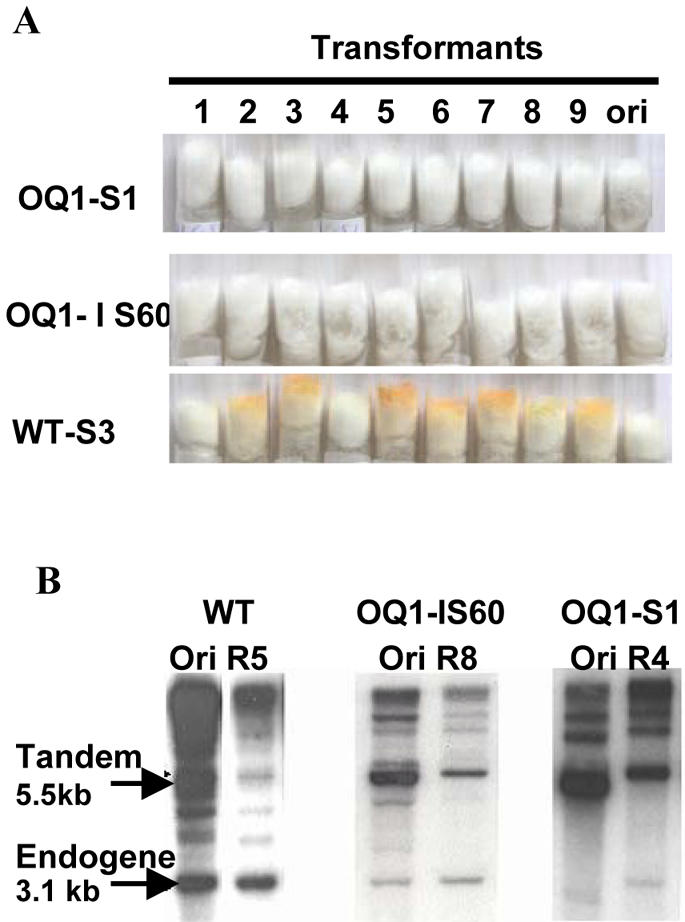
Reversion of silencing in OQ1 and wild-type al-1 transformants. (A) Phenotype of the original culture (‘ori’) with respect to nine single colonies of a silenced OQ1 transformant (OQ1-S1), an inducibly silenced OQ1 transformant (OQ1-IS60) and a silenced wild-type al-1 transformant (WT-S3). (B) Southern analysis of al-1 copy number in the original culture (ORI) as compared with revertant R5 of WT-S3, R8 of OQ1-IS60 and R4 of OQ1-S1. Around 30% of the descendants showed a characteristic loss of transgenic copies.
DISCUSSION
RNA-dependent RNA polymerases are key components of the RNAi machinery. Two different roles have been postulated for these enzymes: first, RdRPs may be involved in converting aberrant RNA molecules produced from silencer loci into dsRNA, or secondly, use siRNAs as primers in the conversion of the target ssRNA into dsRNA, leading to both the degradation of the target RNA by dicer and the accumulation of secondary siRNAs, thus increasing the strength of silencing. Interestingly, we have found that QDE-1 was no longer required upon the direct expression of dsRNA (22). Likewise, in plants, the RdRP SGS2 is not required when PTGS is induced by an inverted repeat (23). These results suggest that the main role of RdRP in transgene-induced gene silencing in Neurospora is the conversion of transgenic RNA into dsRNA. In line with this result, in this work we found that QDE-1 overexpression resulted in an increase in the production of siRNAs. QDE-1 is thought to recognize transgenic RNAs due to some intrinsic ‘aberrancy’ rendering them substrates for RdRPs. Previous reports have suggested that in fungi and plants, a prerequisite for efficient PTGS is the presence of multiple tandemly inserted copies of a transgene and that the abRNA that results from the transcription of such a multi-copy transgenic locus is the trigger of PTGS (11,12). Such tandems are also proposed to be able to maintain silencing through primed polymerization by RdRP, regenerating dsRNA, where single copies would be exhausted by the sequential use of downstream primers (24). We found that the overexpression of the qde-1 gene leads to a dramatic increase in the frequency of transgenic strains showing silencing. In fact, we observed that the silencing frequency of al-1 is ∼92% when QDE-1 is overexpressed, compared with 22% observed in a wild-type background, indicating that almost all transgenic strains are able to support silencing. Importantly, we found that one or two copies can elicit a silencing response when qde-1 is overexpressed, suggesting that rather than a tandem repeat being essential for triggering silencing, every transgenic/repetitive locus possesses the ability to activate silencing. This is in line with early experiments in fungi and plants, which showed that the presence of a tandem repeat per se is not sufficient to elicit silencing (13,14). The fact that only a limited number of transgenic strains (those with the highest number of transgenes) show silencing in the presence of a wild-type level of QDE-1 suggests that silencing in Neurospora is only activated if a given threshold of transgenic/repetitive copies is reached. In reality, such a threshold may reflect ‘quantitative effects’, i.e. silencing may be a gradient from weak to strong, rather than on or off, with the overexpression of qde-1 allowing detection of the silenced phenotype where less transgenic copies are present. Visual inspection of conidial colour in our system does not allow these two models to be differentiated.
Such high levels of silencing are reminiscent of a mutant phenotype seen in Caenorhabditis elegans, in which rrf-3 mutants show hypersensitivity to RNAi (25). The authors suggest that the RRF-3 protein competes with the other required RdRPs RRF-1 and EGO-1 for components or intermediates in the RNAi pathway. We could envision a similar situation here, i.e. that QDE-1 outcompetes the other RdRPs for its RNA substrate, resulting in a higher silencing efficiency. However, mutants in the other two RdRP paralogs of Neurospora sad-1 and RdRP-3 show normal (wild type) silencing efficiencies (our unpublished data), indicating that such a mechanism is not at work in Neurospora.
Quelling is known to revert by excision of tandemly arranged copies during vegetative growth, therefore suggesting that a threshold number of transgenic copies is required not only to activate, but also to maintain silencing. The fact that we found that the phenotypic stability of silencing is strongly increased when qde-1 is overexpressed, even though copies are lost at the same rate as in wild type, indicates that high levels of QDE-1 allow the maintenance of silencing even when the number of transgenic copies are reduced by increasing the production of dsRNA and in turn siRNA molecules. This is further demonstrated by the forced heterokaryon experiment, where the same amount of transgenic RNA produced by al-1 transformed nuclei is able to support silencing only in the presence of the nuclei that overexpress QDE-1. Together our results suggest that in Neurospora, silencing activation and maintenance appear to rely on both the cellular amount of QDE-1 and the amount of transgenic copies producing RNA molecules that act as a substrate for the RdRP, implicating QDE-1 as a rate-limiting factor in PTGS.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Annette Pickford for critical reading, suggestions and revision of the manuscript, and the Whitehead Institute for providing access to the Neurospora genome database. This work was supported by grants from the European Community (QLK3-CT-2000-00078), the Instituto Pasteur Fondazione Cenci Bolognetti, FIRB-MIUR 2001 (RBNEO15MPB_001/RBNE01KXC9_006) and CNR 2003 (Progetto Strategico MIUR: legge 449/97).
REFERENCES
- 1.Pickford A.S. and Cogoni,C. (2003) RNA-mediated gene silencing. Cell. Mol. Life Sci., 60, 871–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hammond S.M., Bernstein,E., Beach,D. and Hannon,G.J. (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature, 404, 293–296. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein E., Caudy,A.C., Hammond,S.M. and Hannon,G.J. (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature, 409, 363–336. [DOI] [PubMed] [Google Scholar]
- 4.Catalanotto C., Azzalin,G., Macino,G. and Cogoni,C. (2002) Involvement of small RNAs and role of the qde genes in the gene silencing pathway in Neurospora. Genes Dev., 16, 790–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fire A., Xu,S., Montgomery,M.K., Kostas,S.A., Driver,S.E. and Mello,C.C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- 6.Voinnet O. (2002) RNA silencing: small RNAs as ubiquitous regulators of gene expression. Curr. Opin. Plant Biol., 5, 444–451. [DOI] [PubMed] [Google Scholar]
- 7.Cogoni C. and Macino,G. (2000) Post-transcriptional gene silencing across kingdoms. Curr. Opin. Genet. Dev., 10, 638–643. [DOI] [PubMed] [Google Scholar]
- 8.Cogoni C. and Macino,G. (1999) Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature, 399, 166–169. [DOI] [PubMed] [Google Scholar]
- 9.Makeyev E.V. and Bamford,D.H. (2002) Cellular RNA-dependent RNA polymerase involved in posttranscriptional gene silencing has two distinct activity modes. Mol. Cell, 10, 1417–1427. [DOI] [PubMed] [Google Scholar]
- 10.Vaucheret H., Beclin,B. and Fagard,M., (2001) Post-transcriptional gene silencing in plants. J. Cell Sci., 114, 3083–3091. [DOI] [PubMed] [Google Scholar]
- 11.Que Q. and Jorgensen,R.A. (1998) Homology-based control of gene expression patterns in transgenic petunia flowers. Dev. Genet., 22, 100–109. [DOI] [PubMed] [Google Scholar]
- 12.Romano N. and Macino,G. (1992) Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol. Microbiol., 6, 3343–3345. [DOI] [PubMed] [Google Scholar]
- 13.Cogoni C., Irelan,J.T., Schumacher,M., Schmidhauser,T.J., Selker,E.U. and Macino,G. (1996) Transgene silencing of the al-1 gene in vegetative cells of Neurospora is mediated by a cytoplasmic effector and does not depend on DNA–DNA interactions or DNA methylation. EMBO J., 15, 3153–3163. [PMC free article] [PubMed] [Google Scholar]
- 14.Sijen T., Wellink,J., Hiriart,J.B. and Van Kammer,A. (1996) RNA-mediated virus resistance: role of repeated transgenes and delineation of targeted regions. Plant Cell, 12, 2277–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis R.H. and De Serres,F.J. (1970) Genetic and microbiological research techniques for Neurospora crassa. Methods Enzymol., 17, 79–143. [Google Scholar]
- 16.Campbell J.W., Enderlin,C.S. and Selitrennikoff,C.P. (1994) Vectors for expression and modification of cDNA sequences in Neurospora crassa. Fungal Genet. Newslett., 41, 20–21. [Google Scholar]
- 17.Staben C., Jensen,B., Singer,M., Pollock,J., Schechtman,M., Kinsey,J. and Selker,E. (1989) Use of a bacterial Hygromycin B resistance gene as a dominant selectable marker in Neurospora crassa transformation. Fungal Genet. Newslett., 36, 79–81. [Google Scholar]
- 18.Cogoni C. and Macino,G. (1997) Isolation of quelling-defective (qde) mutants impaired in posttranscriptional transgene-induced gene silencing in Neurospora crassa. Proc. Natl Acad. Sci. USA, 94, 10233–10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vollmer L.J and Yanofsky,C. (1986) Efficient cloning of genes of Neurospora crassa.Proc. Natl Acad. Sci. USA, 83, 4869–4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nelson M.A., Morelli,G., Carrattoli,A., Romano,N. and Macino,G. (1989) Molecular cloning of a Neurospora crassa carotenoid biosynthetic gene (albino-3) regulated by blue light and the products of white collar genes. Mol. Cell. Biol., 9, 1271–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ebbole D. and Sachs,M.S. (1990) A rapid and simple method for isolation of Neurospora crassa homokaryons using microconidia. Neurospora Newslett., 37, 17–18. [Google Scholar]
- 22.Catalanotto C., Pallotta,M., ReFalo,P., Sachs,M.S., Vayssie,L., Macino,G. and Cogoni,C. (2004) Redundancy of the two dicer genes in transgene-induced posttranscriptional gene silencing in Neurospora crassa. Mol. Cell. Biol., 24, 2536–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beclin C., Boutet,S., Waterhouse,P. and Vaucheret,H. (2002) A branched pathway for transgene-induced RNA silencing in plants. Curr. Biol., 12, 684–688. [DOI] [PubMed] [Google Scholar]
- 24.Martienssen R.A. (2003) Maintenance of heterochromatin by RNA interference of tandem repeats. Nature Genet., 35, 213–214. [DOI] [PubMed] [Google Scholar]
- 25.Simmer F., Tijsterman,M., Parrish,S., Koushika,S.P., Nonet,M.L., Fire,A., Ahringer,J. and Plasterk,R.H. (2002) Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr. Biol., 12, 1317–1319. [DOI] [PubMed] [Google Scholar]