Abstract
Adenosine to inosine editing of mRNA from the human 5-HT2C receptor gene (HTR2C) occurs at five exonic positions (A–E) in a stable stem–loop that includes the normal 5′ splice site of intron 5 and is flanked by two alternative splice sites. Using in vitro editing, we identified a novel editing site (F) located in the intronic part of the stem–loop and demonstrated editing at this site in human brain. We have shown that in cell culture, base substitutions to mimic editing at different combinations of the six sites profoundly affect relative splicing at the normal and the upstream alternative splice site, but splicing at the downstream alternative splice site was consistently rare. Editing combinations in different splice variants from human brain were determined and are consistent with the effects of editing on splicing observed in cell culture. As RNA editing usually occurs close to exon/intron boundaries, this is likely to be a general phenomenon and suggests an important novel role for RNA editing.
INTRODUCTION
Recent findings that the number of genes in the human genome may be as low as 30 000 (1,2) have indicated that variation of gene products by post-transcriptional modifications such as RNA splicing, editing and alternative polyadenylation are essential for the generation of genetic diversity. A common type of RNA editing involves the deamination of adenosine to inosine. This modification is catalyzed by three adenosine deaminases and their isoforms (ADAR1, 2a to 2d and 3) (3–9), which require local double stranded RNA (10–13). Inosine, which base pairs with cytidine, is read as guanosine both by reverse transcriptase and by the cell’s translational machinery. Most examples of RNA editing were identified as discrepancies between genomic and cDNA sequences and consequently there is a strong bias for detecting exonic editing (12–15). Greater than expected levels of inosine (1 per 17 000 ribonucleotides) have been described in rat brain mRNA (16). Since this is more than can be accounted for by the known RNA editing sites, it is likely that many more such sites remain to be discovered. Intronic editing sites have also been described (15,17,18) and may indicate a role for RNA editing in other post-transcriptional processes.
Editing in the 5-HT2C receptor gene (HTR2C) takes place at five positions on exon 5 (13,19–21). All five sites (A–E) are situated in a region that base pairs with an exon-complementary sequence (ECS) in intron 5 to form a stable stem–loop (Fig. 1). Editing results in adenosine to inosine changes in the region encoding the second intracellular loop of the receptor and can generate up to 24 receptor isoforms, ranging from the unedited Ile156 Asn158 Ile160 (INI) to the fully edited Val156 Gly158 Val160 (VGV) (19,20). Edited isoforms exhibit reduced G protein coupling and reduced affinity for serotonin and atypical antipsychotic drugs (13,19–22). Variations in exonic editing have been described in patients with psychiatric disorders (23–27), especially suicide victims with a history of major depression.
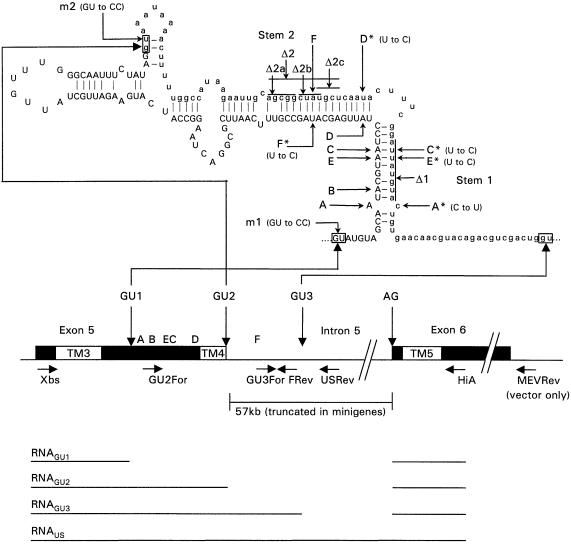
Figure 1.
Region of HTR2C around exons 5 and 6. Positions of the three 5′ splice sites (GU1–3), 3′ splice site (AG) and editing sites A to F are indicated. Exons are represented by blocks, with positions of regions encoding transmembrane domains (TM3-5) indicated (not to scale). Intron 5 is truncated to 0.8 kb in pCR3.1-HTR2C0.8 or 1.2 kb in pCR3.1-HTR2C1.2. Primer locations and predicted RNA transcripts are shown below map. RNA stem–loop, predicted using RNA Draw (46), is shown above map with intronic sequence shown in lower case. Watson–Crick base-pairs are indicated by short lines. An asterisk against an editing site denotes the position of a compensatory mutation. The lines against stem 1 and stem 2 indicate the positions of the deletion mutants.
HTR2C is also subject to alternative splicing between exons 5 and 6, with two alternative 5′ splice sites described (13,21,28,29). Splicing at either site results in truncated proteins that are unlikely to retain receptor function. Alternative splicing of HTR2C may serve to regulate the levels of functional receptor molecules. The normal 5′ splice site GU2 is situated within the stem–loop that contains the five exonic editing sites, with alternative sites GU1 and GU3 located on either side (Fig. 1). The close proximity of the three 5′ splice sites to the editing sites suggests that these two post-transcriptional processes may be causally linked. This can only be in the direction of editing influencing splicing as splicing at GU1 or GU2 destroys the predicted stem–loop and RNA editing is dependent on double stranded RNA (10–13).
In this study, we have investigated the hypothesis that RNA editing modulates selection of splice sites. We have studied splicing in cells transfected with HTR2C minigenes and have shown that base substitutions to mimic adenosine to inosine editing alter the relative amount of splicing at GU1 and GU2. Comparisons of editing patterns between different RNA splice variants from human post-mortem brains are consistent with the results in cell culture.
MATERIALS AND METHODS
Preparation of HTR2C expression constructs
Intron 5 sequence was obtained from BAC clones bA111B4, bA308A10, bA404C21, bA810O3 (Roswell Park) and confirmed on database (AL590097). Truncated HTR2C from exon 5 to 6 (Fig. 1) was cloned into the mammalian expression vector pCR3.1Uni (Invitrogen). Two minigenes pCR3.1-HTR2C0.8 and pCR3.1-HTR2C1.2 (Fig. 1) were constructed and verified by sequencing. All mutant derivatives were performed on pCR3.1-HTR2C0.8 using the QuikChange Site-Directed Mutagenesis kit (Stratagene) and confirmed by sequencing.
In vitro editing
PCR3.1-HTR2C1.2 was transcribed with T7 RNA polymerase using MEGAscript transcription kit (Ambion). Recombinant ADAR2a was prepared from transfected HEK-293 cells and purified using an anti-FLAG antibody (Sigma). In vitro editing was as described previously (9,30). The RNA was amplified by RT–PCR and sequenced to detect editing events.
Cell lines
SH-SY5Y (CRL-2266) and PC-12 (CRL-1721) were purchased from ATCC; COS-7 and HEK-293 cells were generously provided by Professor S. Lovestone and Dr U. D’Souza.
Transient transfections
Transfections of pCR3.1-HTR2C0.8, and derived minigenes, were performed a minimum of three times each using Lipofectamine 2000 (Invitrogen), subculture passages 2–3 and 1 µg DNA per well (24 well plate). Transfection efficiencies were in the range of 30–35% (β-galactosidase assay, data not shown). RNA was prepared at 48 h post-transfection.
Human brain samples
Human brains were from five control individuals: three male, two female; age range, 65–96 years; post-mortem interval range, 16–29 h; causes of death, cardiovascular-associated.
RNA extraction and RT–PCR
For both cell culture and post-mortem brain samples, RNA was extracted using the Absolutely RNA Miniprep kit (Stratagene) and analysed by RT–PCR using MMLV-RT (Promega). For cell culture RNA, to study RNA between exons 5 and 6 (Fig. 1), the reverse transcriptase step used a vector-specific primer MEVRev (CTGGATATCTGCA GAATTCG) to exclude endogenous transcripts. In the PCR step, HiA (CAATAAGAACGAAATTTGGGTCG) was used with primer Xbs (CTGGCCACTACCTAGATAT) over 30 cycles at 93°C/30 s, 65°C/30 s, 72°C/2 min. To exclude amplification of RNAGU1-specific sequences, Xbs was replaced by GU2For (GAGCATAGCCGTTTCAATTCG) as 5′ primer in the PCR step. To exclude amplification of both RNAGU1- and RNAGU2-specific sequences, primer GU3For (GCTCAATACTTTCGGATTATGTAC) was used as 5′ primer.
To study brain RNA between exons 5 and 6, the conditions were as for cell culture RNA except that primer HiA was used in both the RT and PCR steps. To amplify only RNAGU3 and RNAUS (RNAGU3/US), primer FRev (CGTTGTTCACAG TACATAATCC) was used as 3′ primer in both RT and PCR steps. For specific amplification of RNAUS, primer USRev (CCTTTCATTATTGTTAACC) was used as 3′ primer and two sequential 40-cycle amplifications were performed at 93°C/30 s, 49°C/30 s, 72°C/2 min.
Analysis of RNA editing
Products from RT–PCR were separated by electrophoresis on agarose gels. With RT–PCR from brain RNA, the appropriate band was excised, DNA extracted and purified (Qiagen), and cloned into pcDNA2.1 (Invitrogen). RNA editing frequencies were estimated by sequencing individual clones, which varied between 16 and 41 clones per band. Where no clear band was observed (as in four samples amplified from RNAUS, Fig. 4B), the corresponding region of the gel was excised. Clones derived from RNAUS were successfully obtained from three of these four samples (135/CP, 93/T and 293/CP), but not from 135/T (nor from 93/CP or 283/T, not shown). Editing combination frequency comparisons were made between RNAGU2 (seven samples, excluding 135/T; 167 clones) and RNAUS (seven samples; 203 clones) for each brain region and significance was assessed by a stratified chi-square test.
Figure 4.
HTR2C RNA splice variants in human brain. RT–PCR products from RNA from eight brain samples of either thalamus (T) or choroid plexus (CP). (A) Using Xbs and HiA at 30 cycles, with ratios of RNAGU2/RNAGU1 indicated above each lane, from mean of three determinations by fluorescent PCR at 21 cycles and calibrated by standard curve, intraclass correlation coefficient 0.99. (B) Using Xbs and USRev at two rounds of 40 cycles, in the presence (+) or absence (–) of reverse transcriptase. (C) Using Xbs and FRev at 30 cycles, in the presence (+) or absence (–) of reverse transcriptase.
Assay of RNAGU2 to RNAGU1 ratios
Fluorescent PCR for both cell culture and brain-derived cDNA was for 21 cycles of 93°C/30 s, 65°C/30 s, 72°C/2 min with HiA-FAM and unlabelled Xbs. Different numbers of limited cycles were performed prior to fluorescent analysis to ensure quantitative measurements were within the linear range. Products of all fluorescently labelled PCR reactions were resolved on an ABI3100 capillary sequencer with ROX500 size markers (ABI) and the ratio of RNAGU2/RNAGU1 peak heights calculated using Genotyper v3.7. To control for bias due to differential product size, values were calibrated at RNAGU2/RNAGU1 ratios ranging from 7:1 to 1:7 using plasmids (derived from pcDNA2.1) containing either RNAGU2 or RNAGU1 RT–PCR product.
Model for RNAUS
The simple model [RNAUS = q RNAGU2 + (1 – q)RNAGU1] resulted in negative values for RNAGU1, where q is derived from the observed overall RNAGU2/RNAGU1 ratio = q/(1 – q). A new model [RNAUS = q RNAGU2 + (1 – q) RNAGU1 + x] was postulated and using non-linear programming, x2 was minimized across the distribution so that RNAGU1 ≥ 0 and ΣRNAGU1 = 1.
RESULTS
Intronic editing at site F
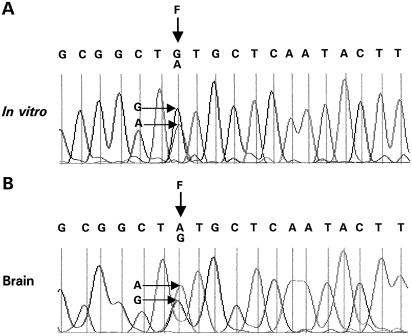
In order to look for novel editing sites in the stem–loop region, mRNA generated from a minigene with a truncated intron 5 (Fig. 1) was incubated with recombinant ADAR2a. After in vitro editing, sequence analysis of cDNA derived from the RNA revealed editing at a novel site (F) in intron 5 (Fig. 2A). Because of its intronic location (Fig. 1), confirmation of editing at site F in human brain requires analysis of RNA spliced at GU3 (RNAGU3) or unspliced RNA (RNAUS). This was achieved by RT–PCR of RNA from several brain regions (prefrontal cortex, parahippocampus, thalamus and choroid plexus) using primers Xbs and FRev which amplify both RNA species (Fig. 1). An example of a sequence from parahippocampus is shown in Figure 2B.
Figure 2.
Detection of intronic editing site F. Sequencing traces of cDNA from (A) RNA from in vitro editing assay, (B) RNAGU3/US from human parahippocampus. The positions of the A and G peaks at site F are indicated by horizontal arrows.
Splicing in minigene RNA in cell culture
To investigate splicing of HTR2C RNA in cell culture, a minigene (Fig. 1) similar to that used for in vitro editing was transfected into several cell lines (COS-7, PC-12, SH-SY5Y, HEK-293). In every cell line examined, when RT–PCR was performed between Xbs and HiA (Fig. 1) only a single band due to splicing at GU1 (RNAGU1) was observed on agarose gel electrophoresis (e.g. in SH-SY5Y cells, Fig. 3A, lane 1), which was confirmed by sequencing. However, some splicing at GU2 did occur as a band due to RNAGU2, also confirmed by sequencing, was observed when GU2For (Fig. 1) was used as the 5′ RT–PCR primer to exclude amplification of RNAGU1 (data not shown). RNAGU3 was only detected when GU3For was used as the 5′ primer to exclude both RNAGU1 and RNAGU2. These experiments suggest that for this minigene there is a hierarchy of 5′ splice site preferences: GU1 > GU2 > GU3.
Figure 3.
Effect of mutations in HTR2C minigenes on RNA splicing. RT–PCR products from RNA in SH-SY5Y cells transfected with minigenes. Wt, wild-type; m1-2, Δ1-2, as indicated in Figure 1; letters alone, editing site with A > G mutation; subscript T, A > T; subscript C, A > C; asterisk, compensatory mutation. The positions of PCR products corresponding to RNAGU1, RNAGU2, RNAGU3 and RNAGU1/RNAGU2 hybrid are as indicated.
When SH-SY5Y cells were transfected with a construct containing a GT to CC substitution to prevent splicing at GU1 (Fig. 1, m1), RT–PCR between Xbs and HiA amplified a single product corresponding to RNAGU2 and confirmed by sequencing (Fig. 3A, lane 2). Prevention of splicing at both GU1 and GU2 by GT to CC mutations at both 5′ splice sites (Fig. 1, m1 and m2) resulted in RNAGU3 (Fig. 3A, lane 3). Transfections with these two mutant minigenes confirm that all the sequence information and splicing factors required for successful splicing at GU2 and GU3 are present and also confirm the hierarchy of 5′ splice site preferences.
A to G mutations to mimic editing
Endogenous cell line HTR2C RNA was edited in both SH-SY5Y and PC-12 cells, but editing of minigene RNA was undetectable in SH-SY5Y cells and infrequent in PC-12 cells (data not shown). To investigate the effects of RNA editing on splicing, minigenes carrying A to G base substitution mutations to mimic RNA editing were constructed. These mutant minigenes were transfected into SH-SY5Y cells to exclude any contribution from endogenous editing. As shown in Figure 3B, splicing at GU2 was clearly observed after single editing mutations at A, E and D, with lower amounts after mutations at B and F. Only the editing mutation at C failed to produce any detectable RNAGU2. Identical results were obtained in PC-12 cells (data not shown). To investigate the effects of editing mutations in combination, cells were transfected with minigenes containing each of the 15 possible pairwise combinations. Five combinations (AC, AD, AF, CF and DF), clearly showed synergy for stimulating splicing at GU2, although others, e.g. AB and CD, did not (Fig. 3C, lanes 2–5, and D, lanes 6, 9, 12). Of the higher combinations, we examined mainly those combinations that were frequent in brain (see below). Generally, splicing increasingly favoured GU2 over GU1 as the number of editing mutations increased (Fig. 3C, lanes 6–12). The relative abundance of RNAGU2 and RNAGU1 in many of these editing mutation combinations was estimated by fluorescent RT–PCR under limited amplification cycles (Materials and Methods) and presented in Table 1. We also found that inclusion of the mutation at F increased editing at GU2 when compared to the corresponding combination in its absence (e.g. Fig. 3C, lanes 5, 12–14).
Table 1. Comparison of RNA editing/splicing relationship between cell culture and brain.
| Major editing combinationsa | Cell culture | Brain | ||||
|---|---|---|---|---|---|---|
| RNAGU2/RNAGU1b | Preferred donor site | RNAGU2 (%) observedc | RNAGU1 (%) estimate 1d | RNAGU1 (%) estimate 2e | Preferred donor sitef | |
| None | 0.0 | GU1 | 10.2 | 60.3 | 56.4 (31.3) | GU1 |
| A | 0.2 | GU1 | 1.8 | 16.1 | 12.5 (6.9) | GU1 |
| C | 0.0 | GU1 | 3.6 | –3.7 | 0.0 (0.0) | GU2 |
| D | 0.4 | GU1 | 13.2 | –11.3 | 0.0 (0.0) | GU2 |
| AC | 1.5 | GU2 | 4.2 | 2.1 | 0.0 (0.0) | GU2 |
| AD | 2.3 | GU2 | 7.8 | 1.2 | 0.0 (0.0) | GU2 |
| CD | 0.1 | GU1 | 3.6 | 11.5 | 7.9 (4.4) | GU1/2 |
| ABD | >40.0 | GU2 | 6.6 | 6.1 | 2.6 (1.4) | GU2 |
| AEC | 4.3 | GU2 | 3.6 | –0.9 | 0.0 (0.0) | GU2 |
| ACD | 0.5 | GU1 | 3.0 | 23.6 | 19.9 (11.1) | GU1 |
| ABED | 5.2 | GU2 | 3.6 | –2.3 | 0.0 (0.0) | GU2 |
| ABCD | 2.1 | GU2 | 20.4 | –2.2 | 0.0 (0.0) | GU2 |
| AECD | 1.3 | GU2 | 3.0 | –1.3 | 0.0 (0.0) | GU2 |
| ABECD | 0.5 | GU1 | 3.6 | 0.4 | 0.0 (0.0) | GU2 |
aExcluding editing combinations with RNAGU2 and RNAGU1 < 3% in brain.
bMean of 2–4 determinations by fluorescent PCR at 21 cycles and calibrated by standard curve, intraclass correlation coefficient 0.99.
cFrom data in Figure 5A.
dEstimated from simple model assuming overall RNAGU2/ RNAGU1 of 1.8.
eEstimated as for d, followed by non-linear programming to remove negative values; numbers in brackets show values after dividing by 1.8 to allow direct comparison with RNAGU2.
fFrom comparison of RNAGU2 with RNAGU1 (estimate 2 divided by 1.8).
In some RT–PCR amplifications of RNA from transfected cell lines a minor product larger than RNAGU2 was observed which approximated to the size predicted for RNAGU3. It was not associated with any particular minigenes, nor was it observed in any of the fluorescent RT–PCR amplifications where limited cycles are used. Sequencing of this product from several different transfections showed that it contained RNAGU1 and RNAGU2 sequences but not RNAGU3. It appears to be a PCR artefact, as previously reported by Wang et al. (21), due to hybrids between RNAGU1 and RNAGU2 presumably formed during later amplification cycles when the primers may no longer be in excess. It should be noted that the only minigene where RT–PCR between exons 5 and 6 generated a confirmed product due to RNAGU3 was m1m2 (Fig. 3A).
How specific are the editing mutations?
The specificity of the editing mutations was investigated for sites E and D by comparing base substitutions by guanosine with substitutions by cytidine or thymidine (compare Fig. 3B, lanes 4, 8, 9 and 6, 10, 11). At both sites, only adenosine to guanosine substitutions affected splicing, with the other base changes having effects that are indistinguishable from wild type. This suggests that the presence of guanosine rather than the absence of adenosine is the more important factor in promoting splicing at GU2. As all three base changes destabilize the stem–loop (Fig. 1), changes in secondary structure are therefore unlikely to play a major role in the effects of editing at E or D on splicing.
Involvement of secondary structure
To further investigate secondary structure involvement, mutations were introduced to compensate for potential base-pair changes induced by editing mutations. Generally, editing mutations destabilize either stem 1 (B, E, C) or 2 (D, F) by replacing an AU base pair by a weak non-Watson–Crick GU base-pair (Fig. 1). However, editing at site A stabilizes stem 1 by replacing an unpaired AC by a GC base-pair. To reverse these predicted effects, compensatory mutations were introduced on the opposite strand (as indicated by asterisks, Fig. 1). For site D, the compensatory mutation removed the effect on splicing of the original mutation (compare Fig. 3B, lanes 6, 14). However, compensatory mutations had little effect at sites A and E (compare Fig. 3B, lanes 2, 12 and lanes 4, 13). Compensatory mutation at F was investigated in double mutants that exhibited strong synergy. For both AF and DF, the compensatory mutation at F restored the splicing pattern to that seen without editing at F (compare Fig. 3D, lanes 2, 6, 7 and lanes 4, 12, 13). In contrast, A and D compensatory mutations had little effect (compare lanes 6, 8 and lanes 12, 14). For CF the F compensatory mutation had a weaker effect than with AF or DF (compare lanes 3, 9, 10), while the C compensatory mutation had little effect (compare lanes 9, 11).
These experiments implicate the stability of stem 2 in the effect on splicing of editing at site F. The evidence for its involvement in editing at site D is weaker, but, because of its position (Fig. 1), editing at D is likely to have only a minor destabilizing effect on stem 2. In contrast, stem 1 does not appear to play a role in the effects of editing at sites A, E and C. To confirm the different roles of stems 1 and 2 in splice site selection, SH-SY5Y cells were transfected with minigenes where either stem was destroyed by a large deletion in the ECS region (Fig. 1, Δ1 and Δ2). The large deletion in stem 2 resulted in increased splicing at GU2, while the large deletion at stem 1 had no effect (Fig. 3E, lanes 1 and 2). Smaller deletions in stem 2 (Fig. 1, Δ2a–c) also increased splicing at GU2 (Fig. 3E, lanes 3 and 4).
Splicing in human brain RNA
We have demonstrated that mimicking editing in HTR2C RNA in cell culture clearly influences the relative amount of RNA splicing at GU1 and GU2, but with no evidence that it alters the minimal amount of splicing at GU3. We examined RNA splicing and editing in HTR2C RNA in human brain to look for evidence that a similar relationship occurs there. We selected two regions, thalamus and choroid plexus, as other studies have shown that these regions express readily detectable HTR2C RNA with alternative splicing at GU1 (21,28,29). Splicing of RNA from thalamus (T) and choroid plexus (CP) in five post-mortem brains (patient IDs: 135, 93, 393, 283 and 189) was investigated by RT–PCR. Primers in exons 5 and 6 (Xbs and HiA, Fig. 1) efficiently amplified two products due to RNAGU1 and RNAGU2 (Fig. 4A), confirmed by sequencing. The relative abundance of these two products was estimated in separate fluorescent RT–PCR experiments (Materials and Methods) and is given above each lane. While some differences between brain samples were apparent, there was no evidence that these were region-specific. As in cell culture, product due to RNAGU3 was not observed. Amplification of RNAUS by these primers is impossible due to the large intron, but could be achieved with difficulty in some samples using primer USRev with Xbs (Fig. 1), requiring two rounds of 40 cycles (Fig. 4B). The identity of the product of predicted size observed in four samples was confirmed by sequencing, while the smaller product amplified in all eight samples was shown to be due to contaminating mitochondrial RNA. In contrast, primer FRev (Fig. 1), which with Xbs is predicted to amplify both RNAUS and RNAGU3 (RNAGU3/US), gave an easily detectable and confirmed product with all samples after 30 cycles of PCR (Fig. 4C). Similar yields of RNAUS and RNAGU3/US were obtained using alternative 3′ primers to USRev and FRev (data not shown). It is therefore unlikely that the observed difficulty in amplifying RNAUS compared to RNAGU3/US is due to different primers used in RT–PCR. This suggests that RNAGU3 is much more abundant than RNAUS and that RNAGU3/US might approximate to RNAGU3, especially in the four samples which contained very little RNAUS. To control for theoretical DNA contamination, RT–PCR without reverse transcriptase was performed for RNAUS and RNAGU3/US. As expected, no product was observed at the appropriate size (Fig. 4B and C).
Editing in human brain RNA
RNA editing was investigated in RNAGU2, RNAUS and RNAGU3/US by sequencing of individual clones derived from RT–PCR products excised from agarose gels. Of the 10 original brain samples only seven could be analyzed for editing at RNAUS as no clones could be obtained from RNAUS for samples 93/CP, 283/T and 135/T. For RNAGU2 and RNAGU3/US, editing was examined in these seven samples plus 135/T, although this sample was excluded from comparisons of editing between RNAGU2 and RNAUS. Intronic site F is not retained in RNAGU2, but is extensively edited in RNAGU3/US as was seen in the hippocampus (Fig. 2B). However, F was rarely edited in RNAUS (data not shown).
The distribution of the 32 possible editing combinations (Fig. 5A), excluding editing at F, was examined in RNAGU2 (167 clones from seven samples; 185 clones from eight samples) and RNAUS (203 clones from seven samples). There was no evidence for region-specific differences, but there were differences between RNAGU2 and RNAUS. In a stratified analysis of each combination in the seven samples common to both, unedited, A and ACD were significantly more frequent in RNAUS, while D and ABCD were significantly more frequent in RNAGU2. For RNAGU3/US, two distinct patterns of editing were observed (Fig. 5B). In the four samples that contained very low amounts of RNAUS (102 clones, upper histogram), two combinations (A and D) were very frequent and accounted for 81% of the total. Editing frequencies in the other group of four samples (110 clones, lower histogram) show a pattern with features intermediate between that of the first group of RNAGU3/US and of RNAUS. This is consistent with the first group of RNAGU3/US approximating to RNAGU3 and the second group reflecting the presence of both RNAGU3 and RNAUS molecules. When editing at F was included in the analysis in the upper histogram, most of the two frequent combinations were found to be also edited at F (92% AF and 100% DF). Only 20% of the other frequent combination (unedited) were edited at F, suggesting that this small group may mainly include the very few RNAUS molecules present in RNAGU3/US in these four samples.
Figure 5.
Editing combination frequencies in different splice variants in human brain. (A) RNAGU2 from eight brain samples (185 clones) and RNAUS from seven brain samples (203 clones). In comparison, with 135/T excluded from RNAGU2 (167 clones), sites significantly more (asterisk) or less (hash) frequently edited are as indicated. Significance by stratified chi-square test: unedited (P = 0.00001), A (P = 0.05), D (P = 0.005), ACD (P = 0.01) and ABCD (P = 0.05). (B) RNAGU3/US from four brain samples (102 clones) with very little RNAUS and approximating to RNAGU3 (upper histogram) and from four brain samples (110 clones) with more RNAUS (lower histogram).
Editing mutation at site F inhibits editing at site D
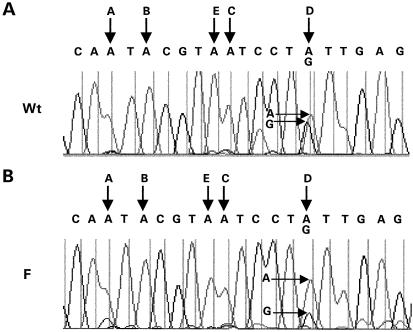
The presence of mainly AF and DF combinations in RNAGU3 in the brain is surprising, especially as after splicing at GU3 (unlike after splicing at GU1 and GU2) the stem–loop is maintained and editing can continue. This suggests that editing at F might inhibit editing at exonic sites and was investigated in RNAGU3/US in PC-12 cells after co-transfection with ADAR2a, where endogenous editing of minigene RNA at site D is frequent. For the wild-type minigene (Fig. 6), the adenosine and guanosine peak heights at site D were similar (G/A = 0.8), whereas for the minigene mutated at F the guanosine peak height was much lower (G/A = 0.3). A similar inhibitory effect on D of editing mutation at F was obtained when minigenes mutated at A and AF were compared (G/A = 0.8 and 0.5 respectively, data not shown). Similar results were obtained in SH-SY5Y and COS-7 cells, also co-transfected with ADAR2a. The inhibition of editing at D by editing at site F is probably due to its destabilization of stem 2, and may also result in inhibition of editing at other exonic sites.
Figure 6.
Inhibition of editing at site D by editing at site F. Sequencing traces of cDNA from RNAGU3/US in PC-12 cells transfected with (A) Wt and (B) F minigenes. The positions of the A and G peaks at site D are indicated by horizontal arrows.
Model to predict splicing fates of frequent RNA editing combinations in human brain
The important issue is whether the observed effects of editing mutations on RNA splicing of minigenes are relevant to the human brain. In order to address this it is necessary to be able to compare the editing combinations in the subsets of RNA molecules subjected to each splicing fate. It is likely that RNAUS represents a very small subset that has been captured in transition before splicing, mainly at GU2 or GU1. The distribution of editing combinations observed in RNAUS may therefore reflect the aggregate of the distributions in RNAGU2 and RNAGU1. This simple model enables a distribution of exonic editing combinations for RNAGU1 to be estimated, provided that the overall ratio of RNAGU2 to RNAGU1 is known. The RNAGU2/RNAGU1 ratios range from 1.0 to 3.6 (Fig. 3A) and, using the mean ratio of 1.8, the frequency distribution predicted by the model for RNAGU1 was calculated and shown for the major editing combinations (Table 1, estimate 1). However, several had negative values. This is not surprising given the underlying complexity, and probably reflects inaccuracies in several assumptions implicit in the simple model. For example, not all combinations are likely to be spliced at the same rate, leading to under-representation in RNAUS if rapidly spliced and over-representation if slowly spliced. To obtain a better estimate for the RNAGU1 editing frequency distribution, we used non-linear programming to apply a minimum adjustment to the RNAUS distribution that is just sufficient to remove the negative values (Table 1, estimate 2). The resulting predicted RNAGU1 frequencies were compared with the observed RNAGU2 frequencies, after taking the overall RNAGU2/RNAGU1 ratio (1.8) into account. Three editing combinations were predicted to be more frequent in RNAGU1 (unedited, A and ACD), one to have a similar frequency (CD) and 10 to be more frequent in RNAGU2 (C, D, AC, AD, ABD, AEC, ABED, ABCD, AECD, ABECD). Most of these 14 predictions are supported by the results in cell culture (Table 1), with four exceptions (C, D, CD, ABECD). Similar results were obtained assuming the extremes of the range (1.0 and 3.6) of RNAGU2 to RNAGU1 ratios observed in brain (data not shown).
DISCUSSION
The central purpose of the investigation described here was to examine the hypothesis that RNA editing in HTR2C RNA modulates selection of alternative 5′ splice sites. Since RNAGU1 lacks all editing sites and RNAGU2 lacks the novel intronic site F, no editing can be measured in RNA after splicing at GU1 and only editing at exonic sites can be measured after splicing at GU2. For these reasons, the approach of transfecting SH-SY5Y cells (where RNA editing is not observed) with mutant minigenes that contain A to G base substitutions to mimic editing was adopted. These experiments showed that many editing mutations, alone and in combination, shift the balance from splicing at GU1 to GU2, but splicing at GU3 was not observed and is probably rare. In human brain, RNA editing in two regions (thalamus and choroid plexus) was studied, but no region-specific differences were observed. The distribution of editing combination frequencies was similar between RNAGU2 and the very rare RNAUS, excluding editing at site F, which cannot be determined in RNAGU2 and was very infrequent in RNAUS. The distribution in the uncommon RNAGU3 appears to be completely different, with very frequent editing at F, and almost entirely consisting of two combinations, AF and DF.
In a model based on the assumption that RNAUS represents molecules awaiting splicing, we estimated the distribution in RNAGU1, excluding editing at site F. Comparison of this distribution with that observed in RNAGU2 showed that most of the editing combinations exhibited the same preference for splicing as was seen with editing mutations in minigenes expressed in cell culture. We need now to consider editing at site F. Since editing at F is infrequent in RNAUS, it follows from the model that F should also be infrequently edited in RNA destined for the two major alternatives: splicing at GU1 and GU2. However, the low frequency of editing at F in RNAUS is not inconsistent with its observed high frequency in RNAGU3 as only a very small fraction of RNAUS appears to be spliced at GU3. This suggests that in brain, editing at F is almost exclusively associated with RNAGU3; yet in cell culture, editing at F is associated with RNAGU2 and RNAGU1, while RNAGU3 appears to be rare.
How might this apparent contradiction be reconciled? Our data have shown that editing at F destabilizes the stem–loop, thereby inhibiting editing at site D and possibly at all exonic sites. Consequently, F is likely to be the last site edited and therefore the most vulnerable to underestimation in RNAUS. Furthermore, by destabilizing the stem–loop, editing at F might increase access of GU2 to U1snRNP, other splicesome components and trans-acting factors. This would cause editing combinations involving F to be more rapidly spliced and would also result in their underestimation. Without F, the model predicts the same preferred splice site for 10 of the frequent combinations in the brain as observed in cell culture, but there are four discrepancies (C, D, CD and ABECD). However, if editing at F is allowed, these four predictions need no longer be inconsistent with cell culture results, as CF, DF, ABECDF favour splicing at GU2 and CDF is spliced at both sites with similar efficiency (Fig. 3C and D; Table 1). Therefore, all the brain and cell culture data can be reconciled if the plausible assumption is made that editing at F is underestimated in RNAUS.
It is not clear how RNA editing might influence the preference for splicing at GU2. Secondary structure may play a role, as many studies show that splicing signals are not recognized when sequestered in a stable hairpin loop (31–34). The predicted stem–loop (Fig. 1) might therefore impede access of U1snRNP and other splicesome components, which is supported by the significant increase in splicing at GU2 observed when stem 2 is destabilized by deletions. However, although the strong secondary structure may inhibit splicing at GU2, we found clear evidence only at intronic site F for its involvement in modulation of splicing by editing. Editing at the exonic sites may influence splicing by altering binding site(s) for trans-acting factors, as has been observed for single base mutations that interfere with splicing (35,36). Intriguingly, HB11-52 sno-RNA has an 18-nucleotide sequence complementary to the exonic region of the stem (37) and binding of this molecule might affect splicing or editing. Many sno-RNA molecules O-methylate a specific target adenosine in rRNA and the position of the equivalent adenosine in HTR2C mRNA is at RNA editing site C. O-methylation of this adenosine inhibits RNA editing at this site (38), but its biological significance is unclear.
It is likely that RNA editing and alternative splicing evolved together in mammalian HTR2C. We have shown that the strongest predictor of splicing at GU1 is unedited mRNA. This encodes the INI isoform, which exhibits constitutive activity (19,39). Consequently, when editing is inefficient, increased splicing at GU1 (probably resulting in an inactive receptor) may act as a control mechanism to decrease biosynthesis of the INI isoform and thereby limit constitutive activity. Interestingly, a recent study found that the cell surface expression of the INI isoform was reduced compared to the fully edited VGV isoform, also limiting constitutive activity (40).
The presence of RNA editing sites close to exon/intron boundaries has previously provoked speculation that editing and splicing might be linked. Supporting this view, GluR-B mRNA molecules in ADAR2 null mice exhibit very little Q/R editing while more RNA molecules are incompletely spliced (41). In malignant gliomas where ADAR2 activity is reduced, both editing and splicing of HTR2C RNA are altered (42). In another tumour, acute myeloid leukaemia, PTPN6 mRNA was hyperedited and aberrantly spliced, possibly due to an edited splice branch site (43). A recent study of R/G editing and splicing in GluR-B showed that the two processes are co-ordinated in vivo despite being mutually inhibitory in vitro (44), although effects on alternative splicing were not addressed. In the only previous study to provide direct evidence for an effect of RNA editing on alternative splicing, ADAR2 edited its own mRNA, creating an alternative 3′ splice site (45). Our results provide further clear evidence for the ability of RNA editing to modulate alternative splicing, and the first in a gene not directly involved in editing. This and the involvement of intronic editing raise the intriguing possibility that this process is far more widespread than previously recognized. Intronic editing is much harder to detect than exonic editing, which has largely relied on serendipity. As most mammalian genes have introns, some very large, there is great potential for many more RNA editing sites to be discovered. Some may have a role in splicing, but others may have different roles in gene expression. Understanding the relationships between RNA editing and other post-transcriptional events is likely to be of great importance.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Professor Kazuko Nishikura for the kind gift of ADAR2a mammalian expression plasmid. We also gratefully acknowledge Dr Nadeem Khan at the London Neurodegenerative Diseases Brain Bank at the Institute of Psychiatry for providing post-mortem human brain samples. Finally we would like to thank Dr Ursula D’Souza for useful comments. This work was funded by the Medical Research Council.
REFERENCES
- 1.Lander E.S., Linton,L.M., Birren,B., Nusbaum,C., Zody,M.C., Baldwin,J., Devon,K., Dewar,K., Doyle,M., FitzHugh,W. et al. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 2.Venter J.C., Adams,M.D., Myers,E.W., Li,P.W., Mural,R.J., Sutton,G.G., Smith,H.O., Yandell,M., Evans,C.A., Holt,R.A. et al. (2001) The sequence of the human genome. Science, 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- 3.Kim U., Wang,Y., Sanford,T., Zeng,Y. and Nishikura,K. (1994) Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proc. Natl Acad. Sci. USA, 91, 11457–11461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Connell M.A., Krause,S., Higuchi,M., Hsuan,J.J., Totty,N.F., Jenny,A. and Keller,W. (1995) Cloning of cDNAs encoding mammalian double-stranded RNA-specific adenosine deaminase. Mol. Cell. Biol., 15, 1389–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dabiri G.A., Lai,F., Drakas,R.A. and Nishikura,K. (1996) Editing of the GluR-B ion channel RNA in vitro by recombinant double-stranded RNA adenosine deaminase. EMBO J., 15, 34–45. [PMC free article] [PubMed] [Google Scholar]
- 6.Melcher T., Maas,S., Herb,A., Sprengel,R., Higuchi,M. and Seeburg,P.H. (1996) RED2, a brain-specific member of the RNA-specific adenosine deaminase family. J. Biol. Chem., 271, 31795–31798. [DOI] [PubMed] [Google Scholar]
- 7.Melcher T., Maas,S., Herb,A., Sprengel,R., Seeburg,P.H. and Higuchi,M. (1996) A mammalian RNA editing enzyme. Nature, 379, 460–464. [DOI] [PubMed] [Google Scholar]
- 8.Bass B.L., Nishikura,K., Keller,W., Seeburg,P.H., Emeson,R.B., O’Connell,M.A., Samuel,C.E. and Herbert,A. (1997) A standardized nomenclature for adenosine deaminases that act on RNA. RNA, 3, 947–949. [PMC free article] [PubMed] [Google Scholar]
- 9.Lai F., Chen,C.X., Carter,K.C. and Nishikura,K. (1997) Editing of glutamate receptor B subunit ion channel RNAs by four alternatively spliced DRADA2 double-stranded RNA adenosine deaminases. Mol. Cell. Biol., 17, 2413–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Higuchi M., Single,F.N., Kohler,M., Sommer,B., Sprengel,R. and Seeburg,P.H. (1993) RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell, 75, 1361–1370. [DOI] [PubMed] [Google Scholar]
- 11.Egebjerg J., Kukekov,V. and Heinemann,S.F. (1994) Intron sequence directs RNA editing of the glutamate receptor subunit GluR2 coding sequence. Proc. Natl Acad. Sci. USA, 91, 10270–10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lomeli H., Mosbacher,J., Melcher,T., Hoger,T., Geiger,J.R., Kuner,T., Monyer,H., Higuchi,M., Bach,A. and Seeburg,P.H. (1994) Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science, 266, 1709–1713. [DOI] [PubMed] [Google Scholar]
- 13.Burns C.M., Chu,H., Rueter,S.M., Hutchinson,L.K., Canton,H., Sanders-Bush,E. and Emeson,R.B. (1997) Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature, 387, 303–308. [DOI] [PubMed] [Google Scholar]
- 14.Sommer B., Kohler,M., Sprengel,R. and Seeburg,P.H. (1991) RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell, 67, 11–19. [DOI] [PubMed] [Google Scholar]
- 15.Bass B.L. (2002) RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem., 71, 817–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paul M.S. and Bass,B.L. (1998) Inosine exists in mRNA at tissue-specific levels and is most abundant in brain mRNA. EMBO J., 17, 1120–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morse D.P. and Bass,B.L. (1999) Long RNA hairpins that contain inosine are present in Caenorhabditis elegans poly(A)+ RNA. Proc. Natl Acad. Sci. USA, 96, 6048–6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morse D.P., Aruscavage,P.J. and Bass,B.L. (2002) RNA hairpins in noncoding regions of human brain and Caenorhabditis elegans mRNA are edited by adenosine deaminases that act on RNA. Proc. Natl Acad. Sci. USA, 99, 7906–7911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niswender C.M., Copeland,S.C., Herrick-Davis,K., Emeson,R.B. and Sanders-Bush,E. (1999) RNA editing of the human serotonin 5-hydroxytryptamine 2C receptor silences constitutive activity. J. Biol. Chem., 274, 9472–9478. [DOI] [PubMed] [Google Scholar]
- 20.Fitzgerald L.W., Iyer,G., Conklin,D.S., Krause,C.M., Marshall,A., Patterson,J.P., Tran,D.P., Jonak,G.J. and Hartig,P.R. (1999) Messenger RNA editing of the human serotonin 5-HT2C receptor. Neuropsychopharmacology, 21, 82S–90S. [DOI] [PubMed] [Google Scholar]
- 21.Wang Q., O’Brien,P.J., Chen,C.X., Cho,D.S., Murray,J.M. and Nishikura,K. (2000) Altered G protein-coupling functions of RNA editing isoform and splicing variant serotonin-2c receptors. J. Neurochem., 74, 1290–1300. [DOI] [PubMed] [Google Scholar]
- 22.Herrick-Davis K., Egan,C. and Teitler,M. (1997) Activating mutations of the serotonin 5-HT2C receptor. J. Neurochem., 69, 1138–1144. [DOI] [PubMed] [Google Scholar]
- 23.Sodhi M.S., Burnet,P.W.J., Makoff,A.J., Kerwin,R.W. and Harrison,P.J. (2001) RNA editing of the 5-HT2c receptor is reduced in schizophrenia. Mol. Psychiatry, 6, 373–379. [DOI] [PubMed] [Google Scholar]
- 24.Niswender C.M., Herrick-Davis,K., Dilley,G.E., Meltzer,H.Y., Overholser,J.C., Stockmeier,C.A., Emeson,R.B. and Sanders-Bush,E. (2001) RNA editing of the human serotonin 5-HT2C receptor. alterations in suicide and implications for serotonergic pharmacotherapy. Neuropsychopharmacology, 24, 478–491. [DOI] [PubMed] [Google Scholar]
- 25.Gurevich I., Tamir,H., Arango,V., Dwork,A.J., Mann,J.J. and Schmauss,C. (2002) Altered editing of serotonin 2C receptor pre-mRNA in the prefrontal cortex of depressed suicide victims. Neuron, 34, 349–356. [DOI] [PubMed] [Google Scholar]
- 26.Iwamoto K. and Kato,T. (2003) RNA editing of serotonin 2C receptor in human postmortem brains of major mental disorders. Neurosci. Lett., 346, 169–172. [DOI] [PubMed] [Google Scholar]
- 27.Schmauss C. (2003) Serotonin 2C receptors: suicide, serotonin and runaway RNA editing. Neuroscientist, 9, 237–242. [DOI] [PubMed] [Google Scholar]
- 28.Xie E., Zhu,L., Zhao,L. and Chang,L.S. (1996) The human serotonin 5-HT2C receptor: complete cDNA, genomic structure and alternatively spliced variant. Genomics, 35, 551–561. [DOI] [PubMed] [Google Scholar]
- 29.Canton H., Emeson,R.B., Barker,E.L., Backstrom,J.R., Lu,J.T., Chang,M.S. and Sanders-Bush,E. (1996) Identification, molecular cloning and distribution of a short variant of the 5-hydroxytryptamine2C receptor produced by alternative splicing. Mol. Pharmacol., 50, 799–807. [PubMed] [Google Scholar]
- 30.Liu Y., George,C.X., Patterson,J.B. and Samuel,C.E. (1997) Functionally distinct double-stranded RNA-binding domains associated with alternative splice site variants of the interferon-inducible double-stranded RNA-specific adenosine deaminase. J. Biol. Chem., 272, 4419–4428. [DOI] [PubMed] [Google Scholar]
- 31.Solnick D. and Lee,S.L. (1987) Amount of RNA secondary structure required to induce an alternative splice. Mol. Cell. Biol., 7, 3194–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blanchette M. and Chabot,B. (1997) A highly stable duplex structure sequesters the 5′ splice site region of hnRNP A1 alternative exon 7B. RNA, 3, 405–419. [PMC free article] [PubMed] [Google Scholar]
- 33.Muro A.F., Caputi,M., Pariyarath,R., Pagani,F., Buratti,E. and Baralle,F.E. (1999) Regulation of fibronectin EDA exon alternative splicing: possible role of RNA secondary structure for enhancer display. Mol. Cell. Biol., 19, 2657–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Varani L., Hasegawa,M., Spillantini,M.G., Smith,M.J., Murrell,J.R., Ghetti,B., Klug,A., Goedert,M. and Varani,G. (1999) Structure of tau exon 10 splicing regulatory element RNA and destabilisation by mutations of frontotemporal dementia and parkinsonism linked to chromosome 17. Proc. Natl Acad. Sci. USA, 96, 8229–8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cartegni L., Chew,S.L., and Krainer,A.R. (2002) Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet., 3, 285–298. [DOI] [PubMed] [Google Scholar]
- 36.Caceres J.F. and Kornblihtt,A.R. (2002) Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet., 18, 186–193. [DOI] [PubMed] [Google Scholar]
- 37.Cavaille J., Buiting,K., Kiefmann,M., Lalande,M., Brannan,C.I., Horsthemke,B., Bachellerie,J.P., Brosius,J. and Huttenhofer,A. (2000) Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organisation. Proc. Natl Acad. Sci. USA, 97, 14311–14316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yi-Brunozzi H.Y., Easterwood,L.M., Kamilar,G.M. and Beal,P.A. (1999) Synthetic substrate analogues for the RNA-editing adenosine deaminase ADAR-2. Nucleic Acids Res., 27, 2912–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herrick-Davies K., Grinde,E. and Niswender,C.M. (1999) Serotonin 5-HT2C receptor RNA editing alters receptor basal activity: implications for serotonergic signal transduction. J. Neurochem., 73, 1711–1717. [DOI] [PubMed] [Google Scholar]
- 40.Marion S., Weiner,D.M. and Caron,M.G. (2004) RNA editing induces variation in desensitization and trafficking of 5-hydroxytryptamine 2c receptor isoforms. J. Biol. Chem., 279, 2945–2954. [DOI] [PubMed] [Google Scholar]
- 41.Higuchi M., Maas,S., Single,F.N., Hartner,J., Rozov,A., Burnashev,N., Feldmeyer,D., Sprengel,R. and Seeburg,P.H. (2000) Point mutation in an AmpA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature, 406, 78–81. [DOI] [PubMed] [Google Scholar]
- 42.Maas S., Patt,S., Schrey,M. and Rich,A. (2001) Underediting of glutamate receptor GluR-B mRNA in malignant gliomas. Proc. Natl Acad. Sci. USA, 98, 14687–14692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beghini A., Ripamonti,C.B., Peterlongo,P., Roversi,G., Cairoli,R., Morra,E. and Larizza,L. (2000) RNA hyperediting and alternative splicing of hematopoiteic cell phosphatase (PTPN6) gene in acute myeloid leukemia. Hum. Mol. Genet., 9, 2297–2304. [DOI] [PubMed] [Google Scholar]
- 44.Bratt E. and Öhman,M. (2003) Coordination of editing and splicing of glutamate receptor pre-mRNA. RNA, 9, 309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rueter S.M., Dawson,T.R. and Emeson,R.B. (1999) Regulation of alternative splicing by RNA editing. Nature, 399, 75–80. [DOI] [PubMed] [Google Scholar]
- 46.Matzura O. and Wennborg,A. (1996) RNAdraw: an integrated program for RNA secondary structure calculation and analysis under 32-bit Microsoft Windows. Comput. Appl. Biosci., 12, 247–249. [DOI] [PubMed] [Google Scholar]