Abstract
Telomerase is a reverse transcriptase that uses an integral RNA molecule to add de novo G-rich repeats onto telomeric DNA, or onto nontelomeric DNA generated during chromosome fragmentation and breakage events. A telomerase-mediated DNA substrate cleavage activity has been reported in ciliates and yeasts. Nucleolytic cleavage may serve a proofreading function, enhance processivity or ensure that nontemplate telomerase RNA sequences are not copied into DNA. We identified and characterized a human telomerase-mediated nucleolytic cleavage activity using enzyme reconstituted in a rabbit reticulocyte lysate in vitro transcription/translation system and native enzyme extracted from cells. We found that telomerase catalyzed the removal of nucleotides from DNA substrates including those that can form a mismatch with the RNA template or that contain nontelomeric sequences located 3′ to a telomeric sequence. Unlike Tetrahymena telomerase, human telomerase catalyzed the removal of more than one nucleotide (up to 13) from telomeric primers. DNA substrates predicted to align at the 3′-end of the RNA template were not cleaved, consistent with cleavage being dictated by the template 5′-end. We also found some differences in the nuclease activity between RRL-reconstituted human telomerase and native enzyme.
INTRODUCTION
Telomeres form a nucleoprotein complex at the ends of linear eukaryotic chromosomes that serves as a protective cap conferring chromosome stability (1). Telomeric DNA is composed of G-rich repeated sequences (TTAGGG in vertebrates) and is a substrate for the ribonucleoprotein (RNP) enzyme telomerase. Telomerase mediates de novo addition of telomeric TTAGGG DNA repeats onto the 3′-ends of chromosomes allowing the maintenance of telomere length and the long-term proliferation of eukaryotic cells (2,3). In addition to elongating pre-existing telomere tracts, telomerase can also catalyze the synthesis of telomeric DNA onto nontelomeric DNA sequences generated by chromosome fragmentation and breakage events (4–7).
The human telomerase enzyme is minimally composed of a protein catalytic subunit, the telomerase reverse transcriptase (hTERT), and the telomerase RNA (hTR) that is used as a template for telomeric DNA synthesis (8). The expression of hTERT and hTR in a rabbit reticulocyte lysate (RRL) in vitro transcription/translation system is sufficient to reconstitute enzyme activity (9,10). hTR contains a short template sequence of 11 nucleotides complementary to the telomeric repeats (8,11). Telomerase elongation and substrate specificity have been characterized in vitro for a wide range of organisms including protozoa, yeast and human (4,12–16). Short single-stranded oligonucleotides containing bases complementary to the template region can serve as telomerase substrates in a DNA primer extension (conventional) assay (14,17). During telomere repeat synthesis, the 3′-end of the DNA substrate anneals to the telomerase RNA template. Nucleotides are added to the DNA until the 5′ terminus of the template sequence is reached; subsequently, the newly synthesized 3′-end of the substrate is repositioned at the beginning (3′ terminus) of the template (18). Repeat addition processivity, defined as successive rounds of nucleotide addition and primer translocation, is a key feature of most telomerase enzymes. The 5′-end of the substrate is also thought to bind an anchor site contributing to telomerase processivity (4,12,13,16, 19–21).
Like a number of DNA and RNA polymerases, telomerase exhibits a DNA substrate cleavage activity in addition to its polymerase function (22,23). In Euplotes crassus, Tetrahymena thermophila, Schizosaccharomyces pombe and Saccharomyces cerevisiae, telomerase cleaves DNA substrates bound via the template sequence of the telomerase RNA (20,24–27) and the cleavage can occur by an endonucleolytic mechanism (25,27). There is some evidence that this cleavage activity is inherent to telomerase. Nuclease activity remains associated with the telomerase complex after extensive purification (28). Cleavage patterns are also altered in specific ways when the telomerase enzyme contains a telomerase RNA mutated in the template sequence (29). RNA sequences outside the template can also influence telomerase cleavage activity (30). Moreover, expression of Tetrahymena TERT and TR in RRL is sufficient to reconstitute cleavage activity (31). Several primers have been tested and reported to be substrates for cleavage. Primers containing bases that can form mismatches with the RNA template sequence are cleaved; cleavage occurs preferentially at the junction of match-mismatch between the primer and the template (20,25,27,28). In E.crassus, telomerase can also remove 3′-terminal nontelomeric DNA from a primer to expose telomeric sequences for elongation (25,28).
The nucleolytic cleavage activity of vertebrate telomerase has not been characterized. In this study, we report the characterization of a human telomerase-mediated nucleolytic cleavage activity using enzyme reconstituted in RRL and native enzyme extracted from cells. We demonstrated that telomerase catalyzes the removal of 1 to 13 nucleotides from telomeric substrates that can align at, or beyond, the 5′-end of the RNA template sequence, that can form a mismatch with the RNA template, or that contain nontelomeric sequences located 3′ to a telomeric sequence. DNA substrates predicted to align at the 3′-end of the RNA template are not cleaved, consistent with cleavage being dictated by the template 5′-end. We also found some differences in the nuclease activity between RRL-reconstituted human telomerase and native enzyme partially purified from 293 cells.
MATERIALS AND METHODS
Oligonucleotides
Gel-purified 5′-biotinylated oligonucleotides were obtained from Operon (Alameda, CA) and resuspended in water. To generate the radiolabeled size markers carrying a 3′-terminal dideoxynucleotide, 5′-biotinylated oligonucleotides were 3′-end labeled using terminal deoxynucleotidyltransferase (Invitrogen) and [α-32P]dATP (cordycepin: NEN Life Science Products) for 30 min at 37°C, followed by DNA precipitation. The modification ‘revdT’ is a non-nucleosidic group linked at the 3′ terminus of the oligonucleotide that blocks any polymerase extension (Operon).
Plasmid constructions
The construction of the pET28b-hTERT and phTR+1 expression plasmids has been described previously (32,33).
In vitro transcription/translation
The RRL in vitro T7-coupled transcription/translation system (Promega) was used as described previously (17,34). Full length hTERT was synthesized in RRL in the presence of purified hTR. hTR was synthesized and purified on RNeasy spin columns (Qiagen) from FspI-linearized phTR+1 plasmid (34).
DNA primer extension assay
A non-PCR-based telomerase elongation assay was performed (14,17). hTERT protein expressed in 20 µl RRL in the presence of hTR was assayed for telomerase activity in a 40 µl final volume reaction using a gel-purified 5′-biotinylated oligonucleotide (Operon). Standard reaction conditions have been described previously (17). The proteinase K solution was 10 mM Tris–HCl pH 7.5, 0.5% SDS, 0.3 mg/ml proteinase K. The elongation products immobilized on magnetic beads were washed twice with buffer A (10 mM Tris–HCl pH 7.5, 1 M NaCl, 0.5 mM EDTA), once with buffer B (10 mM Tris–HCl pH 7.5) and analyzed by 8 or 10% polyacrylamide- urea gel electrophoresis. The relative amount of cleavage-derived products, expressed by the ratio of cleavage products/elongation products, was determined by densitometric analysis of the autoradiographs (ImageQuant software, Molecular Dynamics). The total counts for each signal were normalized to the amount of transcripts by dividing each product signal by the number of dGTPs it contains. The ratio was calculated using the normalized signals of the major elongation products above the input primer and the major cleavage products below the input primer.
Human telomerase extracts
The preparation of partially purified 293 cell extracts has been described previously (32). DNA primer extension assays were performed with 20 µg of this cellular extract as described above.
RESULTS
3′-end cleavage of telomeric DNA substrates
Nucleolytic cleavage of DNA primer substrates can be reconstituted by expressing Tetrahymena TERT in the presence of TR in a RRL in vitro transcription/translation system, and detected using a DNA primer extension (conventional) assay (31). This result suggests that the DNA cleavage may be dependent on the minimal components required for telomerase activity, TERT and TR. To identify and characterize a human telomerase-mediated nucleolytic cleavage activity, we first reconstituted human telomerase in RRL by expressing hTERT in the presence of hTR, and performed a DNA primer extension assay using different 5′-biotinylated telomeric oligonucleotides (Fig. 1C). As previously reported, RRL-reconstituted human telomerase assayed by the primer extension method generates a ladder of elongation products with a six nucleotide pausing pattern (Fig. 1C, lanes 1, 2) (17,35). Processive elongation of DNA primers by the telomerase enzyme is observed as the addition of multiple telomeric repeats (Fig. 1C, lanes 1, 2).
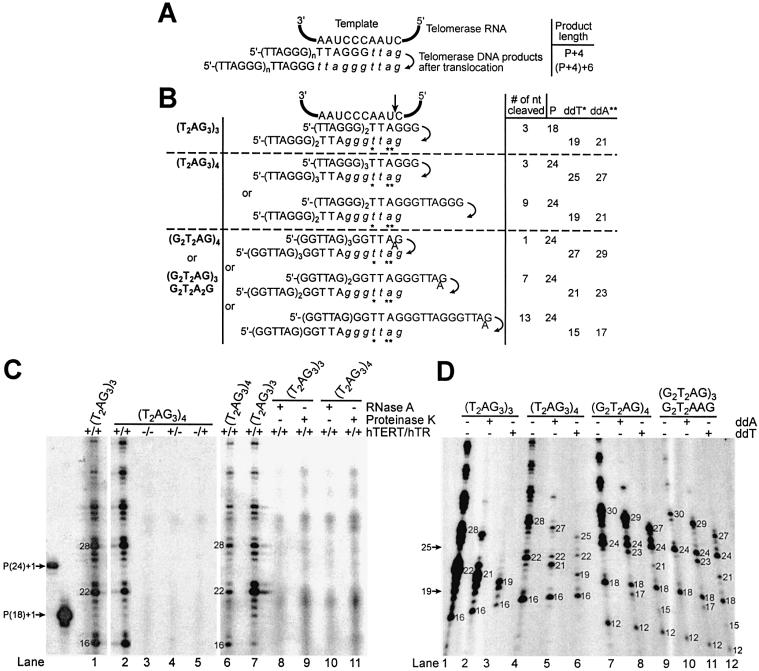
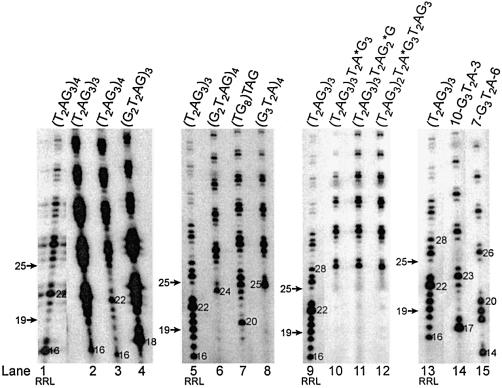
Figure 1.
Cleavage of telomeric 3′-ends. (A and B) Potential alignments of various telomeric oligonucleotides (tested in C and D) with the hTR template, before and after translocation. Compared to (G2T2AG)4, (G2T2AG)3G2T2A2G has an extra A predicted to form a mismatch with the RNA template. Italicized letters represent nucleotides added by the telomerase enzyme onto the 3′-end of the primer during the elongation step. The diagram implies that cleavage and nucleotide addition to the RNA template 5′-end precede translocation. P: primer length. ddA and ddT: dideoxyATP and dideoxyTTP. Numbers in A–D represent the lengths of the primers, elongation or chain termination products. The proposed cleavage site is shown by the vertical arrow. Asterisks indicate the positions of the first ddA or ddT incorporated during the DNA primer extension assay. (C and D) DNA primer extension assays were performed with the indicated oligonucleotides using RRL-reconstituted telomerase enzyme. The elongation products were resolved on 8% polyacrylamide-urea gels and subjected to autoradiography. The 3′-end radiolabeled 5′-biotinylated primers, (T2AG3)3 and (T2AG3)4, migrate, respectively, as 19 and 25mers [P(18)+1 and P(24)+1, respectively].
The Tetrahymena and Euplotes telomerase enzymes generate products that are shorter than the original oligonucleotide substrates, detectable as radiolabeled products that migrate below the input primers (20,25). We first assayed telomeric oligonucleotide substrates [(T2AG3)3 and (T2AG3)4], and two others primers [(G2T2AG)4 and (G2T2AG)3G2T2A2G] that mimic oligonucleotides cleaved by Tetrahymena telomerase (Fig. 1D). (G2T2AG)3G2T2A2G is predicted to form a mismatch with the RNA template (Fig. 1B). Under standard reaction conditions, we observed radiolabeled primer-sized products or products below the input primers (Fig. 1C, lanes 1, 2, 6, 7 and Fig. 1D, lanes 1, 4, 7, 10). This product profile results from the removal of one or several terminal primer nucleotides followed by incorporation of [α-32P]dGTP during elongation by the telomerase enzyme. These short products were not visualized when 5′-end radiolabeled (T2AG3)3 or 3′-end radiolabeled 5′-biotinylated (T2AG3)3 primers were added to RRL reactions in the absence of human telomerase components, even in the presence of dNTPs (data not shown) (17). These radiolabeled primers were neither cleaved nor elongated. These observations suggest that short products are not generated by random nucleases that might be present in the lysate, and are consistent with cleavage being a telomerase-mediated process. Both telomerase elongation activity and the generation of products shorter than the input primer were hTERT and hTR dependent (Fig. 1C, lanes 3–5), and RNase A and proteinase K sensitive (Fig. 1C, lanes 8–11).
The first major product generated by telomerase-mediated elongation of (T2AG3)4 is expected to be 28 nucleotides (nt) in length [primer(P)+4] (Fig. 1A). This product results from enzyme pausing or dissociation upon reaching the 5′-end of the RNA template (Fig. 1A and D, lane 4). Interestingly, telomerase reactions with this primer also led to the formation of cleavage-derived products 16 and 22 nt in length due to the removal of 9 and 3 nt, respectively (Fig. 1B and D, lane 4). The length of shorter products suggested that cleavage occurred 3′ to the A residue within the primer (Fig. 1B). To characterize the short products and identify the position of the cleavage within (T2AG3)4, DNA primer extension reactions were performed in the presence of the chain terminators ddATP or ddTTP. Substitutions of dATP with ddATP or TTP with ddTTP in the conventional assays generated specific chain termination products of different sizes (Fig. 1B and D, lanes 5, 6). In the absence of cleavage, chain termination products would be undetectable since the incorporation of ddATP or ddTTP prevents the incorporation of [α-32P]dGTP (Fig. 1A). Therefore the chain termination products we detected were generated by cleavage followed by subsequent addition of radiolabeled dGTP and ddATP or ddTTP onto the primer (Fig. 1B). These reactions also generated cleavage-derived products 16 and 22 nt in length resulting only from the incorporation of [α-32P]dGTP (Fig. 1D, lanes 5, 6). The results of this experiment indicated that 3 or 9 nt could be cleaved from the 3′-end of (T2AG3)4 depending on the potential alignments of this oligonucleotide with the RNA template, including alignment of the primer beyond the RNA template 5′-end (Fig. 1B).
Primer extension reactions were also performed with (T2AG3)3, (G2T2AG)4 or (G2T2AG)3G2T2A2G (Fig. 1D, lanes 1–3, 7–12). Reactions conducted with (T2AG3)3 generated cleavage-derived products 16 nt in length consistent with the removal of 3 nt from the 3′-end of the primer (Fig. 1B and D, lane 1). Substitutions of dATP with ddATP or TTP with ddTTP generated chain termination products of 21 or 19 nt, respectively, consistent with the incorporation of [α-32P]dGTP subsequent to cleavage and prior to chain termination (Fig. 1B and 1D, lanes 2, 3). These reactions also generated cleavage-derived products 16 nt in length resulting only from the incorporation of radiolabeled dGTP (Fig. 1D, lanes 2, 3). Reactions with (G2T2AG)4 or (G2T2AG)3G2T2A2G (which forms a mismatch with the RNA template) generated cleavage-derived products 24 (primer-size), 18 and 12 nt in length (Fig. 1D, lanes 7, 10), consistent with the cleavage of 1, 7 or 13 nt, depending on the primer alignments with the RNA template (Fig. 1B). Substitutions of dATP with ddATP or TTP with ddTTP in the telomerase assays using (G2T2AG)4 or (G2T2AG)3G2T2A2G as substrates generated chain termination products of 29, 23 and 17 nt (with ddATP: Fig. 1D, lanes 8, 11) or 27, 21 and 15 nt (with ddTTP: Fig. 1D, lanes 9, 12), as well as cleavage-derived products 24, 18 and 12 nt in length resulting only from the incorporation of [α-32P]dGTP (Fig. 1D, lanes 8, 9, 11, 12). The length of cleavage-derived and chain termination products suggests that cleavage occurred 3′ to the TTA residues within the telomeric sequence primers (Fig. 1B).
Effects of different reaction conditions on telomerase-mediated primer cleavage
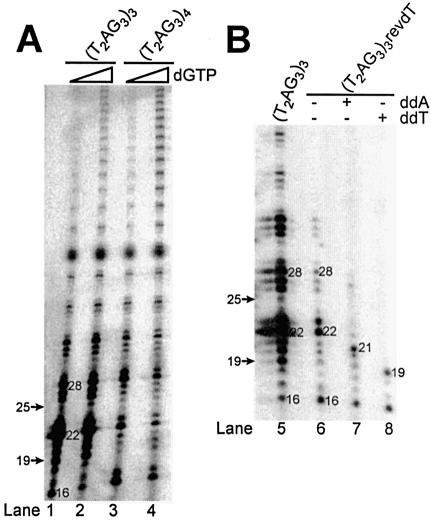
Previous studies in Tetrahymena, Euplotes, yeast and human have shown that an increase in either dGTP or primer concentration in the telomerase assay affects the nucleolytic primer cleavage activity as well as the processivity of the telomerase enzyme (14,27,31,36,37). To determine whether the extent of cleavage is affected by these reaction conditions, we varied either the concentration of dGTP or primer in the telomerase reactions using RRL-reconstituted human telomerase. Unlabeled dGTP was used to increase the total concentration of dGTP from 1.25 to 13.75 µM, which did not affect the relative amount of cleavage-derived products, but promoted the generation of longer products, consistent with an increase in enzyme processivity (Fig. 2A). A similar increase in processivity also occurred by varying the (T2AG3)4 concentration from 0.25 to 5 µM, but the relative amount of cleavage-derived products was also not affected (data not shown).
Figure 2.
Effects of different parameters on telomerase-mediated primer cleavage. (A) Effect of dGTP concentration on telomerase-mediated cleavage activity. DNA primer extension assays were conducted using different concentrations of dGTP. In addition to 1.25 µM [α-32P]dGTP, unlabeled dGTP was added. Lanes 1 and 3 contain no unlabeled dGTP. Lanes 2 and 4 contain 12.5 µM of unlabeled dGTP. (B) Cleavage of a telomeric primer containing a chain terminator (revdT) at the 3′-end. Direct DNA primer extension assays were performed with the indicated oligonucleotides. The 3′-end radiolabeled 5′-biotinylated primers, (T2AG3)3 and (T2AG3)4, migrate, respectively, as 19 and 25mers at the positions indicated by the arrows. ddA and ddT: dideoxyATP and dideoxyTTP. dGTP: deoxyGTP. Numbers in (A) and (B) represent the lengths of the elongation or chain termination products.
We speculated that cleavage could be due to pyrophosphorolysis. This reaction can occur when the base at the 3′ terminus of a DNA chain reacts with pyrophosphate ions in solution to generate a free dNTP molecule and a DNA chain that is one base shorter. As we observed the removal of 3 or 9 nt from (T2AG3)4, the cleavage activity we have described is not likely to be due to pyrophosphorolysis. Moreover, the addition of 5 U of pyrophosphatase, an enzyme used to inhibit pyrophosphorolysis, did not abolish the cleavage of (T2AG3)3 (data not shown).
To further characterize the cleavage reaction, we assayed a telomeric oligonucleotide substrate containing a modification that prevents chain extension [(T2AG3)3revdT]. This oligonucleotide can only be elongated by the telomerase enzyme if the chain terminating modification is removed. Primer extension assays performed with (T2AG3)3revdT in the presence of either dNTPs or ddNTPs generated cleavage-derived products 16 nt in length, consistent with the removal of 3 nt prior to elongation (Fig. 2B, lanes 6–8).
Telomerase-mediated primer cleavage is specified by primer alignments with the hTR template sequence
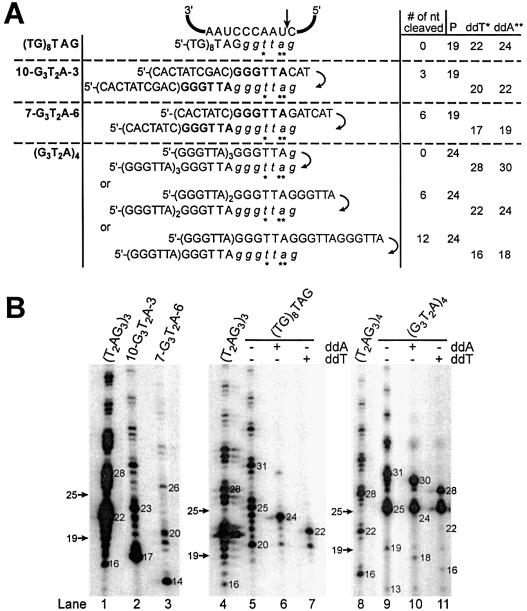
In Tetrahymena and Euplotes, primer–RNA template interactions regulate telomerase-mediated nucleolytic cleavage activity (20,25,28). In general, mismatches between the primer and the RNA template sequence promote primer cleavage at the junction of match-mismatch. As previously described in Figure 1, cleavage of (G2T2AG)3G2T2A2G, a primer which can form a mismatch with the RNA template, produced products 24 nt in length. Such products would be generated by the removal of the last two 3′ nucleotides prior to the incorporation of [α-32P]dGTP during DNA synthesis (Fig. 1D, lane 10). Moreover, certain primers containing nontelomeric sequences that are predicted to form mismatches with the RNA template are also good cleavage and subsequent elongation substrates for Euplotes telomerase (25,28). These primers consist of an internal telomeric cassette flanked by nontelomeric DNA sequences. Euplotes telomerase can remove the 3′ nontelomeric sequences to expose the telomeric sequences for elongation. RRL-reconstituted human telomerase was assayed using primers [CACTATCGAC-G3T2A-CAT] and [CACTATC-G3T2A-GATCAT] abbreviated as (10-G3T2A-3) and (7-G3T2A-6), respectively (the telomeric cassettes are indicated in bold). These primers are predicted to align with the RNA template as indicated in Figure 3A. Telomerase reactions performed with (10-G3T2A-3) or (7-G3T2A-6) led to the formation of products that were shorter than the input primer at 17 and 14 nt, respectively (Fig. 3B, lanes 2, 3). Product size was consistent with primer cleavage occurring 3′ to the telomeric cassette to remove the 3′ nontelomeric sequences (3 or 6 nt). Substitutions of dATP with ddATP or TTP with ddTTP in telomerase assays confirmed that 3 or 6 nt were removed, eliminating the nontelomeric DNA before the initiation of DNA synthesis (Fig. 3A and data not shown).
Figure 3.
Cleavage is specified by the primer alignment relative to the RNA template sequence. (A) Potential alignments of various oligonucleotides (tested in B) with the hTR template, before and after translocation. Italicized letters represent nucleotides added by the telomerase enzyme onto the 3′-end of the primer during the elongation step. P: primer length. ddA and ddT: dideoxyATP and dideoxyTTP. The proposed cleavage site is shown by the vertical arrow. Asterisks indicate the positions of the first ddA or ddT incorporated during the DNA primer extension assay. (B) Direct DNA primer extension assays were performed with the indicated oligonucleotides. 10-G3T2A-3: CACTATCGAC-GGGTTA-CAT; 7-G3T2A-6: CACTATC-GGGTTA-GATCAT, where the bold nucleotides represent a telomeric cassette. The 3′-end radiolabeled 5′-biotinylated primers, (T2AG3)3 and (T2AG3)4, migrate, respectively, as 19 and 25mers at the positions indicated by the arrows. Numbers in (A) and (B) represent the lengths of the primers, elongation or chain termination products.
To determine whether nucleolytic cleavage activity is dictated by the alignment of the primer substrate at the 5′-end of the RNA template, we tested an oligonucleotide [(TG)8TAG] that mimics a primer predicted to align at the 3′-end of the Tetrahymena telomerase RNA template (Fig. 3A) (20). Primer extension reactions with RRL-reconstituted human telomerase did not generate primer-sized products (19 nt) or products below the input primer, but favored the direct extension pathway leading to the formation of products ≥20 nt in length (Fig. 3B, lanes 5–7). This primer generated a six nucleotide pausing pattern starting at 25 nt as expected if it is aligned with the template RNA 3′-end 3′-(A)AUC-5′ (Fig. 3A). These results are comparable to those obtained with Tetrahymena telomerase and a similar primer [(TG)8TTG], supporting the predicted alignment of the (TG)8TAG with the 3′-end of the RNA template (20).
The cleavage activity does not appear to fulfill a classical proofreading function since entirely telomeric substrates are cleaved (Fig. 1) (20,25). In Euplotes, de novo telomere formation is extremely precise, where all new telomeres start with the sequence 5′-GGGGTTTT-3′ (7). We speculated that the preferred substrate for elongation might be one ending with GGGTTA, and predicted that such a substrate might not be cleaved. We assayed (G3T2A)4 and found that RRL-reconstituted human telomerase favored the direct extension pathway leading predominantly to the formation of products ≥25 nt in length (Fig. 3B, lane 9). The generation of shorter products 13 and 19 nt in length derived from the cleavage of 12 and 6 nt from (G3T2A)4 was reduced compared to the generation of shorter products derived from the cleavage of (T2AG3)4 (Fig. 3B, compare lane 8 with lane 9). The sizes of the cleavage-derived products and the chain termination products generated when substituting dATP with ddATP or TTP with ddTTP were consistent with the potential alignments of the (G3T2A)4 with the RNA template (Fig. 3A), and a nucleolytic cleavage activity that occurs 3′ to the A residue. We also found that the cleavage of the telomeric primer [(AG3T2)3] was reduced compared to the cleavage of (T2AG3)4 (data not shown). The reduced cleavage of certain telomeric primers may suggest that these primers are preferred elongation substrates. Alternatively, these primers may preferentially align such that their 3′ sequences do not extend beyond the 5′-end of the template (Fig. 3A).
Primer length requirement of the telomerase-mediated cleavage reaction
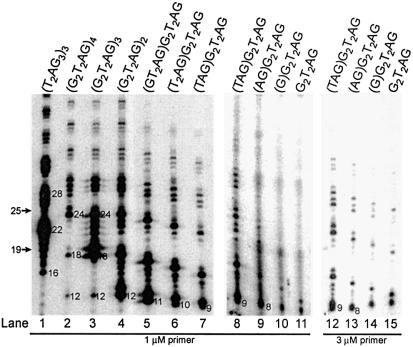
Previous studies have shown that the efficient extension of different length oligonucleotides by human and Tetrahymena telomerases is dependent on telomerase assay reaction conditions (12,15,20,31). We determined the primer length requirement of RRL-reconstituted human telomerase- mediated nucleolytic cleavage activity under our standard reaction conditions. We tested primers containing different numbers of the same telomeric repeat permutation: [(G2T2AG)4, (G2T2AG)3, (G2T2AG)2, (G2T2AG)] and primers shorter than (G2T2AG)2 designed by removing one to six nucleotides from the 5′-end (Fig. 4). We found that primer-sized cleavage-derived products could be visualized in telomerase reactions with primers 9 nt in length or longer (Fig. 4, lanes 2–7). Primers 6, 7 and 8 nt in length were weakly elongated under standard reaction conditions (Fig. 4, lanes 9–11). Upon a 3-fold increase in the primer concentration, it was possible to detect primer-sized products with the 8 nt primer (Fig. 4, lane 13). However, under the same reaction conditions, we could not confirm cleavage of the 6 and 7 nt primers due to their less efficient elongation by telomerase and to the altered mobility of the lowest products derived from these primers (Fig. 4, compare lanes 10, 11 with lanes 14, 15).
Figure 4.
Primer length requirement of the telomerase-mediated cleavage activity at different primer concentrations. DNA primer extension assays were performed with the indicated oligonucleotides. The elongation products were resolved on 10% polyacrylamide-urea gels. The 3′-end radiolabeled 5′-biotinylated primers, (T2AG3)3 and (T2AG3)4, migrate, respectively, as 19 and 25mers at the positions indicated by the arrows. Numbers represent the lengths of the elongation products.
Mechanism of telomeric substrate cleavage
Characterization of E.crassus and S.cerevisiae telomerases indicates that the primer cleavage proceeds by an endonucleolytic mechanism. Since human telomerase-mediated primer cleavage activity eliminates nucleotides that are complementary to the template residues, it did not seem to use a classical proofreading mechanism. Therefore we were also interested to determine whether the cleavage activity proceeds by an endonucleolytic mechanism. We designed oligonucleotides containing a phosphorothioate group (represented by an asterisk), a nuclease-resistant phosphodiester substitution that replaces an oxygen of the phosphodiester bond with a sulfur group. Primer extension telomerase assays performed with (T2AG3)4 resulted in the removal of 3 and 9 nt, generating cleavage-derived products 22 and 16 nt in length as described previously (Fig. 5, lane 1). (T2AG3)4 was modified to contain a phosphorothioate substitution three nucleotides [(T2AG3)3T2A*G3] or one nucleotide [(T2AG3)3T2AG2*G] from the 3′ terminus, respectively. An endonucleolytic cleavage reaction eliminating 3 nt would be affected by the nuclease-resistant substitution located three nucleotides from the 3′-end of (T2AG3)3T2A*G3. However, the endonucleolytic cleavage removing 3 nt should still proceed if the same modification is located one nucleotide from the 3′-end of (T2AG3)3T2AG2*G. Primer cleavage was similarly abolished by the modifications at either position and led to the formation of products ≥28 nt in length (Fig. 5, lanes 2, 3). Cleavage-derived products 16 nt in length generated by the removal of 9 nt from (T2AG3)4 were also inhibited by the modifications at the same positions, suggesting that the cleavage proceeds by an exonucleolytic mechanism. Substitutions of dATP with ddATP or TTP with ddTTP in telomerase assays generated no detectable chain termination products due to the incorporation of ddATP or ddTTP prior to the incorporation of [α-32P]dGTP (data not shown). These results were consistent with an inhibition of primer cleavage prior to the elongation.
Figure 5.
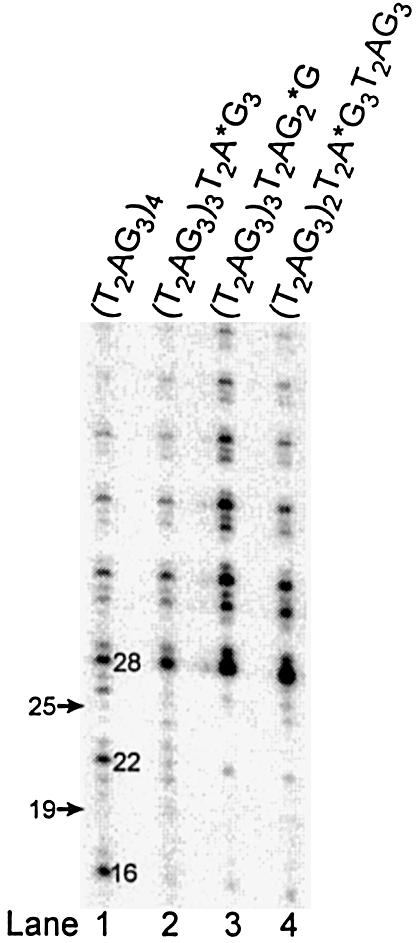
Cleavage of telomeric substrates by human telomerase may proceed by an exonucleolytic mechanism. DNA primer extension assays were performed with the indicated oligonucleotides. The 3′-end radiolabeled 5′-biotinylated primers, (T2AG3)3 and (T2AG3)4, migrate, respectively, as 19 and 25mers at the positions indicated by the arrows. Numbers represent the lengths of the elongation products. The asterisks indicate the positions of the phosphorothioate modifications within the primers.
To determine whether the telomerase-mediated cleavage activity proceeds by an exonucleolytic mechanism, we designed an oligonucleotide carrying a phosphorothioate substitution located nine nucleotides from the 3′-end of (T2AG3)2T2A*G3T2AG3. We predicted that the removal of 9 nt but not 3 nt from the 3′-end of (T2AG3)2T2A*G3T2AG3 would be affected by the nuclease-resistant substitution if the cleavage was exonucleolytic. However, the internal modification in (T2AG3)2T2A*G3T2AG3 abolished the cleavage and inhibited the removal of both 9 and 3 nt and the generation of both shorter products 16 and 22 nt in length (Fig. 5, lane 4). If the cleavage activity is exonucleolytic, we might also expect to see products derived from the removal of fewer than 3 nt for both (T2AG3)3T2A*G3 and (T2AG3)2T2A*G3T2AG3 and products derived from the removal of fewer than 9 nt for the latter primer. However, such products were not detected. These results do not confirm that the cleavage mechanism is exonucleolytic. However, they do not support an endonucleolytic mechanism. Our results raise the possibility that cleavages at multiples sites, whether exonucleolytic or endonucleolytic, might not be independent.
Cleavage activity of endogenous human telomerase
RRL-reconstituted human telomerase functions similarly to endogenous telomerase with respect to the basic in vitro elongation. However, a comparison of RRL-reconstituted Tetrahymena telomerase to endogenous enzyme has revealed some differences, including differences in repeat addition processivity (31,37,38). To identify and characterize endogenous human telomerase-mediated nucleolytic cleavage activity, we performed primer extension assays using native telomerase enzyme partially purified from 293 cell extracts. As shown in Figure 6, endogenous telomerase catalyzed nucleolytic primer cleavage under standard reaction conditions. We tested a series of oligonucleotides that we had characterized for cleavage using RRL-reconstituted human telomerase. (T2AG3)3 and (T2AG3)4 were cleaved and native telomerase generated cleavage-derived products of the same length (16 and 22 nt) as those generated by RRL-reconstituted telomerase (Fig. 6, compare lane 1 with lanes 2, 3). Telomerase reactions were also performed with (T2AG3)4 using different concentrations of dGTP or primer as those we used with RRL-reconstituted human telomerase. The relative amount of cleavage-derived products was not affected (data not shown).
Figure 6.
Telomerase-mediated cleavage activity of endogenous human telomerase. DNA primer extension assays were performed with the indicated oligonucleotides using RRL-reconstituted telomerase enzyme (lanes 1, 5, 9, 13) or enzyme partially purified from 293 cell extracts (lanes 2–4, 6–8, 10–12, 14, 15). The 3′-end radiolabeled 5′-biotinylated primers, (T2AG3)3 and (T2AG3)4, migrate, respectively, as 19 and 25mers at the positions indicated by the arrows. Numbers represent the lengths of the elongation products.
Similar to our previous results using RRL-reconstituted human telomerase, (TG)8TAG and (G3T2A)4 were not cleaved and the native enzyme favored the direct extension pathway leading to the formation of products ≥20 or 25 nt in length, respectively (compare Fig. 3B, lanes 5, 9 with Fig. 6, lanes 7, 8). However, (G2T2AG)3 and (G2T2AG)4 were also cleaved, but generated only primer-sized products (compare Fig. 4, lanes 2, 3 with Fig. 6, lanes 4, 6). These results differ from those obtained using RRL-reconstituted telomerase enzyme where more than one nucleotide were cleaved from these primers. To determine whether primers containing nontelomeric sequences are also good cleavage substrates for endogenous human telomerase, we performed telomerase reactions with (10-G3T2A-3) or (7-G3T2A-6). Such reactions also generated products at 17 and 14 nt, respectively, corresponding to the cleavage of the nontelomeric sequences (3 or 6 nt) by the enzyme (compare Fig. 3B, lanes 2, 3 with Fig. 6, lanes 14, 15). Lastly, to determine whether native human telomerase mediates telomeric primer cleavage via an endonucleolytic mechanism like other endogenous telomerase enzymes, we performed telomerase reactions with telomeric primers containing phosphorothioate substitutions. Native telomerase generated similar products as the RRL-reconstituted telomerase enzyme with the modified primers, suggesting that human telomerase may mediate cleavage by an exonucleolytic mechanism (compare Fig. 5, lanes 2–4 with Fig. 6, lanes 10–12).
DISCUSSION
Properties of the nuclease activity described in our studies suggest that the cleavage reaction is dependent on human telomerase and not a nonspecific nuclease. First, our data indicate that the cleavage reactions occurring with RRL-reconstituted human telomerase and native enzyme share some common features. Similar to RRL-reconstituted Tetrahymena telomerase (31), the human TERT and TR expressed in RRL appear sufficient to cleave a primer DNA substrate. Second, end-radiolabeled primers that are incubated with RRL in the absence of telomerase components are neither cleaved nor elongated. Third, the sites of cleavage within the primer DNA substrates are nonrandom, occurring 3′ to the A residue in the human telomeric sequence repeat, GGTTAG, and appear to be dependent on the primer alignment with the 5′-end of the template. Lastly, though we did not extensively purify human telomerase from 293 cells, primer cleavage and DNA synthesis appear to be functionally coupled following a partial purification, suggesting that it is either an inherent property of the enzyme or is catalyzed by an associated factor.
Regulation of human telomerase-mediated cleavage by primer alignments with the RNA template
Our studies indicate that primer–RNA interactions affect cleavage consistent with results of studies with yeast and ciliate telomerase-mediated nuclease activity (13,20,25,28, 31,39,40). The potential alignments of each primer with the RNA template allowed us to propose a potential cleavage site relative to the RNA template, between the U and C in the sequence 3′-AAUCCCAAUC-5′. The pausing pattern generated by the elongation of the cleavage-derived products is consistent with the addition of radiolabeled dGTP at the 5′-end of the template after a cleavage event. A primer [(TG)8TAG] predicted to align at the 3′-end of the RNA template is not cleaved by human telomerase, leading us to propose that primer cleavage is dependent on primer alignment with the RNA template 5′-end. Similar locations in the Tetrahymena and E.crassus telomerase RNA template were reported to dictate cleavage (20,25), though alternate internal sites have also been observed to mediate cleavage in E.crassus (28). Primers would be cleaved when they are aligned at the 5′-end or beyond the 5′-end of the template to ensure that only residues within the RNA template are copied during telomere synthesis.
Consistent with the potential alignments of primers with the 5′-end of the template, the proposed cleavage site and a possible proofreading function, RRL-reconstituted and endogenous human telomerase catalyzed the removal of nucleotides that form a mismatch with the template similarly to Euplotes and Tetrahymena telomerase (20,25). Consistent with studies of Euplotes and yeast telomerase, our results indicate that human telomerase mediated cleavage at the junction of the telomeric and nontelomeric sequences (25,27). Thus the removal of mismatched sequences from telomerase substrates is consistent with a proofreading function for telomerase-mediated cleavage and the enzyme’s high fidelity to retain only residues that are complementary to the template.
Role of telomerase-mediated cleavage activity in the generation of preferred substrates
Cleavage of telomeric primers by Tetrahymena telomerase has been reported, but appears to be restricted to the removal of one nucleotide (20). Unlike Tetrahymena telomerase, RRL-reconstituted human telomerase catalyzed the removal of more than one nucleotide from primers containing permutations of 5′-GGTTAG-3′. We proposed above that several alignments of the (T2AG3)4 and (G2T2AG)4 primers could occur relative to the RNA template to generate two or three different sizes of cleavage-derived products, respectively. Some of the shorter products would thus be generated by cleavage of sequences that extend past the 5′-end of the RNA template, as has been documented for Tetrahymena telomerase (20). However, the function for the cleavage of telomeric primers and their cleavage at multiple sites is unclear. Cleavage of telomeric primers by S.cerevisiae telomerase to generate labeled products shorter than the input primer by up to eight nucleotides has also been reported though a function for the cleavage of telomeric substrates has not been determined (24). We propose that a requirement for the cleavage of telomeric primers in vivo might be to generate a preferred substrate for human telomerase. For example, de novo telomere formation in E.crassus is extremely precise, with new telomeres initiating with the sequence 5′-GGGGTTTT-3′ (41). S.cerevisiae also initiate telomere formation with TG1–3 repeats (42). As cleavage of the primer ending in 5′-GGGTTA-3′ is reduced, we suggest that new telomeres in human chromosomes might initiate with the sequence 5′-GGGTTA-3′. However, cleavage of the telomeric primer (AG3T2)3 was also reduced. Human telomerase can also add different permutations of the human telomeric sequence to primers derived from the chromosome 16 breakpoint sequence (43). Thus the reduced cleavage of certain telomeric primers may not indicate that they are preferred substrates for elongation by telomerase. Alternatively, such primers may preferentially align such that their 3′ sequences do not extend beyond the 5′-end of the template.
Characterization of the human telomerase-dependent nuclease of telomeric substrates revealed that cleavage may occur by an exonucleolytic mechanism. Our findings were somehow unexpected as primer cleavage by endogenous E.crassus and S.cerevisiae telomerases occurs by an endonucleolytic mechanism (25,27). We obtained similar results whether we used RRL-reconstituted or partially purified native human telomerase enzyme. Though our results suggest that cleavage occurs by an exonucleolytic mechanism, our findings using the primer (T2AG3)2T2A*G3T2AG3 were also surprising, as an internal nuclease-resistant modification should not have affected the cleavage that can occur at the 3′-end of the primer. The modification does not appear to alter the primer alignment with the RNA template, since all the modified primers were efficiently elongated by RRL-reconstituted and endogenous telomerases to generate the expected six nucleotide product pausing pattern. However, one possibility may be that (T2AG3)2T2A*G3T2AG3 aligns only with the modification opposite to the 5′-end of the template that dictates cleavage. Another possibility may be that cleavages at the multiple sites are not independent, and abolishing cleavage at the internal site may affect cleavage at the more 3′-terminal site. Alternatively the modified primers may preferentially align at the 3′-end of the template and are not substrates for a cleavage-mediated reaction that is dependent on primer alignment at the RNA template 5′-end. If either of these scenarios are indeed occurring, we must also consider the possibility that such events may be consistent with an endonucleolytic mode of action.
Difference between RRL-reconstituted telomerase enzyme and native enzyme
In our study, we found that processivity and cleavage differed between RRL-reconstituted human telomerase and native enzyme. The reconstituted enzyme complex could be lacking required associated regulatory factors and/or be assembled in a different conformation. As for Tetrahymena telomerase, RRL-reconstituted human enzyme was less processive than endogenous telomerase (31,37,38). Using the primers (G2T2AG)3 or (G2T2AG)4, the native enzyme generated mainly primer-sized products, consistent with cleavage of similar substrates ending in G3T2G by endogenous Tetrahymena telomerase (20). These results suggest a preferential binding of these primers at the 5′-end of hTR in the native enzyme, and do not support the alignment of such primers beyond the 5′-end of the template. This is in contrast to the results we obtained using RRL-reconstituted enzyme where cleaved products were consistent with the primers aligning beyond the template 5′-end. Moreover, the relative amount of cleaved versus elongated products derived from (T2AG3)3 and (T2AG3)4 is less for the native enzyme than the RRL-reconstituted enzyme. The cleavage, particularly marked for the RRL-reconstituted complex, may result from a high frequency of stalling by the human telomerase at several positions along the RNA template. We speculate that the nuclease activity may be an error-correcting mechanism or a way of rescuing pause/arrested complexes, which facilitates the elongation of DNA substrates (44). The native enzyme or telomerase-associated factors may more stringently regulate the alignment and binding of DNA substrates with the RNA template. Further studies will be required to better characterize the human telomerase-mediated cleavage activity and the dependence of this activity on the minimal telomerase components (sequence or structure of hTERT or hTR) and/or additional protein components.
Acknowledgments
ACKNOWLEDGEMENTS
We thank M. A. Cerone, T. J. Moriarty, D. Marie-Egyptienne and M. A. S. Taboski for comments on the manuscript. C.A. is the recipient of a Fonds de la Recherche en Santé du Québec Chercheur Boursier and a Boehringer Ingelheim (Canada) Inc. Young Investigator Award. S.H. is the recipient of a Faculty of Medicine Internal Studentships of McGill University. This work was supported by grant MOP-14026 from the Canadian Institutes of Health Research (CIHR) to C.A.
REFERENCES
- 1.Blackburn E.H. (2001) Switching and signaling at the telomere. Cell, 106, 661–673. [DOI] [PubMed] [Google Scholar]
- 2.Harrington L. and Robinson,M.O. (2002) Telomere dysfunction: multiple paths to the same end. Oncogene, 21, 592–597. [DOI] [PubMed] [Google Scholar]
- 3.Harrington L. (2003) Biochemical aspects of telomerase function. Cancer Lett., 194, 139–154. [DOI] [PubMed] [Google Scholar]
- 4.Harrington L.A. and Greider,C.W. (1991) Telomerase primer specificity and chromosome healing. Nature, 353, 451–454. [DOI] [PubMed] [Google Scholar]
- 5.Melek M. and Shippen,D.E. (1996) Chromosome healing: spontaneous and programmed de novo telomere formation by telomerase. BioEssays, 18, 301–308. [DOI] [PubMed] [Google Scholar]
- 6.Bednenko J., Melek,M., Greene,E.C. and Shippen,D.E. (1997) Developmentally regulated initiation of DNA synthesis by telomerase: evidence for factor-assisted de novo telomere formation. EMBO J., 16, 2507–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang H. and Blackburn,E.H. (1997) De novo telomere addition by Tetrahymena telomerase in vitro. EMBO J., 16, 866–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng J., Funk,W.D., Wang,S.-S., Weinrich,S.L., Avilion,A.A., Chiu,C.-P., Adams,R.R., Chang,E., Allsopp,R.C., Yu,J., Le,S., West,M.D., Harley,C.B. Andrews,W.H., Greider,C.W. and Villeponteau,B. (1995) The RNA component of human telomerase. Science, 269, 1236–1241. [DOI] [PubMed] [Google Scholar]
- 9.Weinrich S.L., Pruzan,R., Ma,L., Ouellette,M., Tesmer,V.M., Holt,S.E., Bodnar,A.G., Lichtsteiner,S., Kim,N.W., Trager,J.B., Taylor,R.D., Carlos,R., Andrews,W.H., Wright,W.E., Shay,J.W., Harley,C.B. and Morin,G.B. (1997) Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nature Genet., 17, 498–502. [DOI] [PubMed] [Google Scholar]
- 10.Beattie T.L., Zhou,W., Robinson,M.O. and Harrington,L. (1998) Reconstitution of human telomerase activity in vitro. Curr. Biol., 8, 177–180. [DOI] [PubMed] [Google Scholar]
- 11.Chen J.-L., Blasco,M.A. and Greider,C.W. (2000) Secondary structure of vertebrate telomerase RNA. Cell, 100, 503–514. [DOI] [PubMed] [Google Scholar]
- 12.Morin G.B. (1989) The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell, 59, 521–529. [DOI] [PubMed] [Google Scholar]
- 13.Lue N.F. and Peng,Y. (1998) Negative regulation of yeast telomerase activity through an interaction with an upstream region of the DNA primer. Nucleic Acids Res., 26, 1487–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun D., Lopez-Guajardo,C.C., Quada,J., Hurley,L.H. and Von Hoff,D.D. (1999) Regulation of catalytic activity and processivity of human telomerase. Biochemistry, 38, 4037–4044. [DOI] [PubMed] [Google Scholar]
- 15.Baran N., Haviv,Y., Paul,B. and Manor,H. (2002) Studies on the minimal lengths required for DNA primers to be extended by the Tetrahymena telomerase: implications for primer positioning by the enzyme. Nucleic Acids Res., 30, 5570–5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wallweber G., Gryaznov,S., Pongracz,K. and Pruzan,R. (2003) Interaction of human telomerase with its primer substrate. Biochemistry, 42, 589–600. [DOI] [PubMed] [Google Scholar]
- 17.Huard S., Moriarty,T.J. and Autexier,C. (2003) The C terminus of the human telomerase reverse transcriptase is a determinant of enzyme processivity. Nucleic Acids Res., 31, 4059–4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greider C.W. (1996) Telomere length regulation. Annu. Rev. Biochem., 65, 337–365. [DOI] [PubMed] [Google Scholar]
- 19.Greider C.W. (1991) Telomerase is processive. Mol. Cell. Biol., 11, 4572–4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collins K. and Greider,C.W. (1993) Tetrahymena telomerase catalyzes nucleolytic cleavage and nonprocessive elongation. Genes Dev., 7, 1364–1376. [DOI] [PubMed] [Google Scholar]
- 21.Hammond P.W., Lively,T.N. and Cech,T.R. (1997) The anchor site of telomerase from Euplotes aediculatus revealed by photo-cross-linking to single- and double-stranded DNA primers. Mol. Cell. Biol., 17, 296–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shevelev I.V. and Hübscher,U. (2002) The 3′-5′ exonucleases. Nat. Rev. Mol. Cell Biol., 3, 1–12. [DOI] [PubMed] [Google Scholar]
- 23.Arndt K.M. and Kane,C.M. (2003) Running with RNA polymerase: eukaryotic transcript elongation. Trends Genet., 19, 543–550. [DOI] [PubMed] [Google Scholar]
- 24.Cohn M. and Blackburn,E.H. (1995) Telomerase in yeast. Science, 269, 396–400. [DOI] [PubMed] [Google Scholar]
- 25.Melek M., Greene,E.C. and Shippen,D.E. (1996) Processing of nontelomeric 3′ ends by telomerase: default template alignment and endonucleolytic cleavage. Mol. Cell. Biol., 16, 3437–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lue N.F. and Peng,Y. (1997) Identification and characterization of a telomerase activity from Schizosaccharomyces pombe. Nucleic Acids Res., 25, 4331–4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niu H., Xia,J. and Lue,N.F. (2000) Characterization of the interaction between the nuclease and reverse transcriptase activity of the yeast telomerase complex. Mol. Cell. Biol., 20, 6806–6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greene E.C., Bednenko,J. and Shippen,D.E. (1998) Flexible positioning of the telomerase-associated nuclease leads to preferential elimination of nontelomeric DNA. Mol. Cell. Biol., 18, 1544–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ware T.L., Wang,H. and Blackburn,E.H. (2000) Three telomerases with completely non-telomeric template replacements are catalytically active. EMBO J., 19, 3119–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhattacharyya A. and Blackburn,E.H. (1997) A functional telomerase RNA swap in vivo reveals the importance of nontemplate RNA domains. Proc. Natl Acad. Sci. USA, 94, 2823–2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collins K. and Gandhi,L. (1998) The reverse transcriptase component of the Tetrahymena telomerase ribonucleoprotein complex. Proc. Natl Acad. Sci. USA, 95, 8485–8490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Autexier C., Pruzan,R., Funk,W.D. and Greider,C.W. (1996) Reconstitution of human telomerase activity and identification of a minimal functional region of the human telomerase RNA. EMBO J., 15, 5928–5935. [PMC free article] [PubMed] [Google Scholar]
- 33.Bachand F. and Autexier,C. (1999) Functional reconstitution of human telomerase expressed in Saccharomyces cerevisiae. J. Biol. Chem., 274, 38027–38031. [DOI] [PubMed] [Google Scholar]
- 34.Moriarty T.J., Huard,S., Dupuis,S. and Autexier,C. (2002) Functional multimerization of human telomerase requires an RNA interaction domain in the N terminus of the catalytic subunit. Mol. Cell. Biol., 22, 1253–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen J.-L. and Greider,C.W. (2003) Determinants in mammalian telomerase RNA that mediate enzyme processivity and cross-species incompatibility. EMBO J., 22, 304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hammond P.W. and Cech,T.R. (1997) dGTP-dependent processivity and possible template switching of Euplotes telomerase. Nucleic Acids Res., 25, 3698–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hardy C.D., Schultz,C.S. and Collins,K. (2001) Requirements for the dGTP-dependent repeat addition processivity of recombinant Tetrahymena telomerase. J. Biol. Chem., 276, 4863–4871. [DOI] [PubMed] [Google Scholar]
- 38.Bryan T.M., Goodrich,K.J. and Cech,T.R. (2000) A mutant of Tetrahymena telomerase reverse transcriptase with increased processivity. J. Biol. Chem., 275, 24199–24207. [DOI] [PubMed] [Google Scholar]
- 39.Prescott J. and Blackburn,E.H. (1997) Telomerase RNA mutations in Saccharomyces cerevisiae alter telomerase action and reveal nonprocessivity in vivo and in vitro. Genes Dev., 11, 528–540. [DOI] [PubMed] [Google Scholar]
- 40.Autexier C. and Greider,C.W. (1994) Functional reconstitution of wild-type and mutant Tetrahymena telomerase. Genes Dev., 8, 563–575. [DOI] [PubMed] [Google Scholar]
- 41.Klobutcher L.A., Swanton,M.T., Donini,P. and Prescott,D.M. (1981) All gene-sized DNA molecules in four species of hypotrichs have the same terminal sequence and an unusual 3′ terminus. Proc. Natl Acad. Sci. USA, 78, 3015–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kramer K.M. and Haber,J.E. (1993) New telomeres in yeast are initiated with a highly selected subset of TG1–3 repeats. Genes Dev., 7, 2345–2356. [DOI] [PubMed] [Google Scholar]
- 43.Morin G.B. (1991) Recognition of a chromosome truncation site associated with α-thalassaemia by human telomerase. Nature, 353, 454–456. [DOI] [PubMed] [Google Scholar]
- 44.Sastry S.S. and Ross,B.M. (1997) Nuclease activity of T7 RNA polymerase and the heterogeneity of transcription elongation complexes. J. Biol. Chem., 272, 8644–8652. [DOI] [PubMed] [Google Scholar]