Abstract
The infrastructure for data collection implemented by the National Consortium on Alcohol and NeuroDevelopment in Adolescence (N-CANDA) for data collection comprises several innovative features: (a) secure, asynchronous transfer and persistent storage of collected data via a revision control system; (b) two-stage import into a longitudinal database; and (c) use of a script-controlled web browser for data retrieval from a third-party, web-based neuropsychological test battery. The asynchronous operation of data transmission and import is of particular benefit, as it has allowed the consortium sites to begin data collection before the receiving database infrastructure had been deployed. Records were collected within 86 days of funding, 35 days after finalizing the collected instruments. Final instruments were added to the database import 225 days after instrument selection, with up to 173 records already collected at that time. Thus, the concepts implemented in N-CANDA’s data collection system helped reduce project start-up time by several months.
Keywords: informatics, longitudinal data collection, data integration, revision control system
Introduction
The National Consortium on Alcohol and NeuroDevelopment in Adolescence (N-CANDA; http://ncanda.org/), funded by the National Institute on Alcohol Abuse and Alcoholism, is a consortium tasked with investigating the extent to which structural and functional deficits in neurodevelopmental maturation are caused or exacerbated by adolescent alcohol use.
Here, we describe the data collection infrastructure for the N-CANDA Consortium, implemented wherever possible using freely available, open-source software components. All software tools and documentation materials created by us are also freely available at http://nitrc.org/projects/ncanda-datacore/.
The following requirements drove the selection of components for the infrastructure and their specific setup:
1. Automation: transfer of data from the collection sites to the consortium server should require as little human interaction as possible.
2. Security: all storage and transmission systems must use state-of-the-art encryption and access control.
3. Persistence: safeguard against loss of data due to overwriting or deletion.
4. Accountability: changes to existing data must create an audit trail with user names and time stamps.
5. Incremental deployment: due to a tight project schedule, data collection had to begin before the infrastructure (eg, database representations of acquired instruments) was fully in place.
As it turns out, using a networked revision control system for data transmission naturally satisfies all of these demands. Automation is ensured by the ability to drive all data transfers through scripts. Security is provided by access control to the repository and encryption of all data transfers. Persistence and Accountability result from the principle of revision control to track changes to all files (and their authors). Incremental deployment, finally, takes advantage of the ability to ‘play back’ the history of a repository, one change at a time, either globally or for selected files.
Based on these insights, we implemented N-CANDA's data collection, transmission, and integration system using a networked Subversion (https://subversion.apache.org/) repository and a moderate number of custom Perl and Python scripts. The system is ‘asynchronous’ in the sense that data transmission and database import do not have to occur hand-in-hand, but can be separated by an arbitrary time interval.
By reporting the time course of data collected over the first year of funding for the N-CANDA project, we demonstrate the benefits of our system: N-CANDA data collection sites successfully collected data within less than 3 months from the beginning of funding, within about 1 month of finalizing the experimental protocol, and up to 9 months before the database infrastructure was fully in place. For several instruments, over 150 records were collected before the database was deployed to receive these data.
Materials and methods
Data capture uses a client–server infrastructure. Mac and PC laptops with software applications for a variety of neuropsychological tests were centrally prepared and deployed to the collection sites. These laptops submit all collected data to a central server. A partial list of instruments collected by the laptops is provided in table 1, which includes each instrument's details, the data collection software used, and the necessary tools for data conversion. In addition, a number of computerized neuropsychological tests are administered using the Web-based Computerized Neuropsychological Testing (WebCNP) system at the University of Pennsylvania School of Medicine.5
Table 1.
Data captured by the N-CANDA data collection laptops; the ‘data conversion’ steps are automatically executed by the consortium server running Linux, with no user interaction
| Instrument | Data collection | Data conversion |
|---|---|---|
| Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA).1 Depending on the age of the subject, the interview is conducted either with the subject directly (‘SSAGA (Youth)’) or with a parent (‘SSAGA (Parent)’) | Legacy survey implemented with the Blaise computer-assisted interviewing system (http://www.blaise.com/) | Proprietary database format; raw data records extracted using Blaise's ‘Manipula’ tool, running in batch mode using the ‘Wine’ Windows Emulator (http://winehq.org). Basic measures are extracted using a Python script, ‘blaise2csv’. SSAGA diagnoses derived using third-party SAS scripts. |
| Delay discounting2 | Third-party Windows application | Simple tabular text file, parsed using a custom Python script, ‘dd2csv’. |
| Paced Auditory Serial-Addition Task (PASAT)3 | Third-party Windows application | Database file in Microsoft Access format is converted to CSV format using open-source mdb-tools (http://mdbtools.sourceforge.net/), driven by a custom Python script, ‘pasat2csv’. |
| Stroop Match-to-Sample (MtS)4 | ePrime (http://www.pstnet.com/eprime.cfm) | Human-readable log file, parsed and scores computed using a custom Python script, ‘eprime2redcap’. |
| Several study-specific instrument batteries (herein referred to as: Youth Report 1, Youth Report 2, Parent Report, MRI Report) | Customized version of LimeSurvey (http://www.limesurvey.org/) | Single-record, comma-separated file, reformatted by a custom Python script, ‘lime2csv’, to remove special characters from field names, etc. |
All custom scripts referenced in this table are available for download at https://www.nitrc.org/frs/?group_id=672.
CSV, comma-separated-value; N-CANDA, National Consortium on NeuroDevelopment in Adolescence.
For central data management, we chose the Research Electronic Data Capture (REDCap; http://project-redcap.org/) system6 for its web-based design of data collection instruments, secure architecture, and data audit trails, which were shown in an independent evaluation to provide a variety of benefits.7 REDCap allows us to provide all consortium sites with secure, interactive access to the consortium database for queries, data entry, and editing. Crucially, REDCap also provides an application programming interface (API) to import and export data automatically, without user interaction.
Laptop data collection: client
Transmission of data from the collection laptops into a revision control system is the first innovation of our infrastructure, illustrated in figure 1. We use a networked Subversion repository, but the benefits outlined below are not specific to Subversion. Any other modern, networked revision control system (eg, Git, Mercurial) would be equally suitable and would also provide the following advantages.
Figure 1.
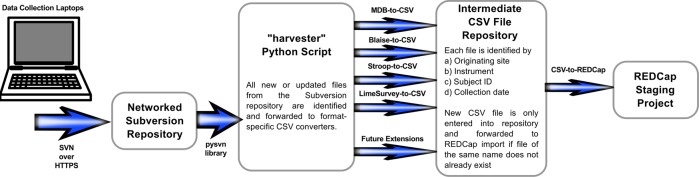
Illustration of the laptop data transmission and import procedure using a networked Subversion repository and the ‘harvester’ Python script.
First, repeated upload of a file under the same name will create a sequence of revisions, but will not remove any existing contents. Thus, no data can be lost due to overwriting or deletion. Any changes can be easily traced through the commit history of the repository. Together, these properties satisfy the design goals Persistence and Accountability, albeit the latter with some limitations in our current setup, where collected data can be traced to the originating laptop, but is not tied to a specific user account on that laptop.
Second, Subversion provides networked operation over a secure, encrypted link (HTTPS; via an Apache web server, http://apache.org/), error handling, and access control, without any custom software components. This satisfies the design goal Security.
The third advantage of using a revision control system is that the history of stored files can be ‘played back’ step by step, from any chosen point in time, either globally or for selected files. This feature allowed the N-CANDA data collection sites to begin transmitting data as soon as the collection laptops were in operation—long before all databases and import scripts were deployed on the server (see ‘Results’ below). This addresses the design goal Incremental deployment.
The final design goal and another important feature of the data transmission is Automation. Multiple output files are created on the laptops for each subject by some of the applications used for clinical interviews and neuropsychology tests. Other applications accumulate their outputs in separate databases. The output paths for all these applications are hard-coded, thus resulting in a large number of files distributed across each laptop's file system on completion of each subject session.
Custom Perl scripts automate the collection, collation, and uploading of the output files and updated databases to the Subversion repository. This eliminates the possibility that research assistants will miss or accidentally overwrite files when preparing to submit data and simplifies the transmission process to the degree that simply clicking an ‘Upload’ desktop icon performs the entire upload procedure.
Laptop data collection: server
On the N-CANDA server, a working copy is kept of the aforementioned Subversion repository and periodically updated by a Python script called ‘harvester’. Any updated or newly added files are forwarded to format-specific scripts that convert them into standardized, comma-separated-value (CSV) files. Unknown file types are silently ignored. These may be files for which database and import procedure have not yet been implemented.
Whenever support for another collected instrument is added to the system, its existing files are removed from the local Subversion working copy on the server. They will then be imported on the next update, as the ‘harvester’ script restores the files and converts them to CSV format. Finally, all CSV files are imported into REDCap using the PyCap library (http://sburns.org/PyCap/), which provides a convenient wrapper for REDCap's API.
Two-stage longitudinal data import
As the N-CANDA study is longitudinal, with a baseline and up to four annual follow-up visits per subject, we configured a longitudinal project in REDCap to integrate all incoming data. This means that a number of ‘events’ were defined, each with a specific subset from a list of data collection instruments.
A problem arises with the assignment of collected data to the correct event (and subject), which requires not only the subject ID but also an event name (eg, ‘baseline’, or ‘1-year follow-up’). If an error was made entering either, then the record would be imported into the wrong subject and/or event in REDCap. Unfortunately, REDCap’s web front-end does not allow for the selective moving of a single instrument from one subject or event to another.
In addition, most third-party software instruments do not allow the entry of an event, but only accept a subject ID. All instruments do, however, record the date of data collection. Thus, subject ID and collection date are two identifying fields that we are able to collect from all instruments.
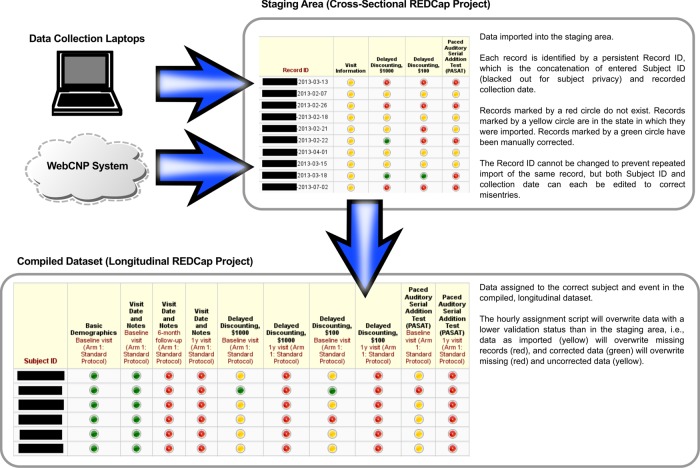
To assign a record to the correct subject and event, we use a two-stage import procedure (figure 2). Every incoming record is imported into a ‘staging area’, which is a standard, cross-sectional REDCap project. Here, each record is identified by a unique, persistent identifier, which is the concatenation of subject ID and collection date as recorded on the laptop (eg, a record of subject A-00001-F-0 collected on April 1, 2013 would receive the persistent identifier ‘A-00001-F-0-2013-04-01’). This identifier cannot be modified, even if ID and/or date are incorrect. Instead, two separate fields for ID and date are provided for corrections, and these are initialized with the recorded values.
Figure 2.
Illustration of the two-stage import procedure for data from N-CANDA data collection laptop or WebCNP system into the longitudinal REDCap project database. N-CANDA, National Consortium on Alcohol and NeuroDevelopment in Adolescence; WebCNP, Web-based Computerized Neuropsychological Testing.
These fields then determine the assignment of each record to the correct subject and event in the longitudinal REDCap project, which is performed by a Python script that periodically runs on the server. It exports records from the staging project, assigns them to the correct subject and event (based on a predefined time window for each event), and imports them into the final, longitudinal project.
WebCNP data retrieval
The WebCNP system (https://webcnp.med.upenn.edu/) stores all test scores and does not require the asynchronous laptop data transmission procedure. However, the WebCNP does not provide an API to automate score retrieval, rather a CSV file must be manually downloaded through the web interface. We were able to automate this process by ‘driving’ a Firefox web browser using the Selenium plug-in (http://seleniumhq.org/) and the ‘selenium’ Python package (https://pypi.python.org/pypi/selenium).
The downloaded files are imported into a REDCap staging project analogous to the one used for data from the collection laptops and then assigned to the correct subject and event. We chose to use a separate staging REDCap project for WebCNP simply for convenience and conceptual separation. Our custom scripts for WebCNP data retrieval and REDCap import are available for download at https://www.nitrc.org/frs/?group_id=672.
Results
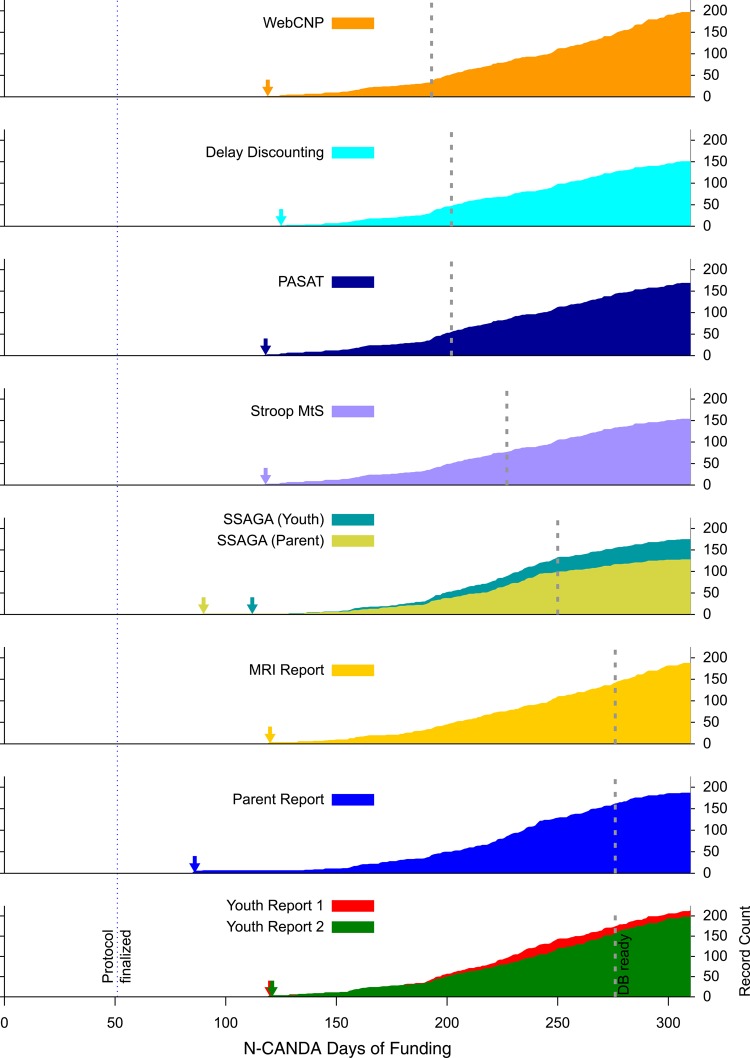
To illustrate the importance of the infrastructure described herein for the N-CANDA data collection, we show the timelines of collected records for several instruments in figure 3. For each instrument, the date when the database and import procedure were ready for that instrument is marked, as is the date when the protocol was finalized (ie, when the final selection of instruments was made). All dates are shown relative to the start of N-CANDA funding.
Figure 3.
Number of records collected over time after funding began (September 15, 2012) for different instruments of the N-CANDA protocol. The protocol was finalized during a consortium meeting November 5 and 6, 2012 (Day 51; marked by dotted line across graphs). In the graph for each instrument, an arrow marks the day when the first record was collected, and the dashed gray line marks the date when the REDCap database and import procedure were ready for that instrument. N-CANDA, National Consortium on Alcohol and NeuroDevelopment in Adolescence.
As figure 3 shows, revision-controlled data transfer has allowed N-CANDA to begin data collection very quickly, as soon as the collection laptops had been configured and deployed to the consortium sites. Implementation of the receiving infrastructure did not require additional delays.
This is further illustrated by table 2, which shows the number of records collected for a selection of instruments by the time the database and import systems were ready for each. While work was ongoing, the N-CANDA data collection sites were able to collect at least 46 records (‘Delay Discounting’), and as many as 173 records (‘Youth Report 1’).
Table 2.
Instruments collected by the N-CANDA consortium (partial list as reported in this article), showing date of first collected record, date when database and import were ready for each instrument, and number of records collected by the time database and import were ready
| Instrument | First record collected | Database and import ready | Records collected prior to database ready |
|---|---|---|---|
| Delay discounting | 2013-01-18 (day 125) | 2013-04-05 (day 202) | 46 |
| PASAT | 2013-01-11 (day 118) | 2013-04-05 (day 202) | 54 |
| Stroop MtS | 2013-01-11 (day 118) | 2013-04-27 (day 224) | 74 |
| SSAGA (parent) | 2012-12-14 (day 90) | 2013-05-23 (day 250) | 98 |
| SSAGA (youth) | 2013-01-05 (day 112) | 2013-05-23 (day 250) | 132 |
| MRI report | 2013-01-13 (day 120) | 2013-06-18 (day 276) | 141 |
| Parent report | 2012-12-10 (day 86) | 2013-06-18 (day 276) | 161 |
| Youth report 1 | 2013-01-13 (day 120) | 2013-06-18 (day 276) | 173 |
| Youth report 2 | 2013-01-14 (day 121) | 2013-06-18 (day 276) | 155 |
‘Day’ is the number of days after project funding was received (September 15, 2012).
MtS, Match to Sample; N-CANDA, National Consortium on Alcohol and NeuroDevelopment in Adolescence; PASAT, Paced Auditory Serial-Addition Task; SSAGA, Semi-Structured Assessment for the Genetics of Alcoholism.
Discussion
We have implemented a data collection and management system for a multi-site, longitudinal neuroscience study of adolescent drinking. The innovative use of a revision control system for data transmission enabled data collection within 3 months of funding, thus eliminating the need to wait until a data management system was fully in place.
To accommodate the longitudinal study design, we implemented an automated system, based on REDCap and a number of custom Python scripts, to collect data identified by subject ID and collection date and assign these data to the correct events.
Third, a script-driven web browser allows us to automatically retrieve data from a third-party, web-based neuropsychological test system that does not offer an API for data downloads.
A potential weakness of our system is that collecting data prior to availability of the receiving infrastructure carries an increased risk of data loss or corruption due to the lack of immediate feedback. Thus, errors in collected data may go undetected for longer than they otherwise would. We have found, however, that the benefits of acquiring data at all greatly outweigh the potential risks.
Acknowledgments
This work was supported by the National Institute on Alcohol Abuse and Alcoholism through the National Consortium for Alcohol and NeuroDevelopment in Adolescence (N-CANDA). The N-CANDA Consortium comprises the following components: Administrative Component at University of California, San Diego (Grant AA021695, PIs: S. Brown and S. Tapert); Data Analysis Component at SRI International (Grant AA021697; PIs: A. Pfefferbaum and T. Rohlfing); N-CANDA: Duke (Grant AA021681, PI: M. De Bellis); N-CANDA: OHSU (Grant AA021691, PI: B. Nagel); N-CANDA: Pittsburgh (Grant AA021690, PI: D. Clark); N-CANDA: San Diego (Grant AA021692, PI: S. Tapert); and N-CANDA: SRI (Grant AA021696, PIs: I. Colrain and F. Baker). The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Study data were collected and managed using REDCap electronic data capture tools hosted at SRI International. REDCap (Research Electronic Data Capture)6 is a secure, web-based application designed to support data capture for research studies, providing: (1) an intuitive interface for validated data entry; (2) audit trails for tracking data manipulation and export procedures; (3) automated export procedures for seamless data downloads to common statistical packages; and (4) procedures for importing data from external sources.
The N-CANDA Software Collection and operating manuals are available from http://nitrc.org/projects/ncanda-datacore/
Footnotes
Contributors: TR designed the overall data management system architecture, implemented the server infrastructure, oversaw system integration, and wrote this article. KC conceived the laptop-based client side of the data capture system and led the implementation of data collection instruments. TH designed and implemented the client-side parts of the laptop-based data collection infrastructure. WC implemented data scoring and file format conversion scripts and contributed to the implementation of the server-side data collection instruments in the REDCap database. BNN contributed to the design and implementation of the overall data management system, suggested critical components, and implemented automated data consistency checks. All authors reviewed, edited, and approved the submitted version of this article.
Funding: TR and WC are investigators with the N-CANDA Data Analysis Component at SRI International (Grant AA021697; PIs: A. Pfefferbaum and T. Rohlfing). KC is an investigator with the N-CANDA Administrative Component at University of California, San Diego (Grant AA021695, PIs: S. Brown and S. Tapert). TH is a paid consultant for the N-CANDA Administrative Component. BNN is a paid consultant for the N-CANDA Data Analysis Component.
Competing interests: None.
Ethics approval: Institutional Review Boards of SRI International, University of California–San Diego, Duke University, University of Pittsburgh, Oregon Health and Sciences University, and Stanford University.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Hesselbrock M, Easton C, Bucholz KK, et al. A validity study of the SSAGA—A comparison with the SCAN. Addiction 1999;94:1361–70 [DOI] [PubMed] [Google Scholar]
- 2.Bickel WK, Yi R, Landes RD, et al. Remember the future: working memory training decreases delay discounting among stimulant addicts. Biol Psychiatry 2011;69:260–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gronwall DMA. Paced auditory serial-addition task: a measure of recovery from concussion. Percept Mot Skills 1977;44:367–73 [DOI] [PubMed] [Google Scholar]
- 4.Schulte T, Mueller-Oehring EM, Rosenbloom MJ, et al. Differential effect of HIV infection and alcoholism on conflict processing, attentional allocation, and perceptual load: evidence from a Stroop match-to-sample task. Biol Psychiatry 2005;57:67–75 [DOI] [PubMed] [Google Scholar]
- 5.Gur RC, Richard J, Hughett P, et al. A cognitive neuroscience-based computerized battery for efficient measurement of individual differences: standardization and initial construct validation. J Neurosci Methods 2010;187:254–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harris PA, Taylor R, Thielke R, et al. Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009;42:377–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Franklin JD, Guidry A, Brinkley JF. A partnership approach for Electronic Data Capture in small-scale clinical trials. J Biomed Inform 2011;44(Suppl 1):S103–8 [DOI] [PMC free article] [PubMed] [Google Scholar]