Abstract
This article describes the patient-centered Scalable National Network for Effectiveness Research (pSCANNER), which is part of the recently formed PCORnet, a national network composed of learning healthcare systems and patient-powered research networks funded by the Patient Centered Outcomes Research Institute (PCORI). It is designed to be a stakeholder-governed federated network that uses a distributed architecture to integrate data from three existing networks covering over 21 million patients in all 50 states: (1) VA Informatics and Computing Infrastructure (VINCI), with data from Veteran Health Administration's 151 inpatient and 909 ambulatory care and community-based outpatient clinics; (2) the University of California Research exchange (UC-ReX) network, with data from UC Davis, Irvine, Los Angeles, San Francisco, and San Diego; and (3) SCANNER, a consortium of UCSD, Tennessee VA, and three federally qualified health systems in the Los Angeles area supplemented with claims and health information exchange data, led by the University of Southern California. Initial use cases will focus on three conditions: (1) congestive heart failure; (2) Kawasaki disease; (3) obesity. Stakeholders, such as patients, clinicians, and health service researchers, will be engaged to prioritize research questions to be answered through the network. We will use a privacy-preserving distributed computation model with synchronous and asynchronous modes. The distributed system will be based on a common data model that allows the construction and evaluation of distributed multivariate models for a variety of statistical analyses.
Keywords: clinical data research network, comparative effectiveness research, patient-centered research, distributed analysis
Participating health systems
The patient-centered Scalable National Network for Effectiveness Research (pSCANNER) is a consortium of three existing networks that together constitute a highly diverse patient population with respect to insurance coverage, socioeconomic status, demographics, and health conditions. Table 1 summarizes key characteristics of the institutions within each existing network.
Table 1.
System characteristics of pSCANNER's participating institutions
| Institution | Number of patients | Number of hospitals/clinics | EHR used |
|---|---|---|---|
| University of California, San Diego (UCSD) | 2.2 million | 4 hospitals 142 clinics |
Epic |
| University of California, Los Angeles (UCLA) | 4.1 million | 3 hospitals 300 clinics |
Epic |
| University of California, San Francisco (UCSF) | 3 million | 1 hospital 463 clinics |
Epic |
| San Francisco General Hospital (SFGH) | 0.5 million | 1 hospital 28 clinics |
Lifetime Clinical Records |
| University of California, Davis (UCD) | 2.2 million | 1 hospital 77 clinics |
Epic |
| University of California, Irvine (UCI) | 1.4 million | 1 hospital 184 clinics |
Allscripts—Sunrise |
| Veterans Affairs (VA) | 8.7 million | 151 hospitals 909 ambulatory care and community-based outpatient clinics |
VistA |
| AltaMed | 0.3 million | 31 clinics | NextGen |
| QueensCare Family Clinics | 19 000 | 6 clinics | Sage |
| The Children's Clinic (TCC) | 24 000 | 5 clinics | Epic |
Each institution is listed with its respective number of patients, number of hospitals/clinics, and the electronic health record (EHR) system.
pSCANNER, patient-centered Scalable National Network for Effectiveness Research.
VA Informatics and Computing Infrastructure (VINCI; http://www.hsrd.research.va.gov/for_researchers/vinci/) is a major informatics initiative of the Veterans Health Administration (VHA) that provides a secure, central platform for performing research and supporting clinical operations activities. In addition to national data, VINCI hosts commercial and custom analytical software for natural language processing, annotation, data exploration, and epidemiological analysis. The VHA treats 8.76 million veterans within an integrated healthcare delivery system, which includes hospitals, outpatient pharmacies, ancillary care facilities, and laboratory and radiology services. Since the 1980s, all VA facilities have used the same electronic health record (EHR) system, named Veterans Information System Technology Architecture (VistA). On a nightly basis, the VA's clinical data warehouse (CDW) is updated from VistA. This CDW stores vast amounts of clinical data dating back to 2000. The CDW and other VHA patient-level data remain behind VA firewalls. However, within pSCANNER, VINCI patient data will be available for consultation through privacy-preserving, distributed computing methodology. The VA team within pSCANNER will continue to develop analytical modules and map its CDW to the Observational Medical Outcomes Partnership (OMOP) common data model.1
The University of California Research exchange (UC-ReX; http://www.ucrex.org) was established in 2010 and has been funded by the University of California Office of the President since 2011 through the UC-Biomedical Research Acceleration, Integration, and Development (UC-BRAID) Initiative, which streamlines operations within the UC system such as institutional review board (IRB) activity, biorepository coordination, and other research activities. UC-ReX is also supported by the National Institutes of Health (NIH) Clinical Translational Science Awards from five UC Health Systems (UC Davis, Irvine, Los Angeles, San Francisco, and San Diego). UC-ReX has EHR data for over 12 million patients, and has used the i2b22 data model for cohort discovery based on a limited set of variables related to demographics, diagnoses, laboratory tests, and medications. In pSCANNER, UC-ReX will map its CDWs to the OMOP model.
Scalable National Network for Effectiveness Research (SCANNER; http://scanner.ucsd.edu) was established in late 2010 with Agency for Healthcare Research and Quality (AHRQ) funds. Its goal was to provide a secure, scalable distributed infrastructure to facilitate comparative effectiveness research3 among widely dispersed institutions, and to provide flexibility to participant sites in the means for data-driven collaboration. SCANNER developed a comprehensive service-oriented framework for mapping policy requirements into network software and data operations,4 and demonstrated ability to interoperate with institutions that were not in its initial member list. SCANNER services were adopted in interventional comparative effectiveness trials led by the University of Southern California (USC) team and funded by the National Institute on Aging (NIA). Four interventional studies (three randomized trials and one quasi-randomized trial) have been carried out on the network. Through these studies, SCANNER's data model was further developed to capture important interventional variables and economic outcomes. Sites included three federally qualified health systems in the Los Angeles area: AltaMed, QueensCare Family Clinics, and The Children's Clinic of Long Beach. The SCANNER team has collaborated with other networks in the development and/or utilization of standards and tools (eg, data model development of OMOP V.4.0 in collaboration with investigators from SAFTINet5).
Use cases and participatory research
Since 2010, components of SCANNER technology have facilitated scaling from the original AHRQ- and NIA-sponsored studies to an additional intervention study involving patient-reported data in an obesity cohort at QueensCare Family Clinics and an independently funded Center for Medicare & Medicaid Innovation (CMMI) study targeting patients with congestive heart failure and patients with high body mass index. The SCANNER team also completed engagement research that included patients (six focus groups,6 California statewide survey of consumers, VA patient surveys, patient navigator preliminary surveys, as well as a national survey). Findings from these studies informed the approach to patient engagement in governance and in prioritization of research questions that will be used in pSCANNER. Initial use cases in pSCANNER will focus on three conditions: (1) congestive heart failure; (2) obesity; (3) Kawasaki disease. Kawasaki disease, a rare disease, is an acute vasculitis that causes heart disease in children and young adults.7 Patients/caregivers, clinicians, researchers, and administrators who represent these conditions will be recruited from participating clinical sites and advocacy and patient organizations to participate at multiple levels from stakeholder input, to advisory board, to national committee. Patient leaders, such as the patient co-chair of the steering committee and patient co-chairs of the advisory board, will be key decision-makers in the development of guidelines that will apply to all three conditions for the whole network. They will also help design the best approaches for recruiting and engaging patients at all levels of governance. One example of our engagement process is the use of a systematic approach to achieve consensus on identification and prioritization of research questions to be studied in the network. We will apply the RAND/UCLA modified Delphi Appropriateness Method, a deliberative and iterative approach to attaining consensus through discussion and feedback.8 While in-person engagement may be desirable, it is often not pragmatic: events can be time- and cost-prohibitive for larger groups of stakeholders. Figure 1 depicts our planned method of engagement. To convene large (n=360) regional or national samples of stakeholders representing patients, clinicians, and researchers across the three different health conditions, we will apply an online Delphi process. Deliberative research has shown that offering data on opinions from subgroups of discussants as well as the overall group9 can facilitate more effective conclusions. We will use RAND's online Delphi consensus management system, called ExpertLens,10 which allows stakeholders with different expertise to weigh in on all questions in order to float issues that might not be considered by a less diverse group. The system has been used in mixed stakeholder groups, including patients and clinicians, and found to facilitate rapid consensus panels for selecting research questions with large groups of stakeholders with much greater efficiency than the traditional Delphi process.11 Using this approach, pSCANNER will have the ability to capture consensus input from a large group of participating stakeholders in a systematic manner to drive research priorities.
Figure 1.
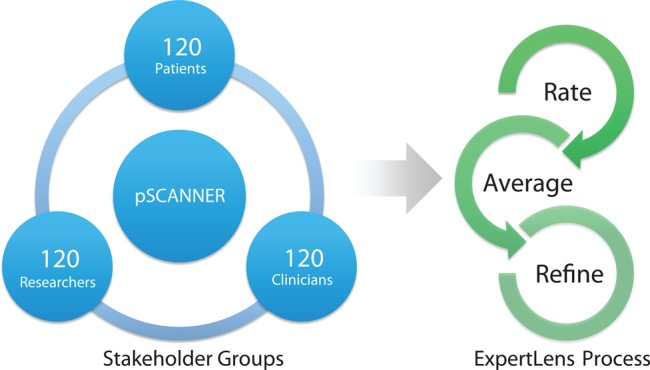
Stakeholder engagement. Stakeholders from across patient-centered Scalable National Network for Effectiveness Research (pSCANNER) sites (360 total) will be recruited to participate in a three-round ExpertLens process, which will prioritize research questions that should be addressed by pSCANNER. In round 1, participants will rate different research priorities and research questions. In round 2, medians and quartiles of group responses to each question will be presented to the participants. In round 3, participants will be asked to modify their round 1 responses based on round 2 feedback and discussion.
Standards for representing data elements and data processing
pSCANNER will adhere to recognized terminologies, including meaningful use or Centers for Medicare & Medicaid Services (CMS) billing terminology standards for diagnosis codes (The International Classification of Diseases, Ninth Revision, Clinical Modification (ICD-9-CM), ICD-10-CM or SNOMED Clinical Terms (SNOMED CT)), procedures and test orders (Current Procedural Terminology (CPT) or Healthcare Common Procedure Coding System (HCPCS)), and medications (National Drug Code (NDC) or RxNorm). In some cases, the native encoding for laboratory information is Logical Observation Identifiers Names and Codes (LOINC); in other cases, these mappings are translated and maintained as part of research data warehouses.
Federally incentivized standards for terminologies and structures have been established for communicating single records; indeed, patients themselves can request these digital ‘continuity of care documents’ and relay them to Clinical Data Research Networks (CDRNs) or Patient-Powered Research Networks (PPRNs). However, multisite PCOR analysis requires communicating rules for processing raw population-level data into prepared analytic datasets. There are emerging standards from Health Level Seven (HL7), the Health Quality Measures Format (HQMF), and Quality Reporting Document Architecture (QRDA) specifications for datasets and abstract data processing rules for electronic quality measures. These standards have not yet become fully integrated into federally incentivized data policy. In SCANNER, we adopted a syntax that is compatible with the HQMF and QDRA because these standards have some preliminary endorsement by the National Quality Forum, CMS, and Office of the National Coordinator as a means of specifying population-level datasets for electronic quality measures. pSCANNER datasets will be specified in this syntax, which can then be interpreted by an adapter to generate executable queries (SCANNER implemented an adapter for OMOP V.4.012). All institutions in pSCANNER have agreed to standardize their data model to OMOP and to install a pSCANNER node to allow distributed computing, which greatly enhances distributed count query capabilities into multivariate analytics.
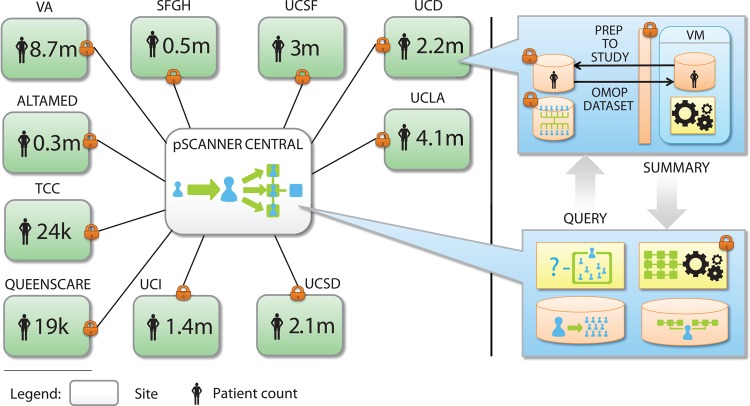
Figure 2 illustrates how pSCANNER will operate. The standard operating procedures for data harmonization will include well-defined steps for data modeling and quality control using tools that have been developed and continue to be developed by the OMOP data management collaborative.13–16 All steps will be published in a standard format to ensure that the data within the network will adhere to standard operating procedures, and that they can be easily shared with other members of PCORnet that adopt OMOP or similar models. Members of pSCANNER are active participants in the Data Quality Assessment collaborative (http://repository.academyhealth.org/dqc/), and have developed additional tools for quality auditing and assessing validity and fitness for use in both research and other secondary uses of population-level data.17–19
Figure 2.
Patient-centered Scalable National Network for Effectiveness Research (pSCANNER) architecture. pSCANNER is a clinical data research network that will integrate over 21 million patients. It will use privacy and security tools to enable distributed analysis of data while keeping data in their host institutions and adhering to all applicable federal, state, and institutional policies. k, thousand; m, million; OMOP, Observational Medical Outcomes Partnership; QueensCare, QueensCare Family Clinics; SFGH, San Francisco General Hospital; TCC, The Children's Clinic of Long Beach; UCD, University of California, Davis; UCI, University of California, Irvine; UCLA, University of California, Los Angeles; UCSD, University of California San Diego; UCSF, University of California, San Francisco; VA, Veterans Affair; VM, Virtual Machine.
Privacy, policy, and technology
pSCANNER addresses institutional policies and patient preferences for data sharing by leveraging recent privacy policy study findings in its technology design and implementation. SCANNER's original partners consisted of institutions with highly diverse policies related to the use of EHRs for research. We produced a comprehensive comparison of the legal requirements and differences among federal and state regulations for states involved in SCANNER.13 We carefully documented health system privacy requirements obtained from institutional documents as well as interviews with system leaders. Additionally, we interviewed patients and clinicians to understand their preferences towards individual privacy.6 20 The SCANNER team described the security and privacy standards used in CDRNs.21 We also conducted a systematic review of privacy technology used in CDRNs.3 We codified data-sharing policies13—each institution specified the policies to which it should adhere. For example, institutions that could share patient-level data were welcome to do so, while institutions that had to keep their patient-level data within their firewalls could share aggregate data such as coefficient estimates, which are equally useful in building multivariate models that span all institutions.22 23 As other institutions joined the network, only new policies were encoded.
We have architected pSCANNER so that authorized users can use a privacy-preserving distributed computation model and research portal that was successfully piloted in SCANNER. This portal includes a study protocol and policy registry—for each study, sites approve specific analytic tools, datasets, and protocols for data privacy and security. Role-based access controls (RBACs) corresponding to federal, state, institutional, and study-specific policies are encoded in the registry and enforced through SCANNER data access services. SCANNER's main goal was to develop a set of highly configurable, computable policies to control data access and to develop effective methods to perform multisite analyses, without necessarily transferring patient-level data.3 22 24 25 Traditional approaches to multisite research involve transferring patient-level data to be pooled for inferential statistics (eg, multivariate regression). While this can be supported, we conduct comparative effectiveness research with increased efficiency because SCANNER allows distributed regressions, thereby avoiding more complex IRB and data use agreements. SCANNER's analytic library was seeded with analysis tools used and validated in multiple publications,26–29 and incorporated into the Observational Cohort Event Analysis and Notification System (OCEANS) (http://idash.ucsd.edu/dbp-tools#overlay-context=idash-software-tools) and Grid Binary Logistic Regression (GLORE),22 which include multivariate analysis methods that allow model fitting, causal inference, and hypothesis testing. When a study policy is created, the site principal investigator specifies the allowable analytic methods and transfer protocols. In particular, each study's site principal investigator will determine if results must be held locally and approved by a delegated representative prior to release for transfer. Our distributed system allows the construction and evaluation of multivariate models that can be used for statistical process control, data safety monitoring in clinical trials, adjustment for confounders, propensity score matching, risk prediction, and other methods used in PCOR (figure 3).
Figure 3.
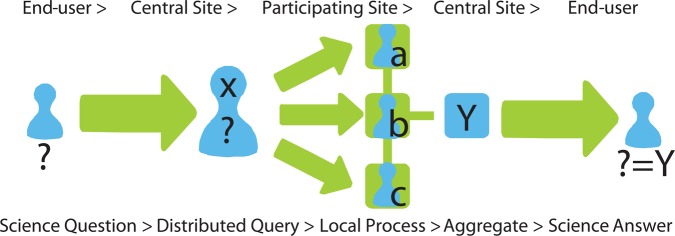
Distributed computing. Patient-centered Scalable National Network for Effectiveness Research (pSCANNER) answers an end-user's scientific question by distributing the corresponding query to each participating site, processing the query locally while preserving each site's stringent data privacy and security requirements, then aggregating the responses into a coherent answer.
Secure access
The pSCANNER network partners have security policies already in place as follows.
The SCANNER Central node is located within a secure Health Insurance Portability and Accountability Act (HIPAA)-compliant environment and will migrate to the platform developed for the NIH-funded integrating data for analysis, anonymization, and sharing center (iDASH), which is now being modified to be Federal Information Security Management Act (FISMA) certified. For studies that require pooling of data, the hub will store the data in iDASH and will use methods that were developed to protect privacy of individuals3 30 31 and institutions from which the data originate.22 23 25 32 Future work in the pSCANNER project will extend these capabilities to include infrastructure necessary for managing randomized clinical trials, including randomization, recruitment, and enrollment tracking systems. These will require modifications to the current RBAC model. Access to this environment is provided through a virtual private network, and we are implementing a two-factor authentication based on the RSA technology, which issues new keys every minute. These one-time keys are displayed on key fobs or through free Apps as soft tokens on smartphones. All protected health information rests behind the firewall of each institution. Requests are received by SCANNER software outside the firewall and transmitted to the Virtual Machine (VM) located inside the firewall according to predefined authorization rules. The SCANNER network software is compliant with National Institute of Standards and Technology RBAC, and applies best practices for RESTful web services to ensure that both data and role-based policy settings are not vulnerable to attack by adversaries.33 34 All communications are fully encrypted end-to-end, and all nodes participating in the network authenticate each other through X.509 certificate exchanges.
UC-ReX uses authentication through the UC campus active directory of the requester. Users have to login through a virtual private network when they are not on campus. Since the network currently only provides results of count queries, all results are provided automatically, with the addition of some noise in the counts35 36 to prevent users from uniquely identifying a specific patient through a series of queries.
VINCI and the VA are transitioning to a two-factor authentication system to authorize users. Currently, centrally managed username and password are required. Users can optionally use personal identification verification cards with passwords to access VA systems. Soon, personal identification verification cards will be required VA wide to access the VA network. Within a protected enclave of the secure VA network, VINCI hosts integrated national data. Access to data is provisioned through an electronic system that matches IRB protocol to datasets. VINCI has 105 high-performance servers and 1.5 petabytes of high-speed data storage with multiple layers of security. The remote computing environment enables data analysis to be performed directly on VINCI servers. Unless explicitly requested and institutionally approved, all sensitive patient data must remain on VINCI project servers. VINCI staff audit for appropriateness all data transfers out of the VINCI enclave.
Summary
Our network of three networks representing multiple health systems and diverse populations embodies the challenges and opportunities that PCORnet itself has to face. While we have based the design of our system on qualitative research and stakeholder engagement, in practice, adoption and success will depend on many factors. For example, pSCANNER will encode a significant portion of policies in software, use a flexible strategy to harmonize data, and use privacy-preserving technology that enables highly diverse institutions to join the network and allow stakeholders to participate. Significant challenges in terms of providing sufficient incentives for patients, clinicians, and health systems to participate and ensuring the sustainability of the network, which were not the focus of this article, will also need to be addressed. The pSCANNER project offers a unique opportunity to make progress toward these objectives, and share results with a community of researchers and representatives from a broader group of stakeholders. It represents a unique opportunity to reaffirm our goals: our health systems have public service as their primary mission, from both a healthcare and an educational perspective. pSCANNER is itself a reflection of this mission, teamwork, and focus on patient outcome research to improve health.
Acknowledgments
The authors would like to thank Gurvaneet Randhawa, Deven McGraw, Erin Holve, and John Mattison for support and guidance provided to the SCANNER project. The authors would also like to thank SCANNER team investigators and staff, Aziz Boxwala, Omolola Ogunyemi, Susan Robbins, Naveen Ashish, Serban Voinea, Grace Kuo, Robert El-Kareh, David Chang, and Natasha Balac.
Footnotes
Collaborators: The pSCANNER team includes the following people: Lahey Clinic: Frederic Resnic; RAND: Dmitry Khodyakov; UC-ReX: Lattice Armstead; UCD: Travis Nagler, Sam Morley, Nicholas Anderson; UCI: Dan Cooper, Dan Phillips; UCLA: David Heber, Zhaoping Li, Michael K Ong, Ayan Patel, Marianne Zachariah; UCSD: Jane C Burns, Lori B Daniels, Son Doan, Claudiu Farcas, Rita Germann-Kurtz, Xiaoqian Jiang, Hyeon-eui Kim, Paulina Paul, Howard Taras, Adriana Tremoulet, Shuang Wang, Wenhong Zhu; UCSF: Douglas Berman, Angela Rizk-Jackson; USC: Mike D'Arcy, Carl Kesselman, Tara Knight, Laura Pearlman; VA Palo Alto and Department of Medicine, Stanford: Paul Heidenreich; VASDHS: Dena Rifkin, Carl Stepnowsky, Tania Zamora; VA SLC: Scott L DuVall, Lewis J Frey, Jeffrey Scehnet, Brian C Sauer, Julio C Facelli, Ram K Gouripeddi; VA TN: Jason Denton, Fern FitzHenry, James Fly, Vincent Messina, Freneka Minter, Lalit Nookala, Heidi Sullivan, Theodore Speroff, Dax Westerman.
Contributors: LO-M wrote the manuscript and edited all versions. ZA, MEM, and JRN wrote the portions related to VHA. DSB, LD, DG, MKK, and MH wrote the portions related to UC-ReX. JND, KKK, and DM wrote the portions related to SCANNER. MED organized the manuscript and edited all versions. The pSCANNER team developed most of the work described in the manuscript, and provided contributions to the cohort description, technical infrastructure, and ELSI components.
Funding: This work is funded by PCORI contract CDRN-1306-04819. Prior work described in this paper was supported by AHRQ grant R01HS019913 and NIA grant 1RC4AG039115-01 as part of the American Recovery and Reinvestment Act, NIH grants U54HL108460, UL1TR000124, UL1TR000004, UL1TR000002, UL1TR000153, UL1TR000100, the EDM Forum, VA grants HSR&D CDA-08-020, HSR&D IIR-11-292, HSR&D HIR 08-204, a grant funded from the University of California Office of the President/UC-BRAID, the Gordon and Betty Moore Foundation grant to the UC Davis Betty Irene Moore School of Nursing, and the California Health and Human Services Agency.
Competing interests: None.
Provenance and peer review: Commissioned; externally peer reviewed.
Contributor Information
Collaborators: Frederic Resnic, Dmitry Khodyakov, Lattice Armstead, Travis Nagler, Sam Morley, Nicholas Anderson, Dan Cooper, Dan Phillips, David Heber, Zhaoping Li, Michael K Ong, Ayan Patel, Marianne Zachariah, Jane C Burns, Lori B Daniels, Son Doan, Claudiu Farcas, Rita Germann-Kurtz, Xiaoqian Jiang, Hyeon-eui Kim, Paulina Paul, Howard Taras, Adriana Tremoulet, Shuang Wang, Wenhong Zhu, Douglas Berman, Angela Rizk-Jackson, Mike D'Arcy, Carl Kesselman, Tara Knight, Laura Pearlman, Paul Heidenreich, Dena Rifkin, Carl Stepnowsky, Tania Zamora, Scott L DuVall, Lewis J Frey, Jeffrey Scehnet, Brian C Sauer, Julio C Facelli, Ram K Gouripeddi, Jason Denton, Fern FitzHenry, James Fly, Vincent Messina, Freneka Minter, Lalit Nookala, Heidi Sullivan, Theodore Speroff, and Dax Westerman
References
- 1.Observational Medical Outcomes Partnership: Common Data Model Specifications Version 4.0. 2012
- 2.Kohane IS, Churchill SE, Murphy SN. A translational engine at the national scale: informatics for integrating biology and the bedside. J Am Med Inform Assoc 2012;19:181–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang X, Sarwate AD, Ohno-Machado L. Privacy technology to support data sharing for comparative effectiveness research: a systematic review. Med Care 2013;51(8 Suppl 3):S58–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim KK, McGraw D, Mamo L, et al. Development of a privacy and security policy framework for a multistate comparative effectiveness research network. Med Care 2013;51(8 Suppl 3):S66–72 [DOI] [PubMed] [Google Scholar]
- 5.Schilling LM, Kwan BM, Drolshagen CT, et al. Scalable Architecture for Federated Translational Inquiries Network (SAFTINet) Technology Infrastructure for a Distributed Data Network. eGEMs 2013;1. http://dx.doi.org/10.13063/2327-9214.1027%5Bpublished Online First: Epub Date [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mamo LA, Browe DK, Logan HM, et al. Patient Informed Governance of Distributed Research Networks: Results and Discussion from Six Patient Focus Groups. Washington, DC: American Medical Informatics Association Annual Symposium, 2013 [PMC free article] [PubMed] [Google Scholar]
- 7.Burns JC, Glode MP. Kawasaki syndrome. Lancet 2004;364:533–44 [DOI] [PubMed] [Google Scholar]
- 8.Dalkey N, Helmer O. An experimental application of the Delphi method to use of experts. Manag Sci 1963;9:428–76 [Google Scholar]
- 9.Rowe G, Wright G. The Delphi technique as a forecasting tool: issues and analysis. Int J Forecast 1999;15:353–75 [Google Scholar]
- 10.Dalal S, Khodyakov D, Srinivasan R, et al. ExpertLens: a system for eliciting opinions from a large pool of non-collocated experts with diverse knowledge. Technol Forecast Soc Change 2011;78:1426–44 [Google Scholar]
- 11.Khodyakov D, Hempel S, Rubenstein L, et al. Conducting online expert panels: a feasibility and experimental replicability study. BMC Med Res Methodol 2011;11:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meeker D, Skeels C, Pearlman L, et al. A System and User Interface for Standardized Preparation of Analytic Data Sets. Bethesda, MD: Observational Medical Outcomes Partnership Annual Symposium, 2013 [Google Scholar]
- 13.Ogunyemi OI, Meeker D, Kim HE, et al. Identifying appropriate reference data models for comparative effectiveness research (CER) studies based on data from clinical information systems. Med Care 2013;51(8 Suppl 3):S45–52 [DOI] [PubMed] [Google Scholar]
- 14.Overhage JM, Ryan PB, Reich CG, et al. Validation of a common data model for active safety surveillance research. J Am Med Inform Assoc 2012;19: 54–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reich C, Ryan PB, Stang PE, et al. Evaluation of alternative standardized terminologies for medical conditions within a network of observational healthcare databases. J Biomed Inform 2012;45:689–96 [DOI] [PubMed] [Google Scholar]
- 16.Ryan PB, Madigan D, Stang PE, et al. Empirical assessment of methods for risk identification in healthcare data: results from the experiments of the Observational Medical Outcomes Partnership. Stat Med 2012;31:4401–15 [DOI] [PubMed] [Google Scholar]
- 17.Kahn MG, Brown J, Dahm L, et al. Pains and palliation in distributed research networks: lessons from the field AMIA summit on clinical research informatics. American Medical Informatics Association, 2013 [PMC free article] [PubMed] [Google Scholar]
- 18.Meeker D, Hussey S, Shetty K, et al. Project Report: Integration of Structured Evidence and Data into eMeasure Development, Office of the National Coordinator for Health Information Technology: Office Of the National Coordinator, 2013
- 19.Meeker D, Pearlman L, Doctor JN. Data quality in multisite research networks. Baltimore, MD: AcademyHealth Annual Research Meeting, 2013 [Google Scholar]
- 20.Kim KK, Browe DK, Logan HC, et al. Data governance requirements for distributed clinical research networks: triangulating perspectives of diverse stakeholders. J Am Med Inform Assoc 2014;21:714–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohno-Machado L, Alipahan N, Day ME, et al. Comprehensive Inventory of Research Networks: Clinical Data Research Networks, Patient-Powered Research Networks, and Patient Registries. 2013. http://www.pcori.org/assets/2013/06/PCORI-Comprehensive-Inventory-Research-Networks-061213.pdf
- 22.Wu Y, Jiang X, Kim J, et al. Grid Binary LOgistic REgression (GLORE): building shared models without sharing data. J Am Med Inform Assoc 2012;19:758–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu Y, Jiang X, Ohno-Machado L. Preserving Institutional Privacy in Distributed binary Logistic Regression. AMIA Annu Symp Proc 2012;2012:1450–8 [PMC free article] [PubMed] [Google Scholar]
- 24.Ohno-Machado L. To share or not to share: that is not the question. Sci Transl Med 2012;4:165cm15. [DOI] [PubMed] [Google Scholar]
- 25.Wang S, Jiang X, Wu Y, et al. EXpectation Propagation LOgistic REgRession (EXPLORER): distributed privacy-preserving online model learning. J Biomed Inform 2013;46:480–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matheny ME, Morrow DA, Ohno-Machado L, et al. Validation of an automated safety surveillance system with prospective, randomized trial data. Med Decis Making 2009;29:247–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matheny ME, Normand SL, Gross TP, et al. Evaluation of an automated safety surveillance system using risk adjusted sequential probability ratio testing. BMC Med Inform Decis Mak 2011;11:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Resnic FS, Gross TP, Marinac-Dabic D, et al. Automated surveillance to detect postprocedure safety signals of approved cardiovascular devices. JAMA 2010;304:2019–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang W, Li P, Wang S, et al. WebGLORE: a web service for Grid LOgistic REgression. Bioinformatics 2013;29:3238–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gardner J, Xiong L, Xiao Y, et al. SHARE: system design and case studies for statistical health information release. J Am Med Inform Assoc 2013;20:109–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohammed N, Jiang X, Chen R, et al. Privacy-preserving heterogeneous health data sharing. J Am Med Inform Assoc 2013;20:462–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Que J, Jiang X, Ohno-Machado L. A collaborative framework for Distributed Privacy-Preserving Support Vector Machine learning. AMIA Annu Symp Proc 2012;2012:1350–9 [PMC free article] [PubMed] [Google Scholar]
- 33.Pautasso C, Zimmerman O, Leymann F. RESTful web services vs. “big” web services: making the right architectural decision. 17th international conference on World Wide Web, New York, NY, 2008 [Google Scholar]
- 34.Serme G, de Oliveira AS, Massiera J, et al. Enabling message security for RESTful services. Web Services (ICWS), 2012 IEEE 19th International Conference on; 24–29 June 2012; Honolulu, HI, 2012 [Google Scholar]
- 35.Boxwala AA, Kim J, Grillo JM, et al. Using statistical and machine learning to help institutions detect suspicious access to electronic health records. J Am Med Inform Assoc 2011;18:498–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murphy SN, Gainer V, Mendis M, et al. Strategies for maintaining patient privacy in i2b2. J Am Med Inform Assoc 2011;18(Suppl 1):i103–8 [DOI] [PMC free article] [PubMed] [Google Scholar]