Abstract
The Greater Plains Collaborative (GPC) is composed of 10 leading medical centers repurposing the research programs and informatics infrastructures developed through Clinical and Translational Science Award initiatives. Partners are the University of Kansas Medical Center, Children's Mercy Hospital, University of Iowa Healthcare, the University of Wisconsin-Madison, the Medical College of Wisconsin and Marshfield Clinic, the University of Minnesota Academic Health Center, the University of Nebraska Medical Center, the University of Texas Health Sciences Center at San Antonio, and the University of Texas Southwestern Medical Center. The GPC network brings together a diverse population of 10 million people across 1300 miles covering seven states with a combined area of 679 159 square miles. Using input from community members, breast cancer was selected as a focus for cohort building activities. In addition to a high-prevalence disorder, we also selected a rare disease, amyotrophic lateral sclerosis.
Keywords: PCORI, Data Warehouse, CTSA, Clinical Research, Comparative Effectiveness, Patient Centered
The Greater Plains Collaborative (GPC) brings together over 10 million covered lives and encompasses over 20 hospitals, 700 clinic locations, and 8000 providers responsible for tertiary and quaternary care in most regions, primary care for specific populations, and comprehensive management and follow-up for patients with rare diseases (such as amyotrophic lateral sclerosis (ALS)) and for those with our selected common disease, breast cancer. Of these, over 6 million have data, such as laboratory results, medications, vital signs and diagnoses, maintained in electronic health records (EHRs). This population covers the spectrum from primary care networks serving rural and small communities to urban populations with significant African-American and Hispanic representation. The centers at Wisconsin, Kansas, Nebraska, and Minnesota also have active liaisons with their respective state’s Native American populations. With one current exception, all of our sites include comprehensive pediatric as well as adult care (Children's Mercy Hospital).
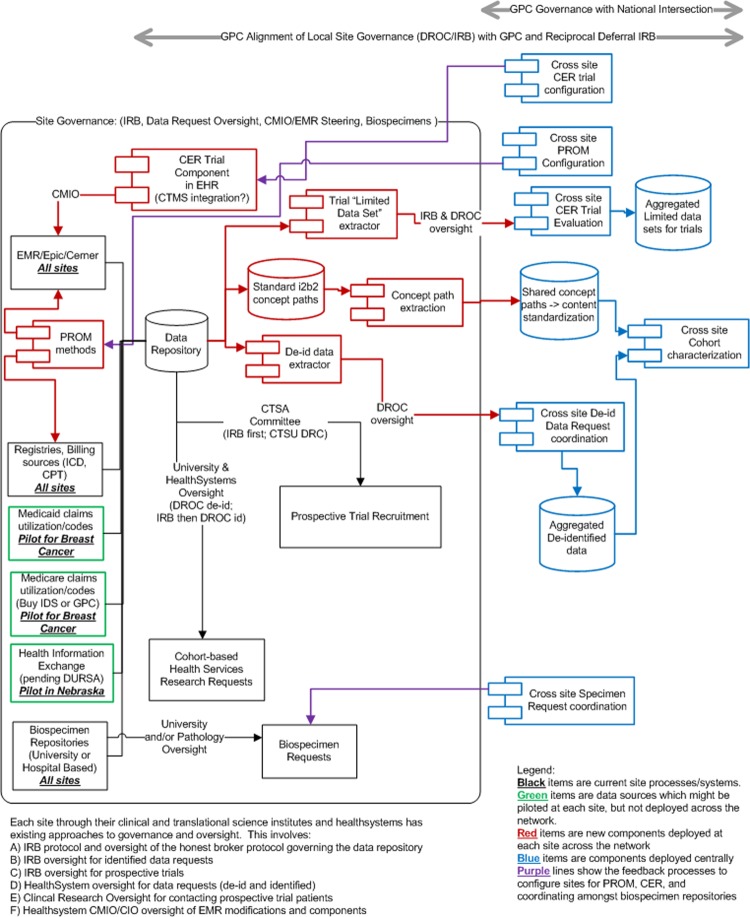
Figure 1 illustrates the GPC's data sources, technical components, and governance. Each site in the GPC has existing processes and governance between their research and healthcare system organizations to support clinical trials and the reuse of health record data for research. Existing resources are shown in black, new site data sources that can supplement longitudinal data capture in green, new components to be deployed at the sites in red, and GPC-level data stores and components in blue.
Figure 1.
Greater Plains Collaborative network components. Existing resources are shown in black, new site data sources that can supplement longitudinal data capture in green, new components to be deployed at the sites in red, and GPC-level data stores and components in blue.
Site-level governance is an essential part of the GPC and involves:
Institutional review boards (IRBs), which oversee identified data requests and prospective trials
Data request oversight committees, which incorporate healthcare system and university oversight of data requests and approve fully deidentified data requests that are classified as being outside the scope of human subject research by the IRB. After data requests are approved, a neutral member of the informatics team, the ‘honest broker’, extracts data from the data repository for the researcher
University- and hospital-based biospecimen resource request processes governing the release of samples
Healthcare system EHR steering committees, which oversee the configuration and standardization of clinical systems
Clinical and translational science committees, which govern the use of registries for prospective trial recruitment
There are three potential additional areas (green in figure 1) for incorporating data: health information exchanges (HIEs), Medicare claims data on care received outside our health systems and available to accountable care organizations, and state Medicaid claims.
To create a highly productive and responsive network, the GPC will integrate the following components (red in figure 1) at each site with our existing EHRs, i2b2 (Informatics for Integrating Biology and the Bedside) data repositories, data-capture systems, and personal health records used for patient registries and engagement:
Data standardization: the concept paths used by i2b2 to describe observations and findings will be harvested along with usage statistics to share at the GPC level
Deidentified dataset extraction: for cohort characterization, we have developed a lightweight i2b2 plug-in to be used by each site's honest broker to extract cohort datasets and securely transfer them to the GPC data store for analysis
Patient-reported outcome measures (PROMs): standardized measures will be deployed using either EHR patient portals or data collection instruments for existing registry and research management systems such as REDCap (Research Electronic Data Capture)
Comparative effectiveness research (CER) trial components: we will configure CER trial components directly in the EHR (preferred) or integrate existing data capture and trial management systems such as REDCap, Velos, and OnCore because of either limited EHR build team capacity or flexibility to efficiently iterate prototypes
Limited dataset extraction: methods similar to 2 above but CER trials will require precise dates and times to monitor accrual and performance.
It is important for a new network to start off by building trust and therefore we will initially limit the data handled at the network level. We will focus attention on establishing governance and interoperability and allow trust among our sites to develop. The first steps involve establishing an overall master data-sharing agreement. It will be based on examples of existing University of Kansas Medical Center (KUMC) data-sharing agreements. We also plan to develop an IRB-reciprocal deferral model for the network. We also will deploy the following activities/components at the GPC level (blue in figure 1):
An i2b21 ontology database: to store the terminologies used at each site, but not patient data. We will harvest the terms used at the sites and statistics of the number of facts and patients observed for each term. This will allow us to measure overall network alignment with national standards, and to map and monitor processes to increase data harmonization in an iterative manner.
Data request oversight tools: based on KUMC REDCap2-based tools for use by the GPC-level oversight of data requests, biospecimen requests, and tracking CER trial approvals. These tools allow an authenticated faculty to sponsor student/staff access and submit data use requests via REDCap surveys, which are then reviewed by hospital, clinic, and university oversight officials, who use organization-specific case report forms to approve access and data requests. A final case report form is used by the honest broker to track data fulfillment, the i2b2 queries used to define the cohort and data elements, and the patients included in the released datasets.
A REDCap data store and RStudioServer3 analysis suite: to maintain aggregate deidentified datasets for cohort characterization.
A development environment: to configure generalized patient-reported outcome modules within EHRs and research registry tools at the sites. For prototyping measures in REDCap, the common instance for deidentified data will be used. For modules deployed using the EHRs, a development environment will be configured with a site's EHR.
A development environment: to configure generalized CER modules to be deployed in the EHRs at the sites. Where applicable, this will use the common REDCap and RStudioServer environments but augmented by GPC development environments for services (web services/interface engines) and stubs for application programming interfaces to site EHRs.
A REDCap data store and RStudioServer analysis suite: to maintain aggregate limited datasets for monitoring prospective CER trials. We will work with the Patient-Centered Outcomes Research Institute (PCORI) and the national coordinating center to align these tools with national objectives and trial design considerations.
Since 2011, the KUMC medical informatics team has distributed the open-source HERON4 framework for migrating transactional data into a vendor-neutral i2b2 data warehouse. This is valuable to collaborators using the Epic EHR, as well as organizations that use standard datasets (eg, North American Association of Central Cancer Registries (NAACCR) tumor registries, the Social Security Administration's Data Master File for mortality, the University HealthSystem Consortium Clinical Data Base). The team has devised methods5 for incorporating locally developed REDCap research registries within the i2b2 environment and for integrating preliminary statistical analysis into the i2b2 framework using the open-source R language.6 This integration will be used for data exchange between GPC sites and GPC data stores.
The GPC sees PCORI's Clinical Data Research Networks (CDRNs) as a test of the nation's multi-billion dollar investment in EHRs. While the Office of the National Coordinator requires attestation that a provider organization has implemented a certified EHR, there has been little quantitative measurement regarding the degree to which the data contained in EHRs are capable of being used to measure clinical effectiveness. While we laud the efforts to create HIEs, there is concern that such exchanges may devolve to the lowest common denominator of interoperability and lack the rich detail and structure required to support research. In contrast, the GPC sites obtain the underlying detailed observations recorded directly in EHR and billing systems, standardized registries, biorepository databases, and supplemental electronic data-capture methods. CDRNs will provide a true test of our emerging national learning healthcare system by developing targeted trials for specific clinical populations and outcomes. The GPC network standards will adhere to national and international data standards specified by the nationwide health information network (NwHIN)7 and subsequent guidance provided by the Office of the National Coordinator for Health Information Technology and outlined by meaningful use stage 2 (MU28) and stage 3 criteria.
We use i2b2 as a common data model to consolidate data from (a) EHRs, (b) administrative ‘billing’ data and derived benchmarking datasets such as the University HealthSystem Consortium Clinical Data Base (UHC CDB),9 (c) research registries (eg, Tumor Registries) and (d) PROMs (prototyped in REDCap). We bind both the internal EHR concept codes and mapped code sets to standard terminologies into i2b2 so we can quantitatively measure MU2 attainment based on both concept coverage and the amount of observed data.
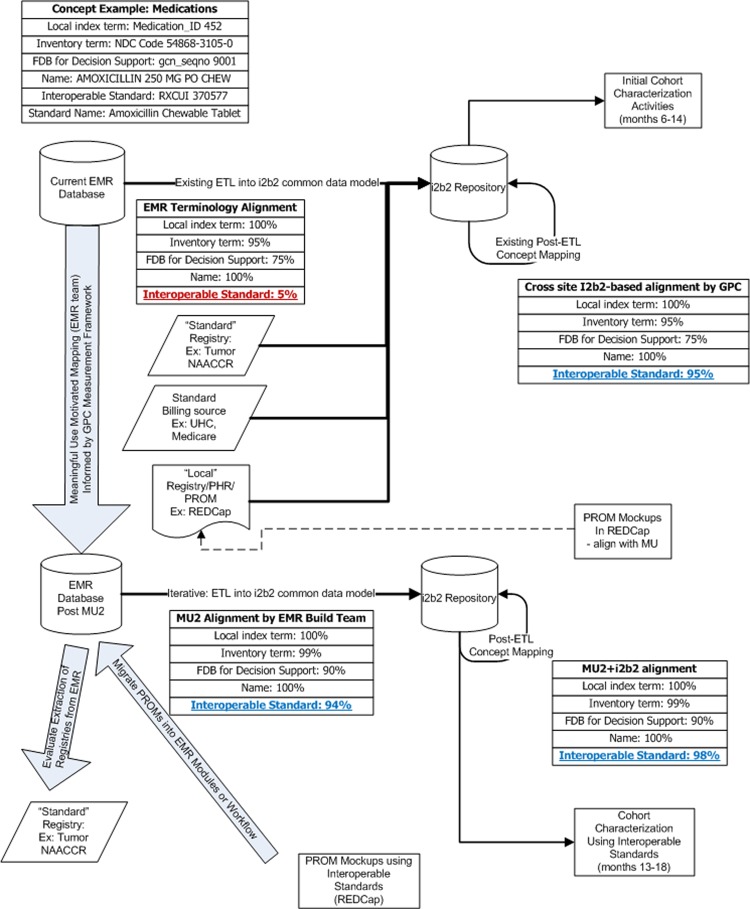
Figure 2 provides on an example of an amoxicillin chewable tablet concept which has an internal code (452). This code has mappings to a First Databank code (gcnseqno 9001) for allergy checking, as well as one to many relationships to various National Drug Codes (NDCs) stocked by the pharmacies (NDC 54868-3105-0 manufactured by Physicians Total Care; NDC 0093-2268-01 manufactured by Teva USA). An EHR may have 5% of its medication formulary aligned with interoperable standard medication terminology. As an interim technique, these data can still be integrated for multisite queries by using the flexibility of the i2b2 data model to map local terminology codes to interoperable standards. Using existing concept mapping techniques, as well as select manual mappings illustrated in figure 3, the i2b2 common data are expected to achieve 95% alignment. In our example, the medication concept is mapped to an RxNorm Semantic Clinical Drug Form (RXCUI 370577),10 facilitating query across different dispense sizes.
Figure 2.
Greater Plains Collaborative (GPC) interoperable standardization measurement framework.
Figure 3.

Flexible terminology mappings in Informatics for Integrating Biology and the Bedside (i2b2).
We will share our findings and measurement framework with the chief medical information officers who must configure the EHRs to comply with MU2. The timing of federal incentive payments will catalyze this activity. Once the MU2 work is complete, we might see 94% alignment natively within the EHR, enabling deployment of standardized CER trial components and PROMs within the clinical workflow. By incorporating existing standard research registries, such as the NAACCR tumor registry, we can also directly evaluate MU2-compliant EHRs’ and billing systems’ abilities to represent existing research information models. Breast cancer provides an ideal exemplar.
This work is possible because of the increasing support of the NwHIN domain model by EHR vendors. The GPC network will allow us to design around Epic EHR considerations and then to generalize our approach to two other EHR systems (Cerner at Children's Mercy and Cattails MD at the Marshfield Clinic). For diagnoses, a vendor, Intelligent Medical Objects, provides mapping for SNOMED CT11 and International Classification of Diseases—Clinical Modification (ICD-*-CM)12 coding of diagnoses and history. However, mapping of family history, allergy records, findings, and procedures to SNOMED CT will be required. For medication orders and prescriptions, Epic partners First Data Bank on mappings to RxNorm. RxNorm is in varying stages of deployment among our sites. All sites are responsible for mapping of immunizations to Centers for Disease Control and Prevention (CDC) Vaccines Administered (CVX) codes.13 14 For laboratory results, LOINC15 is installed within Epic at each site's discretion and requires mapping at the site of laboratory master tables to LOINC for query access to coded LOINC results as well as deployment in any enterprise laboratory information systems. For procedures, Epic provides and supports coding to ICD-*-CM, ICD-*-PCS (Procedure Coding System), CPT (Current Procedural Terminology) and Healthcare Common Procedure Coding System (HCPCS) as part of the model system, with sites responsible for installing yearly updates.
Benchmarking activities derived from administrative/billing data sources and national registries provide additional sources of data for the GPC. Specifically, the UHC CDB provides rich, standardized diagnoses, encounter details, and outcomes derived from billing, while the NAACCR—used by tumor registrars—provides established mechanisms for characterizing the breast cancer population predominantly codified by the International Classification of Diseases for Oncology.16 Targeting these standardized datasets allows us to create an ETL (extract, transform, load) process, which benefits the national network, is vendor agnostic, and will enable direct comparison of EHR-derived network capabilities with administrative data and abstracted research registries traditionally used in health service research.17–20 This also complements efforts at sites to incorporate billing information with that from financial systems. Further, we will develop ETL processes for incorporating Medicare and Medicaid claims data into i2b2 based on standard data formats.
EHRs’ functionality or the capacity of healthcare system information technology teams to collect PROMs via patient portals or to integrate with patients’ personal health records is often a lower priority than documentation during the healthcare encounter. Rare diseases (such as ALS) and even common conditions (such as breast cancer) struggle to have data elements (eg, performance status for cancer) captured discretely in the EHR. We will use REDCap, deployed at all GPC sites, as a simple user interface to prototype codification of data-capture instruments such as PROMs and the National Institute for Neurological Disorders ALS common data elements.
We do not see a requirement for real-time interfaces between sites and the GPC centrally to fulfill the initial objectives. Instead, site honest brokers will use an open-source i2b2 plug-in, RDataBuilder, to extract data into a standardized R data frame. We believe this honest-broker-mediated approach will suffice for initial development of CER trial monitoring, but we remain open to developing more scalable approaches grounded in practical experience from running trials as part of the national network.
Footnotes
Contributors: All authors were involved in contributing to the communication narrative, editing, and the proposal design.
Funding: This project is supported by PCORI contract CDRN-1306-04631 and in part by NIH grant UL1TR000001.
Competing interests: None.
Provenance and peer review: Commissioned; internally peer reviewed.
References
- 1.Murphy SN, Weber G, Mendis M, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2). J Am Med Inform Assoc 2010;17:124–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harris PA, Taylor R, Thielke R, et al. Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009;42:377–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Racine JS. RStudio: a platform-independent IDE for R and Sweave. J Appl Econ 2013;27:167–72 [Google Scholar]
- 4.Waitman LR, Warren JJ, Manos EL, et al. Expressing observations from electronic medical record flowsheets in an i2b2 based clinical data repository to support research and quality improvement. AMIA Annu Symp Proc 2011;2011:1454–63 [PMC free article] [PubMed] [Google Scholar]
- 5.Adagarla B, Connolly DW, Nair M, et al. Integrating REDCap patient registries within an i2b2 integrated data repository, 2013 AMIA clinical research informatics joint summit.
- 6.The R Project for Statistical Computing. http://www.r-project.org/ (accessed 25 Sep 2013)
- 7.Nationwide Health Information Network Policy Researchers & Implementers. http://www.healthit.gov/policy-researchers-implementers/nationwide-health-information-network-nwhin (accessed 25 Sep 2013)
- 8.What is Meaningful Use? Policy Researchers & Implementers. http://www.healthit.gov/policy-researchers-implementers/meaningful-use (accessed 25 Sep 2013)
- 9.UHC Clinical Data Base/Resource Manager. https://www.uhc.edu/11536.htm (accessed 25 Sep 2013)
- 10.U.S. National Library of Medicine. RxNorm. http://www.nlm.nih.gov/research/umls/rxnorm/ (accessed 25 Sep 2013) [DOI] [PubMed]
- 11.International Health Terminology Standards Development Organization. SNOMED CT. http://www.ihtsdo.org/snomed-ct/snomed-ct0/ (accessed 25 Sep 2013)
- 12.ICD—Classifications of Diseases, Functioning, and Disability. http://www.cdc.gov/nchs/icd.htm (accessed 25 Sep 2013)
- 13.Center for Disease Control and Prevention. Immunization Information Systems. http://www.cdc.gov/vaccines/programs/iis/code-sets.html and http://www.cdc.gov/vaccines/programs/iis/code-sets.html (accessed 25 Sep 2013)
- 14.Center for Disease Control and Prevention. HL7 Standard Code Set: CVX—Vaccines Administered. http://www2a.cdc.gov/vaccines/iis/iisstandards/vaccines.asp?rpt=cvx (accessed 25 Sep 2013)
- 15.McDonald CJ, Huff SM, Suico JG, et al. LOINC, a universal standard for identifying laboratory observations: a 5-year update. Clin Chem 2003;49:624–33 [DOI] [PubMed] [Google Scholar]
- 16.World Health Organization. International Classification of Diseases for Oncology, 3rd Edition. http://www.who.int/classifications/icd/adaptations/oncology/en/ (accessed 25 Sep 2013)
- 17.KU Medical Center Medical Informatics. UHC Source. https://informatics.kumc.edu/work/wiki/UHCSource (accessed 25 Sep 2013)
- 18.KU Medical Center Medical Informatics. UHC SQL. https://informatics.kumc.edu/work/browser/heron_load/uhc_i2b2_transform.sql (accessed 25 Sep 2013)
- 19.KU Medical Center Medical Informatics. Tumor Registry. https://informatics.kumc.edu/work/wiki/TumorRegistry (accessed 25 Sep 2013)
- 20.KU Medical Center Medical Informatics. NAACCR SQL. https://informatics.kumc.edu/work/browser/heron_load/naaccr_txform.sql (accessed 25 Sep 2013)