Abstract
The anticancer antibiotic chromomycin A3 (Chro) is a DNA minor groove binding drug belonging to the aureolic family. Chro likely exerts its activity by interfering with replication and transcription. Chro forms a dimer, mediated by a divalent metal ion, which binds to G/C-rich DNA. Herein we report the first crystal structure of Chro bound to d(TTG GCCAA)2 DNA duplex solved by multiwavelength anomalous diffraction (MAD) based on the chelated Co3+ ion. The structure of the Mg2+ complex was subsequently refined at 2.15 Å resolution, which revealed two complexes of metal-coordinated dimers of Chro bound to the octamer DNA duplex in the asymmetric unit. The metal ion is octahedrally coordinated to the O1 and O9 oxygen atoms of the chromophore (CPH), and two water molecules act as the fifth and sixth ligands. The two coordinated water molecules are hydrogen bonded to O2 atoms of C5 and C13 bases. The Chro dimer binds at and significantly widens the minor groove of the GGCC sequence. The long axis of each chromophore lies along and stacks over the sugar–phosphate backbone with the two attached saccharide moieties (rings A/B and C/D/E) wrapping across the minor groove. DNA is kinked by 30° and 36° in the two complexes, respectively. Six G-specific hydrogen bonds between Chro and DNA provide the GGCC sequence specificity. Interestingly, DNA in concert with Chro appears to act as an effective template to catalyze the deamination of Co(NH3)63+, as shown by circular dichroism and crystal structure data. Our results present useful structural information for designing new anticancer drug derivatives in the future.
INTRODUCTION
The anticancer antibiotics chromomycin A3 (Chro) and mithramycin (plicamycin®) (Mith) belong to the aureolic family isolated from Streptomyces griseus and Streptomyces plicatus, respectively (1). Both drugs are DNA-binding antitumor agents and have been used clinically in cancer therapies (2–4). These two drugs contain five sugar rings connected to an aglycon [chromophore (CPH)] via O-glycosidic bonds, with the A-B disaccharide on one side while the C-D-E trisaccharide is on the other (Fig. 1A). Their antitumor properties may be attributed to their inhibitory effects on replication and transcription of tumor cells (3,5). Chro and its analog Mith are potent inhibitors of neuronal apoptosis induced by oxidative stress or the anticancer drug camptothecin, where mechanisms were proposed to be due to the inhibition of the binding of transcription factors Sp1 and Sp3 to their cognate ‘G-C’ box (6). Bianchi et al. demonstrated that the Chro and Mith were powerful inducers of erythroid differentiation of K562 cells by binding to the human γ-globin promoter and altering the pattern of protein binding to this promoter (7). These different cellular responses may be related to the binding mechanisms of these two drugs to DNA.
Figure 1.
(A) Chemical structure of chromomycin A3 (Chro) and mithramycin (Mith). (B and C) CD spectra of Chro in the presence of Mg2+, which varied in concentration from 0 to 8 mM without (B) and with (C) d(TTGGCCAA)2. The drug concentration (monomer) was 40 µM. (D) Comparison of the CD spectrum of d(TTGGCCAA)2 (solid line), and the normalized sum of the CD spectrum of [(Chro)2-Mg2+]–d(TTGGCCAA)2 minus [(Chro)2-Mg2+] (dashed line) in Tris–HCl (pH 7.3) plus 100 mM NaCl at room temperature. The DNA duplex concentration was 20 µM.
Optical spectroscopy revealed that both drugs form a dimer mediated by a single divalent metal ion, such as Mg2+ (8,9). The ion-coordinated drug dimer is bound to a widened minor groove centered around the G/C-rich sequences (10–14). Footprinting studies have also shown that both Chro and Mith require a G-C binding site that is at least three base pairs long (5,10). In addition to Mg2+, other divalent cations with a radius <0.85 Å are candidates for drug dimer formation (15,16). The solution structures of both complexes including the Mg2+-coordinated Chro–DNA complex (two drug equivalents per duplex) have been analyzed by nuclear magnetic resonance (NMR) spectroscopy (17–20). However, the precise coordination geometry around the metal ion remains unclear.
Herein we describe the first crystal structure of Chro bound to DNA d(TTGGCCAA)2 duplex, which reveals that the metal-coordinated dimer of Chro kinks DNA from the minor groove side and induces the local duplex conformational change. The metal ion in the complex has an octahedral coordination, with two water molecules acting as the fifth and sixth ligands. In addition, we show that the binding of Chro to DNA caused the deamination of Co(NH3)63+. Thus, DNA acts as an effective template to catalyze the deamination. Our structural analysis provides a basis upon which to design a novel anticancer drug.
MATERIALS AND METHODS
The synthetic DNA oligonucleotides were purified by gel electrophoresis. Chro and Mith were purchased from Sigma Chemical Co. (St Louis, MO). Absorbance measurements were carried out in a quartz cuvette using a Hitachi U-2000 spectrophotometer. The concentrations of Mith and Chro were estimated using extinction coefficients of 10 000 M–1cm–1 at 400 nm and 8800 M–1cm–1 at 405 nm, respectively. The circular dichroism (CD) spectra were collected between 520 and 200 nm, with bandwidth at a 1 nm interval using a JASCO-720 CD spectropolarimeter. All spectra were the average of five runs. The method of CD spectral analysis was as described previously (21,22).
Crystallizations of Mg2+- and Co3+-chelated Chro–DNA complexes were performed using the vapor diffusion method (23). The Mg2+-form crystals were obtained from solutions containing 2.5 mM DNA (single strand), 5 mM Chro, 25 mM sodium-cacodylate buffer (pH 5.0) and 12.5 mM MgCl2, equilibrated at 4°C against 500 µl of 50% 2-methyl-2,4-pentanediol (MPD). The cobalt-derivative crystals were obtained from solutions containing 0.94 mM DNA (single strand), 1.1 mM Chro, 37.7 mM sodium-cacodylate buffer (pH 6.0), 2.4 mM cobalt(III)-hexaammine, 4.7 mM MgCl2 and 2.8 mM spermine, equilibrated at 4°C against 30 ml 40% PEG400 solution. The crystallographic statistics are listed in Table 1.
Table 1. Crystallographic and refinement data of magnesium- and cobalt-derived from Chro–DNA complexes.
| λ for data collection (Å) | 1.5342 | 1.60652 | 1.60576 | 1.51202 |
|---|---|---|---|---|
| Inflection point | Peak | High remote | ||
| Crystallographic data | ||||
| Metal | Mg2+ | Co3+ | Co3+ | Co3+ |
| a = b (Å) | 42.23 | 42. 01 | 42.01 | 42.01 |
| c (Å) | 246.11 | 240.26 | 240.26 | 240.26 |
| Space group | P6522 | P6522 | P6522 | P6522 |
| Resolution (Å) | 50–2.15 | 50–3.0 | 50–3.0 | 50–3.0 |
| No. of reflections [>0 σ(I)] | 7815 | 6683a | 6712a | 6724a |
| <I/σ(I)> | 6 | 22 | 22 | 22 |
| Rmerge (%) | 5.5 (7.0)b | 8.8 | 10.4 | 8.9 |
| Completeness (%) | 90.5 (80)b | 99.3 | 99.5 | 98.9 |
| Refinement data | ||||
| R/Rfree (5% data) | 0.235/0.278 | |||
| Bond length (Å) | 0.022 | |||
| Bond angle (°) | 2.2 | |||
| No. of DNA/drug/water atoms | 644/339/279 |
aIncluding Friedel’s pairs.
bNumbers in parentheses are from the outermost shell (2.15–2.25 Å) data.
Diffraction data of Chro–DNA complex crystal were measured at 110 K with the Structural Biology Center undulator beamline 19ID at the Advanced Photon Source, using fundamental undulator harmonics and the 3 × 3 mosaic charge coupled device (CCD) detector, as described previously (24,25). Crystallographic data integration and reduction were performed using the program package HKL2000 (26). Multiwavelength anomalous diffraction (MAD) data were collected from four wavelengths using cobalt as the anomalous scattering atom (22,24). Interestingly, two cobalt sites were located and phases were derived as implemented in the crystallographic suite SOLVE (27). The figure-of-merit was 0.60 and 0.91 before and after density modification, respectively. The resulting MAD electron density map at 3.0 Å resolution was used to build the initial models using the program O (28).
There were two independent complexes in the asymmetric unit. The DNA force field parameters of Parkinson et al. were used (29). The force field of Chro was generated using the atomic coordinates of its 0.68 Å resolution crystal structure (unpublished results). The initial model after rigid refinement had an R-factor of 43%, which was improved to 23% after several rounds of simulated annealing using the Crystallo graphy & NMR system (CNS) (30). This refined model from the derivative drug-DNA crystal was transformed to native drug-DNA by substituting two Co3+ with two Mg2+, followed by a subsequent round of rigid body refinement. The Mg2+-chelated structure was refined further by the simulated annealing procedure incorporated in CNS (30). DNA helical parameters were calculated with 3DNA and CURVES (31). The coordinates have been deposited in the Protein Data Bank (accession code 1VAQ).
RESULTS AND DISCUSSION
Interactions of chromomycin A3 with divalent ions in the presence and absence of DNA duplex
Chro is optically active. Here, we characterized the CD spectral changes between the dimer and monomer of Chro in the presence of divalent cations with and without DNA. Figure 1B and C shows the CD spectra (520–220 nm) of Chro in the presence and absence of DNA duplex upon the addition of Mg2+, respectively. The changes in CD spectra are associated with concomitant decreased positive and negative peak intensities at 413 nm and 400 nm in the visible region, respectively. However, owing to DNA binding, the spectral changes showed a slight difference in the ultra-violet (UV) region upon the addition of Mg2+, with increased positive peak intensity at 295 nm, different from the spectra in the absence of DNA, which showed decreased negative peak intensity at 300 nm. In order to compare the chelating affinity of divalent metal ions to Chro in the presence of DNA, we carried out titration studies (metal concentrations versus normalized intensity change at λ275 nm) and calculated the metal:drug ratio at which a complete formation of drug dimer is achieved. Our results showed that the lowest ratio was ∼0.6 in the presence of DNA. Note that Chro shows a higher affinity (∼200 fold) towards divalent metal ions in the presence of DNA than without DNA, suggesting that DNA may acts as an effective template to facilitate the chelation.
CD spectra are good indicators of DNA conformational changes induced by DNA-binding drugs (22). Figure 1D shows a comparison of CD spectra between d(TTGGCCAA)2 and [(Chro)2-Mg2+]–d(TTGGCCAA)2. The induced CD intensities of 287 and 275 nm can be used as markers for the relative binding affinities of Chro towards these DNA oligomer duplexes.
Deamination of cobalt-hexaammine by chromomycin A3 bound to DNA duplex
We obtained crystals of Chro bound with d(TTGGCCAA)2 in the presence of Co(NH3)63+ and Mg2+. Surprisingly, fluorescence scanning of the crystal indicated the presence of cobalt ions in the crystal. Subsequent analysis of diffraction data confirmed the locations of two cobalt ions. We solved the crystal structure of the [(Chro)2-Co3+]–d(TTGGCCAA)2 complex by the MAD method using cobalt ions as the anomalous scattering atoms. We found two cobalt ions that are coordinated to O1 and O9 oxygen atoms of the chromophores of two Chros in cobalt-derivative crystals (vide intra). It is interesting to note that Co(NH3)63+ alone does not bind to either drug, because the six NH3 ligands on Co(NH3)63+ are strongly ligated to Co3+. However, Chro mixed with Co(NH3)63+ was shown to bind to DNA in the form of a [(Chro)2-Co3+] complex, suggesting that the four ammonia molecules from cobalt-hexaammine have been replaced by the two chelating Chro molecules during the crystallization process, in which DNA appeared to act as an effective template to facilitate the deamination. This result agrees with the CD study showing that DNA enhanced the metal-ion-chelating ability of Chro.
The observation that DNA serves as a template for chemical equilibrium may be a general phenomenon. For example, it was shown that the lactone form of camptothecin is favored over the carboxylate form when DNA is present (32).
Overall structure of the Chro–DNA complex
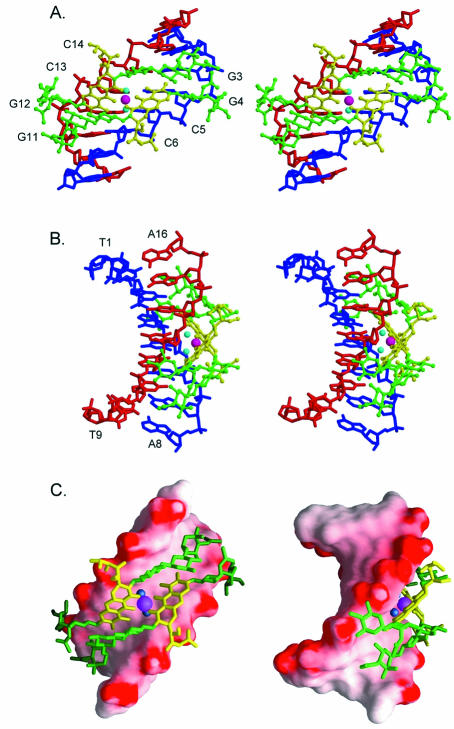
The [(Chro)2-Mg2+]–d(TTGGCCAA)2 complex crystallizes in the hexagonal space group P6522, and the crystals diffract to 2.15 Å resolution. All atoms for both molecules have been modeled in and are well defined, as indicated by the quality of the electron density maps. Figure 1SA and B shows the T1:A9 Watson–Crick base pair and the Chro dimer chelated with the Mg2+ ion, respectively (Supplementary Material is available at NAR Online). Two independent Chro–DNA duplex complexes found in one asymmetric unit are packed through van der Waals contacts between the Chro drugs (Fig. 2A and B). These complexes are labeled as CPXI and CPXII. Even though the two complexes have different packing environments, they are virtually superimposable, with a root mean square deviation (r.m.s.d.) of 1.24 Å (983 atoms). The crystal structure of the Chro–DNA complex (CPXI) showed that two Chro molecules form a dimer mediated by a single Mg2+ ion and that the two Chro monomers are aligned in a head-to-tail orientation with a pseudo 2-fold symmetry (Fig. 3A–C). Comparing the previous NMR structure of [(Chro)2-Mg2+]–d(TTGGCCAA)2 complex, which was determined by nuclear overhauser effect (NOE) and pseudocontact shifts (20), and the present crystal structure, there are differences between the two structures, with the r.m.s.d. of the overall structure being 2.04 Å.
Figure 2.
Two independent Chro–DNA complexes (CPXI and CPXII) were found in one asymmetric unit. (A) Chro dimer in van der Waals drawings and DNA in skeletal drawing. (B) Chro in ball-and-stick representation and DNA in van der Waals drawings.
Figure 3.
Stereoscopic drawings of the Mg2+-coordinated Chro-(TTGGCCAA)2 complex (CPXI) viewed from the minor groove (A) and backbone (B) direction, related by a 2-fold symmetry operation (Chro in ball-and-stick representation and DNA in skeletal line drawing). (C) Surface representation of the [(Chro)2-Mg2+]–d(TTGGCCAA)2 complex shown with DNA electrostatic potentials viewed from the minor groove (left) and backbone (right).
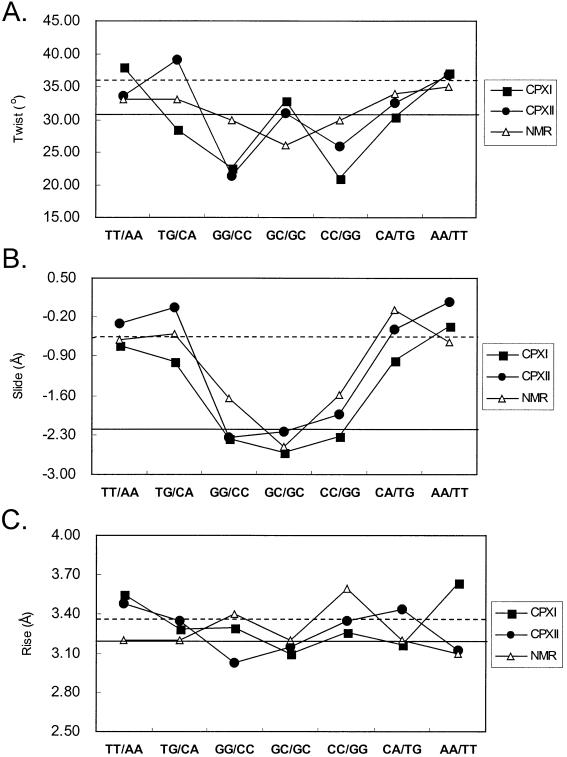
The DNA structure of the complexes maintains a right-handed helical structure, with the central 4-bp segment adopting a B-like character with a pronounced kink. The central GpGpCpC segment surrounding the Chro dimer binding site shows considerable changes in the helical parameters. Of particular interest is the significant unwinding of base pairs at the GpG and CpC steps. This is in contrast to the NMR structure, which shows unwinding at the central GpC step (Fig. 4A). The average helical twist values of the GpGpCpC segment of CPXI and CPXII are calculated to be 25.4° and 26.4°, respectively. This average twist value corresponds to an unwinding angle of ∼10° from B-DNA, which is greater than the unwinding angle of ∼5° derived from the NMR solution structure. A distinct pattern of rise (∼3.1 Å) and slide (approximately –2.3 Å) also occurs at the GpGpCpC segment of DNA duplexes (Fig. 4B and C). In contrast, the two base-pair steps on the 3′ and 5′ side of the DNA duplexes TpT and ApA show a normal B-DNA-like slide (∼0 Å) and rise (∼3.5 Å). In addition, the TpG steps show a positive roll of ∼17° into the major grooves, causing DNA bending.
Figure 4.
Comparison of DNA twist (A), slide (B) and rise (C) parameters in three structures (crystal CPXI, CPXII and NMR structure). Values typical for A-DNA (solid line) and B-DNA (dashed line) are also shown.
Main chain torsion angles of DNA duplex
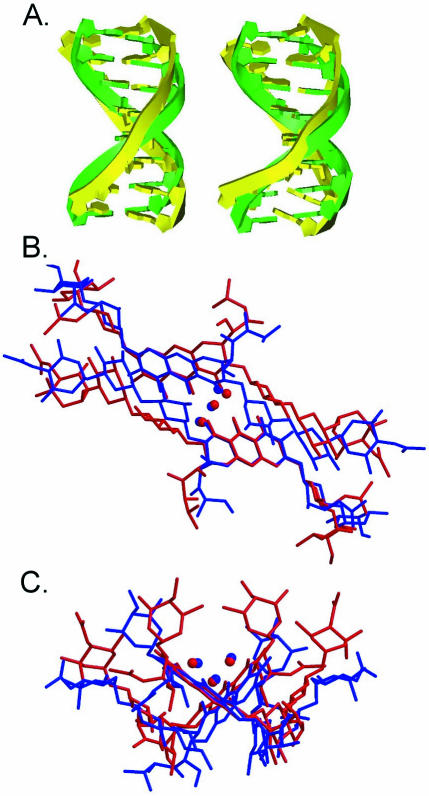
The torsion angles of two DNA duplexes obtained from CPXI and CPXII were analyzed using the CURVE program (31). The glycosidic torsion angles, χ, of all residues fall into the anti conformation (160° to ∼300°). The central guanine residues surrounding the Chro binding site, G4 and G11, uniquely show the trans (χ ≈ 180°) conformation. The difference in conformation of the sugar phosphate chains around the Chro binding site is readily observed in the backbone torsion angles. The torsion angle, γ, of the G4 and G11 residues adopts the trans conformation (∼160°), while the other residues adopt the gauche+ (approximately 30–50°) conformation. The torsion angle, β, of all residues was not affected by the drug binding, which has the canonical value of ∼165°. However, the torsion angle, α, of the G3 and G12 residues around the C-D-E side chains of Chro adopts a unique lower value (∼120°) compared with its canonical A- and B-form value of ∼300°. The overall DNA helical axes of the two complexes, CPXI and CPXII, have ∼30° and ∼36° bends toward the major groove, respectively, due to the binding of the Chro dimer (Fig. 5A). Crystal packing may contribute to the slight difference between the two complexes. This bending is greater than that reported (4°) for the NMR structures (20).
Figure 5.
(A) Comparison of regular B-DNA and kinked DNA obtained from two Chro–DNA complexes [CPXI (left) and CPXII (right)], which are depicted as ribbon drawings in green and yellow, respectively. (B and C) Superposition of two independent Chro dimer structures obtained from two crystal structures, i.e. [(Chro)2-Mg2+]–d(TTGGCCAA)2 (red) and high resolution (0.68 Å) Chro dimer (blue), done by least-square fitting of one of the chromophore rings from the front (B) and side (C) view.
Structure of the Chro dimer and its binding to DNA
The structure of the Chro dimer derived from the X-ray crystal structure differs to that of the NMR analysis, with the r.m.s.d. of the drug itself being ∼2.8 Å. The two Chro chromophores of the complex of the X-ray structure make a dihedral angle of ∼85° between their planes, which is different to that (75°) of the NMR structure (Fig. 3A and B). This probably implies that the pseudocontact shift and NOE-based NMR analysis were not sufficient to define the orientation of the drug dimer.
The C-D-E trisaccharide segments from individual monomers adopt an extended conformation that projects in opposite directions in the dimer. The metal ion shows an octahedral coordination to O1 and O9 atoms of the two chromophores and two water molecules (Wat1 and Wat2) that act as the fifth and sixth ligands (Fig. 6A); this differs to that of the NMR structure, which shows that the metal ion has a tetrahedral coordination geometry. Each water oxygen atom makes an ∼2.0 Å coordination bond to the metal (Fig. 6C). These two water molecules also form specific hydrogen bonds with C5O2(C13O2), C6O4′(C14O4′) and CPHO8 (Fig. 6A and C). The O1(Wat1)–Mg–O1(Wat2) coordination angles are in the 90° (±9°) range (Fig. 1S; Supplementary Material is available at NAR Online). The O9–Mg–O9 angle between intermolecule chromophores has an almost linear value of 174°. Besides these two coordinating water molecules, other bridging water molecules may also play an important mediating role in the interactions between the drug and DNA duplex (Table 2S; Supplementary Material is available at NAR Online).
Figure 6.
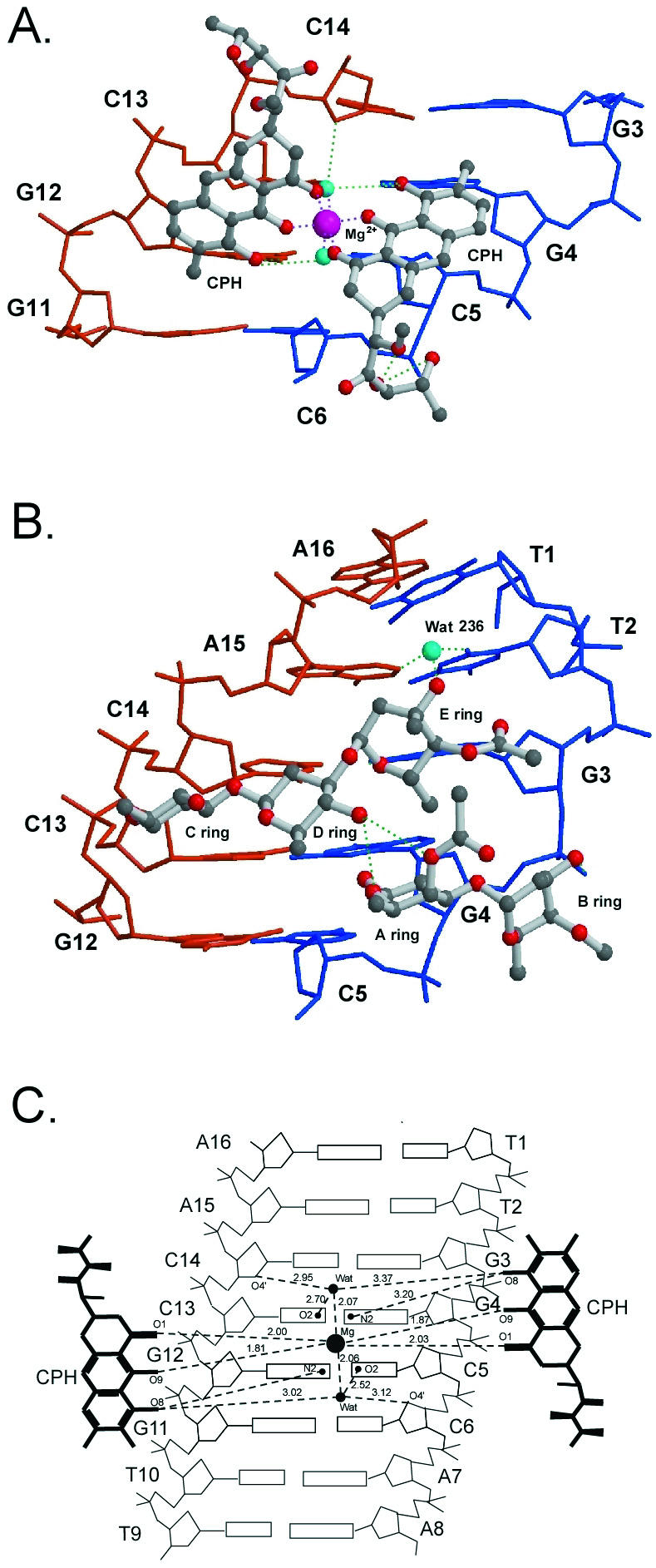
(A) Close-up view of the molecular structures of Chro–DNA complex at central GGCC base pair steps of the refined structure. (B) Close-up view of the molecular structures of the Chro–DNA complex at terminal base pair steps. (C) Schematic diagram illustrates the contact between chromophore and DNA duplex. The dashed line represents direct hydrogen bonds and coordinated bonds.
It is worthy to note that the coordination distances between CPHO9 and Mg2+ range from 1.83 to 1.88 Å, which are significantly shorter than those between CPHO1 and Mg2+ (ranging from 2.01 to 2.11 Å) (Table 1S; Supplementary Material is available at NAR Online). The data suggest that the C9–O9 bond is likely in the anionic phenolate form, which results in a stronger O9––Mg2+ (and thus shorter) coordination bond. Consequently, the (Chro)2-Mg2+ dimer is a neutral species that does not carry any electric charge.
The two Chro monomers within the dimer are stabilized by mutual stacking of the aromatic portion of the chromophore of one monomer with the C-D glycosidic linkage of the other monomer. There are strong intermolecular Chro–Chro hydrogen bonds between the O4 of D-ring and the O1 of A-ring, and the O4 of D-ring and the O4 of A-ring (Table 2). It is interesting that the stacking interaction between two chromophore rings and the central part of the DNA duplexes is similar to the packing interaction resulting from the A-DNA molecules abutting their terminal base pairs onto the shallow and wide minor groove of other A-DNA duplexes in the P212121 crystal lattice (33,34) (Fig. 6A). The saccharide rings of the chromophore interact extensively with DNA (Fig. 6B). The crowding between the methyl groups of the E-ring side chain and the G12 (G4) residue caused the helical twist between G11 (G3) and G12 (G4) to decrease (22° versus 35° in B-form DNA).
Table 2. Hydrogen bonding interactions in the Chro–DNA complex.
| Donor | Acceptor | CPXI (Å) | CPXII (Å) | |
|---|---|---|---|---|
| Chro–Chro | ||||
| D ring-A ring (inter) | DO4 | AO1 | 2.86/3.08 | 2.89/3.08 |
| D ring-A ring (inter) | DO4 | AO4 | 3.31/3.30 | 3.42/3.18 |
| C ring-D ring (intra) | CO4 | DO1 | 2.67/2.88 | 2.68/2.68 |
| CPH-CPH (intra) | CPHO4′ | CPHO1′ | 3.20/– | 2.44/– |
| Chro–DNA | ||||
| CPH-G4/G12 | GN2 | CPHO8 | 3.20/2.89 | 3.14/2.85 |
| E ring-G3/G11 | GN2 | EO1 | 3.05/3.05 | 3.00/3.12 |
| B ring-G4/G12 | BO3 | GO2P | 2.90/2.89 | 3.38/3.46 |
| CPH-C14 | CPHO3′ | CO1P | 2.99 | – |
| CPH-A15 | CPHO2′ | GO2P | 3.28 | – |
| CPH-C6 | CPHO4′ | GO1P | 2.45 | – |
| CPH-C14 | CPHO2′ | CO1P | – | 2.37 |
| CPH-A15 | CPHO4′ | AO1P | – | 2.70 |
| CPH-A15 | CPHO4′ | AO2P | – | 2.97 |
The specificity of the chromomycin dimer for the GpGpCpCp sequence is due to the specific strong intermolecular Chro–DNA hydrogen bonds between the O8 of chromophore and the N2 atom of G4 (G12) base, and the O1 oxygen of E-ring and the N2 atom of G3 (G11) base (Table 2). In addition, the O3 of the B-ring side chain forming a hydrogen bond to the O2P of G4 (G12) is found in both the CPXI and CPXII complexes. A previous NMR structure showed that the O8 group of the chromophore forms hydrogen bonds with the N3 atom of the G4 (G12) base (17,20), but that the distance between these two atoms in the crystal structure is ∼4.5 Å, too far for a hydrogen bond to form. An earlier crystal structure of d(CATGGCCATG) with a central GpGpCpCp region showed a smooth bend of 23° over the central 4 bp, caused by preferential stacking interactions at guanine bases (35). The bend is produced by a roll of base pairs along their long axes, in the direction that compresses the wide major groove of the double helix. Hence, it is likely that this major-groove compression and the concomitant widening of the minor groove partly contribute to the binding preference of the Chro dimer to the GpGpCpCp sequence.
Crystal packing of Chro–DNA complexes
There are two major different packing interactions occurring between two asymmetric units in the crystal lattice (P6522): end-to-end and end-to-side, mediated by π–π stacking and van der Waals contact, respectively (Fig. 2S; Supplementary Material is available at NAR Online). At one interface of the end-to-end packing, the end of one DNA duplex molecule is packed against the end of another molecule, with a base-stacking distance of ∼3.4 Å between the contacting terminal residues of CPXI (T1-A16) [or CPXII (A8-T9)] of independent asymmetric units. The other interface of the end-to-side packing is stabilized by the formation of intramolecular van der Waals contact and hydrophobic contact between the terminal residues of CPXI (A8-T9) [or CPXII (T1-A16)] and the Chro saccharide residues within the independent asymmetric units.
CONCLUSION
Recently, the use of the aureolic family of drugs is re-emerging. Fibach et al. (36) revealed that Mith could induce fetal hemoglobin production in normal and thalassemic human erythroid precursor cells, and it was proposed as a therapeutic agent in several neoplastic diseases. In addition, the new aureolic acid-type compounds, generated through combinatorial biosynthetic methods, were investigated for their application in cancer therapy and their effect on Sp binding to the c-src promoter region (37,38). Although NMR structural information on Chro and Mith bound to DNA duplexes has been available for some time (17–20), some ambiguities still remain. For example, the coordination geometry surrounding the Mg2+ ion was not unequivocally defined. Our X-ray crystallographic studies reported here have firmly established the detailed structural information regarding the DNA binding model of the metal-mediated dimer of the drug. The coordination around the metal ion is in octahedral geometry. The metal ion binds to the two oxygen atoms of each chromophore and two water molecules act as the fifth and sixth ligands. In addition, other bridging water molecules that mediate the interaction between the Chro and DNA in the duplex were also observed in the crystal structure.
We had previously solved the crystal structure of the (Chro)2-Mg2+ complex in the absence of DNA duplex at 0.68 Å resolution by using direct methods (unpublished results). We superimposed these two Chro dimer conformations with/without DNA duplex. They showed that the Chro dimer without DNA exhibits less inter-stacking interaction between the chromophore and the C-D disaccharide than in the Chro dimer bound to DNA (Fig. 5B). Inspection of the Chro dimer conformation in the presence of DNA duplex reveals that the saccharide side chains of Chro protrude in a manner similar to that of a scorpion (Figs 3C and 5C). Our results reveal a useful direction to take when designing new anticancer drugs in the future.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online. This includes three supplementary figures: the refined (2Fo–Fc) Fourier electron density map (Fig. 1S), the crystal packing (P6522) diagram of the Chro–DNA complex (Fig. 2S) and the drawing of the Mg2+-coordinated Chro-(TTGGCCAA)2 complex (CPXII) (Fig. 3S). Five supplementary tables are also available, including the distances and angles at the Mg2+-coordination site (Table 1S), the bridging waters that mediated the Chro–DNA interaction (Table 2S), base pair parameters (Table 3S) and torsion angles of DNA duplexes (Tables 4S and 5S).
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by National Science Council (NSC) grant 91-3112-P-001-019-Y (Taiwan) to A.H.-J.W.
REFERENCES
- 1.Slavik M. and Carter,S.K. (1975) Chromomycin A3, mithramycin and olivomycin: antitumor antibiotics of related structure. Adv. Pharmacol. Chemother., 12, 1–30. [DOI] [PubMed] [Google Scholar]
- 2.Kennedy B.J. (1972) Mithramycin therapy in testicular cancer. J. Urol., 107, 429–432. [DOI] [PubMed] [Google Scholar]
- 3.Foley J.F., Lemon,H.M., Miller,D.M. and Kessinger,A. (1972) The treatment of metastatic testicular tumors. J. Urol., 108, 439–442. [DOI] [PubMed] [Google Scholar]
- 4.Du Priest R.W. Jr and Fletcher,W.S. (1973) Chemotherapy of testicular germinal tumors. Oncology, 28, 147–163. [DOI] [PubMed] [Google Scholar]
- 5.Fox K.R. and Howarth,N.R. (1985) Investigations into the sequence-selective binding of mithramycin and related ligands to DNA. Nucleic Acids Res., 13, 8695–8714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chatterjee S., Zaman,K., Ryu,H., Conforto,A. and Ratan,R.R. (2001) Sequence-selective DNA binding drugs mithramycin A and chromomycin A3 are potent inhibitors of neuronal apoptosis induced by oxidative stress and DNA damage in cortical neurons. Ann. Neurol., 49, 345–354. [PubMed] [Google Scholar]
- 7.Bianchi N., Osti,F., Rutigliano,C., Corradini,F.G., Borsetti,E., Tomassetti,M., Mischiati,C., Feriotto,G. and Gambari,R. (1999) The DNA-binding drugs mithramycin and chromomycin are powerful inducers of erythroid differentiation of human K562 cells. Br. J. Haematol., 104, 258–265. [DOI] [PubMed] [Google Scholar]
- 8.Aich P. and Dasgupta,D. (1995) Role of magnesium ion in mithramycin-DNA interaction: binding of mithramycin-Mg2+ complexes with DNA. Biochemistry, 34, 1376–1385. [DOI] [PubMed] [Google Scholar]
- 9.Chakrabarti S., Aich,P., Sarker,D., Bhattacharyya,D. and Dasgupta,D. (2000) Role of Mg2+ in the interaction of anticancer antibiotic, chromomycin A3 with DNA: does neutral antibiotic bind DNA in absence of the metal ion? J. Biomol. Struct. Dyn., 18, 209–218. [DOI] [PubMed] [Google Scholar]
- 10.Cons B.M. and Fox,K.R. (1989) Interaction of mithramycin with metal ions and DNA. Biochem. Biophys. Res. Commun., 160, 517–524. [DOI] [PubMed] [Google Scholar]
- 11.Gao X.L. and Patel,D.J. (1990) Chromomycin dimer-DNA oligomer complexes. Sequence selectivity and divalent cation specificity. Biochemistry, 29, 10940–10956. [DOI] [PubMed] [Google Scholar]
- 12.Keniry M.A., Banville,D.L., Simmonds,P.M. and Shafer,R. (1993) Nuclear magnetic resonance comparison of the binding sites of mithramycin and chromomycin on the self-complementary oligonucleotide d(ACCCGGGT)2. Evidence that the saccharide chains have a role in sequence specificity. J. Mol. Biol., 231, 753–767. [DOI] [PubMed] [Google Scholar]
- 13.Aich P. and Dasgupta,D. (1990) Role of Mg2+ in the mithramycin–DNA interaction: evidence for two types of mithramycin-Mg2+ complex. Biochem. Biophys. Res. Commun., 173, 689–696. [DOI] [PubMed] [Google Scholar]
- 14.Liu C. and Chen,F.M. (1994) Oligonucleotide studies of sequence-specific binding of chromomycin A3 to DNA. Biochemistry, 33, 1419–1424. [DOI] [PubMed] [Google Scholar]
- 15.Itzhaki L., Weinberger,S., Livnah,N. and Berman,E. (1990) A unique binding cavity for divalent cations in the DNA-metal-chromomycin A3 complex. Biopolymers, 29, 481–489. [DOI] [PubMed] [Google Scholar]
- 16.Weinberger S., Shafer,R. and Berman,E. (1988) On the interaction of chromomycin A3 with calf thymus DNA in the presence of metal cations at different pH values. Biopolymers, 27, 831–842. [DOI] [PubMed] [Google Scholar]
- 17.Gao X.L., Mirau,P. and Patel,D.J. (1992) Structure refinement of the chromomycin dimer-DNA oligomer complex in solution. J. Mol. Biol., 223, 259–279. [DOI] [PubMed] [Google Scholar]
- 18.Sastry M. and Patel,D.J. (1993) Solution structure of the mithramycin dimer–DNA complex. Biochemistry, 32, 6588–6604. [DOI] [PubMed] [Google Scholar]
- 19.Sastry M., Fiala,R. and Patel,D.J. (1995) Solution structure of mithramycin dimers bound to partially overlapping sites on DNA. J. Mol. Biol., 251, 674–689. [DOI] [PubMed] [Google Scholar]
- 20.Gochin M. (2000) A high-resolution structure of a DNA-chromomycin-Co(II) complex determined from pseudocontact shifts in nuclear magnetic resonance. Structure Fold. Des., 8, 441–452. [DOI] [PubMed] [Google Scholar]
- 21.Hou M.H., Lin,S.B., Yuann,J.M., Lin,W.C., Wang,A.H.-J. and Kan L.S. (2001) Effects of polyamines on the thermal stability and formation kinetics of DNA duplexes with abnormal structure. Nucleic Acids Res., 29, 5121–5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hou M.H., Robinson,H., Gao,Y.G. and Wang,A.H.-J. (2002) Crystal structure of actinomycin D bound to the CTG triplet repeat sequences linked to neurological diseases. Nucleic Acids Res., 30, 4910–4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang A.H.-J. and Gao,Y.G. (1990) Crystallization of oligonucleotides and their complexes with antitumor drugs. Methods, 1, 91–99. [Google Scholar]
- 24.Walsh M.A., Evans,G., Sanishvili,R., Dementieva,I. and Joachimiak,A. (1999) MAD data collection—current trends. Acta Crystallogr., D55, 1726–1732. [DOI] [PubMed] [Google Scholar]
- 25.Robinson H., Gao,Y.G., Yang,X., Sanishvili,R., Joachimiak,A. and Wang,A.H.-J. (2001) Crystallographic analysis of a novel complex of actinomycin D bound to the DNA decamer CGATCGATCG. Biochemistry, 40, 5587–5592. [DOI] [PubMed] [Google Scholar]
- 26.Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 27.Terwilliger T.C. and Berendzen,J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr., D55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao Y.G., Robinson,H., Sanishvili,R., Joachimiak,A. and Wang,A.H.-J. (1999) Structure and recognition of sheared tandem G × A base pairs associated with human centromere DNA sequence at atomic resolution. Biochemistry, 38, 16452–16460. [DOI] [PubMed] [Google Scholar]
- 29.Parkinson G., Vojtechovsky,J., Clowney,L., Brunger,A.T. and Berman,H.M. (1996) New parameters for the refinement of nucleic acid containing structures. Acta Crystallogr., D52, 57–64. [DOI] [PubMed] [Google Scholar]
- 30.Brunger A.T., Adams,P.D., Clore,G.M., DeLano,W.L., Gros,P., Grosse-Kunstleve,R.W., Jiang,J.S., Kuszewski,J., Nilges,M., Pannu,N.S. et al. (1998) Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr., D54, 905–921. [DOI] [PubMed] [Google Scholar]
- 31.Lavery R. and Sklenar,H. (1988) The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J. Biomol. Struct. Dyn., 6, 63–91. [DOI] [PubMed] [Google Scholar]
- 32.Yang D., Strode,J., Spielmann,H., Wang,A.H.-J. and Burke,T. (1998) DNA interactions of two clinical camptothecin drugs stabilize their active lactone forms. J. Am. Chem. Soc., 120, 2970–2980. [Google Scholar]
- 33.Gao Y.G., Robinson,H. and Wang,A.H.-J. (1999) High-resolution A-DNA crystal structures of d(AGGGGCCCCT). An A-DNA model of poly(dG) × poly(dC). Eur. J. Biochem., 261, 413–420. [PubMed] [Google Scholar]
- 34.Wang A.H.-J., Fujii,S., van Boom,J.H. and Rich,A. (1982) Molecular structure of the octamer d(G-G-C-C-G-G-C-C): modified A-DNA. Proc. Natl Acad. Sci. USA, 79, 3968–3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goodsell D.S., Kopka,M.L., Cascio,D. and Dickerson,R.E. (1993) Crystal structure of CATGGCCATG and its implications for A-tract bending models. Proc. Natl Acad. Sci. USA, 90, 2930–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fibach E., Bianchi,N., Borgatti,M., Prus,E. and Gambari,R. (2003) Mithramycin induces fetal hemoglobin production in normal and thalassemic human erythroid precursor cells. Blood, 102, 1276–1281. [DOI] [PubMed] [Google Scholar]
- 37.Remsing L.L., Bahadori,H.R., Carbone,G.M., McGuffie,E.M., Catapano,C.V. and Rohr,J. (2003) Inhibition of c-src transcription by mithramycin: structure-activity relationships of biosynthetically produced mithramycin analogues using the c-src promoter as target. Biochemistry, 42, 8313–8324. [DOI] [PubMed] [Google Scholar]
- 38.Remsing L.L., Gonzalez,A.M., Nur-e-Alam,M., Fernandez-Lozano,M.J., Brana,A.F., Rix,U., Oliveira,M.A., Mendez,C., Salas,J.A. and Rohr,J. (2003) Mithramycin SK, a novel antitumor drug with improved therapeutic index, mithramycin SA and demycarosyl-mithramycin SK: three new products generated in the mithramycin producer Streptomyces argillaceus through combinatorial biosynthesis. J. Am. Chem. Soc., 125, 5745–5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.