Abstract
Primate lentiviruses including human immunodeficiency virus type 1 (HIV-1) and simian immunodeficiency viruses (SIVs) evolved through the acquisition of antagonists against intrinsic host restriction factors, such as tetherin. It is widely accepted that HIV-1 has emerged by zoonotic transmission of SIV in chimpanzee (SIVcpz), and that SIVcpz Nef protein antagonizes chimpanzee tetherin. Although Nef of SIVcpz shares a common ancestor with that of SIVrcm, an SIV in red-capped mangabey (Cercocebus torquatus), it remains unclear whether SIVrcm Nef can antagonize tetherin of its natural host. In this study, we determine the sequence of red-capped mangabey tetherin for the first time and directly demonstrate that SIVrcm Nef is the bona fide antagonist of red-capped mangabey tetherin. These findings suggest that SIVrcm Nef is the functional ancestor of SIVcpz Nef. Moreover, molecular phylogenetic analyses reveal that tetherins of the genus Cercocebus have experienced adaptive evolution, which is presumably promoted by primate lentiviruses.
So far, more than 40 primate lentiviruses (PLVs) including human immunodeficiency viruses (HIVs) and simian immunodeficiency viruses (SIVs) have been identified1. Understanding the evolutionary history of PLVs is one of the most important and interesting topics in the field of retrovirology. However, since PLVs have experienced multiple cross-species transmissions and complicated recombination, it is difficult to elucidate how genetic conflicts between the ancient PLVs and their respective host species resulted in evolution and diversification.
One way to approach this issue is to concentrate on the evolution and the diversity of specific accessory genes in PLVs. All PLVs identified so far encode 8 common genes: gag, pol, env, tat, rev, vpr, vif, and nef2. In addition, the PLVs in the HIV-1 lineage encode an additional accessory gene, vpu, whereas those in the HIV-2 lineage encode the other unique gene, vpx2. Interestingly, recent studies focusing on the interplay between viral and host proteins3,4,5 have provided lines of evidence for supporting “Red Queen hypothesis”6 or “evolutionary arms race”, which is an important assumption proposed for the co-evolution of PLVs and their host species. For instance, the accessory proteins such as Vif, Vpx, Vpu, and Nef encoded by PLVs have been acquired and evolved their abilities to antagonize cellular anti-PLV restriction factors such as apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G (APOBEC3G)7, SAM domain and HD domain 1 (SAMHD1)8,9, and bone marrow stromal antigen 2 ([BST2], also known as tetherin, CD317, and HM1.24; hereinafter referred to as “tetherin”)10,11. Tetherin impairs the release of nascent viral particles from virus-producing cells10,11. In contrast to APOBEC3G and SAMHD1, which are antagonized by single accessory proteins Vif and Vpx respectively, PLVs acquired diverse strategies to antagonize tetherin5,12. For instance, in Old World monkeys (OWMs; the family Cercopithecidae) and hominoids (the family Hominidae), most PLVs including SIVcpz from chimpanzee (Pan troglodytes; CPZ), SIVgor from gorilla (Gorilla gorilla; GOR), SIVsm from sooty mangabey (Cercocebus atys; SM), and SIVmac from rhesus macaque (Macaca mulatta; RM) antagonize tetherins of their own hosts by their Nef proteins13,14,15,16. On the other hand, vpu is encoded only in seven out of the more than 40 PLVs identified: SIVgsn from greater-spot nosed monkey (Cercopithecus nictitans), SIVmon from mona monkey (Cercopithecus mona), SIVmus from moustached monkey (Cercopithecus cephus), SIVden from Dent's mona monkey (Cercopithecus denti), SIVcpz, SIVgor, and HIV-1 from human (Homo sapiens; HU)5. Of note, certain vpu-encoding SIVs from OWMs (SIVgsn, SIVmon, SIVmus, and SIVden) and HIV-1 antagonize tetherins of their own hosts by their Vpu proteins10,11,14,17,18,19. These findings strongly suggest that PLVs have adapted to their hosts by acquiring the anti-tetherin factors at each adaptation process, and therefore, that the relationship between tetherin and viral antagonists can be a clue to reveal the evolutionary diversification of PLVs.
It is widely accepted that two HIVs, HIV-1 and HIV-2, have respectively emerged from independent zoonotic transmission of SIVs to HU around 100 years ago20,21,22. HIV-1 has risen out of SIVcpz from CPZ to HU23,24, while HIV-2 has resulted from zoonotic infection of SIVsm from SM25. Also, it has been estimated that SIVcpz has emerged by the recombination of two lineages of SIVs: SIVgsn/mon/mus and SIVrcm from red-capped mangabey (Cercocebus torquatus; RCM)26. Since SIVcpz Nef is phylogenetically similar to SIVrcm Nef, it seems conceivable that the ancestor of SIVcpz Nef is SIVrcm Nef5,14. Moreover, because SIVcpz Nef is able to counteract CPZ tetherin14, it has been hypothesized that SIVrcm Nef is the antagonist of RCM tetherin5,27. However, the sequence of RCM tetherin has not yet determined, and it remains unknown whether SIVrcm Nef is the bona fide antagonist of RCM tetherin. In this study, we determine the sequence of RCM tetherin for the first time and directly demonstrate that SIVrcm Nef counteracts anti-viral activity of RCM tetherin. Moreover, phylogenetic analyses of 47 primate tetherin sequences reveal that tetherins of the genus Cercocebus including RCM and SM are under positive selection.
Results
Phylogeny reconstruction of 47 primate tetherins
In order to directly elucidate the interplay between SIVrcm Nef and RCM tetherin, we set out to determine the sequence of RCM tetherin. The sequencing analyses of genomic DNA isolated from six wild-caught RCMs28,29 revealed that five out of the six open reading frames (ORFs) of RCM tetherin were identical, while a RCM tetherin displayed a double peak at nucleotide position 60 (GAC or GAT; synonymous mutation). These results imply the presence of polymorphism in RCM tetherin.
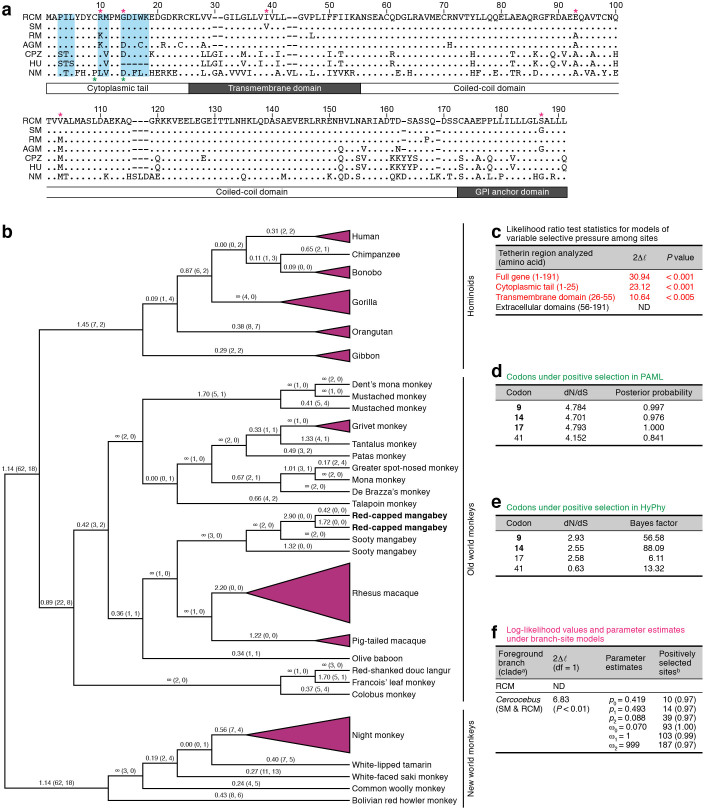
As shown in Figure 1a, compared with the amino acid sequence of RCM tetherin, HU tetherin displayed the 5-amino acid deletion (14GDIWK18) in the cytoplasmic domain, and HU and CPZ tetherins showed the 2-amino acid insertion (30GI31) in the transmembrane domain, both of which are in line with the previous observations13,14,17,18,27,30. The phylogenetic tree of 47 primate tetherins reconstructed by using maximum likelihood (ML) method (Figure 1b) revealed that the two RCM tetherins, which are newly sequenced in this study, are clustered with SM tetherins with stronger bootstrap support (87%), suggesting that RCM tetherin is most closely related to SM tetherin. Because only RCM and SM are the monkeys belonging to the genus Cercocebus among the 27 OWMs (the family Cercopithecidae; Table 1), this result is reasonable.
Figure 1. Positive selection detected in primate tehterin.
(a) Amino acid sequences of primate tetherin. RCM tetherin (GenBank accession number AB907706; determined in this study), SM tetherin (FJ864713), RM tetherin (FJ943432), African green monkey (Chlorocebus aethiops; AGM) tetherin (FJ943430), CPZ tetherin (NM_001190480), HU tetherin (NM_004335), and night monkey (Aotus vociferans; NM) tetherin (FJ638415) are respectively shown. The numbers indicate the amino acid positions in NM tetherin. The two positively selected sites (positioned at 9 and 14), which are determined by both site model of PAML (Figure 1d) and REL method of HyPhy (Figure 1e), are indicated with green asterisks. The six amino acids (positioned at 10, 14, 39, 93, 103, and 187) inferred to be under positive selection in Cercocebus tetherin (the clade of RCM and SM tetherins) (Figure 1f) are indicated with pink asterisks. The amino acids, which are putatively associated with the ability to induce NFκB-dependent signaling36, are indicated with shading in pale blue. (b) Phylogenic tree of 47 primate tetherins reconstructed using ML method. The tree was rerooted with the NWM clade. The dN/dS ratios are shown on each branch and the numbers in parenthesis represent nonsynonymous (left) and synonymous (right) changes, respectively. (c) The positive selection detected in different regions of the tetherin gene. The regions inferred to be under positive selection with statistical significance are represented in red. ND, not detected (2Δl = −0.000002). (d and e) Positively selected sites identified in our analyses. In panel d, the codons under positive selection identified by PAML with posterior probability > 0.95 are shown in bold. In panel e, the codons under positive selection inferred by HyPhy with Bayes factor > 50 are shown in bold. (f) The result obtained from the twobranch-site analyses for RCM and Cercocebus clades. All PAML analyses were performed under two models of codon usage, F61 and F3x4, and they yield consistent results. a, All nodes/branches within RCM and the Cercocebus clades were respectively designated as the foreground branches. b, The number in parenthesis represents posterior probability.
Table 1. GenBank accession numbers of primate tetherins used in this study.
| Family/infraordera | Common nameb | Scientific name | Accession numberc |
|---|---|---|---|
| Hominidae (Hominoids) | Human | Homo sapiens | AK223124 |
| Human | Homo sapiens | NM_004335 | |
| Chimpanzee | Pan troglodytes | NM_001190480 | |
| Bonobo | Pan paniscus | HM136907 | |
| Bonobo | Pan paniscus | XM_003817802 | |
| Gorilla | Gorilla gorilla | GQ925926 | |
| Gorilla | Gorilla gorilla | HM136906 | |
| Gorilla | Gorilla gorilla | XM_004060266 | |
| Orangutan | Pongo pygmaeus | HM136908 | |
| Orangutan | Pongo abelii | NM_001172587 | |
| Gibbon | Hylobates agilis | HM136910 | |
| Gibbon | Nomascus leucogenys | HM136909 | |
| Cercopithecidae (OWMs) | Dent's mona monkey | Cercopithecus denti | HE680870 |
| Mustached monkey | Cercopithecus cephus | GQ864267 | |
| Mustached monkey | Cercopithecus cephus | GQ925925 | |
| Grivet monkey | Chlorocebus aethiops | FJ943430 | |
| Grivet monkey | Chlorocebus aethiops | HM136912 | |
| Tantalus monkey | Chlorocebus tantalus | FJ345303 | |
| Patas monkey | Erythrocebus patas | HM136911 | |
| Greater spot-nosed monkey | Cercopithecus nictitans | GQ925923 | |
| Mona monkey | Cercopithecus mona | GQ925924 | |
| De Brazza's monkey | Cercopithecus neglectus | HE680871 | |
| Talapoin monkey | Miopithecus talapoin | HM136913 | |
| Red-capped mangabey | Cercocebus torquatus | This studyd | |
| Red-capped mangabey | Cercocebus torquatus | This studye | |
| Sooty mangabey | Cercocebus atys | FJ864713 | |
| Sooty mangabey | Cercocebus atys | FJ864714 | |
| Rhesus macaque | Macaca mulatta | FJ943431 | |
| Rhesus macaque | Macaca mulatta | FJ943432 | |
| Rhesus macaque | Macaca mulatta | GQ304749 | |
| Rhesus macaque | Macaca mulatta | HM136914 | |
| Rhesus macaque | Macaca mulatta | HM775182 | |
| Rhesus macaque | Macaca mulatta | NM_001161666 | |
| Pig-tailed macaque | Macaca nemestrina | FJ914988 | |
| Pig-tailed macaque | Macaca nemestrina | FJ914989 | |
| Olive Baboon | Papio anubis | XM_003915138 | |
| Red-shanked douc langur | Pygathrix nemaeus | HM136916 | |
| Francois' leaf monkey | Trachypithecus francoisi | HM136917 | |
| Colobus monkey | Colobus guereza | HM136915 | |
| Platyrrhini (NWMs) | Night monkey | Aotus lemurinus | FJ638414 |
| Night monkey | Aotus vociferans | FJ638417 | |
| Night monkey | Aotus vociferans | FJ638418 | |
| Night monkey | Aotus vociferans | FJ638415 | |
| White-lipped tamarin | Saguinus labiatus | HM136918 | |
| White-faced saki monkey | Pithecia pithecia | HM136920 | |
| Common woolly monkey | Lagothrix lagotricha | HM136922 | |
| Bolivian red howler monkey | Alouatta sara | HM136921 |
aFamily (Hominidae and Cercopithecidae) and infraorder (Platyrrhini) are presented in italic. Popular name of each family/infraorder is presented in parenthesis. OWMs, old world monkeys; NWMs, new world monkeys.
bThe common name of each primate is identical to that in Figure 1B.
cThe GenBank accession numbers (http://www.ncbi.nlm.nih.gov/genbank/) of tetherins are listed.
dAB907706.
eAB907707.
Positive selection detected in the evolution of primate tetherin
The nonsynonymous to synonymous rate ratios (dN/dS) inferred by using a free-ratio model in the PAML package are shown on the ML tree (Figure 1b), which varied among the branches. Since a dN/dS value significantly greater than one is an indicator of positive selection, our result indicates that positive selection probably operated on the tetherin gene episodically during primate evolution. This result is consistent with that obtained in previous studies31,32.
The two pairs of site models in PAML produced similar results and the result obtained from M7 (neutral model) versus M8 (selection model) comparison is shown in Figure 1c. The dN/dS ratio was significantly greater than one for full-length tetherin, cytoplasmic tail, and transmembrane domains. These findings indicate that the functionally important regions of tetherin have evolved under strong positive selection, which agreed with previous reports31,32,33. The site model analysis also identified three codons, 9, 14, and 17, to be under positive selection with posterior probability greater than 0.95 (Figure 1d). Two of them, codons 9 and 14, were also detected by the random effects likelihood (REL) analysis implemented in the HyPhy package with Bayes factor greater than 50 (Figure 1e; these sites are also indicated with green asterisks in Figure 1a). Although these two sites have been reported by a previous study in which 20 primate tetherins were analyzed31, our results further suggest that they may have experienced positive selection in the evolution of many primate lineages, because more primate tetherins were included in our analysis. Moreover, the codon 41, which has been reported to be a positively selected site in a previous study31, was detected in our analysis as well with posterior probability of 0.841 (Figure 1d).
Furthermore, the branch-site tests in PAML revealed that the likelihood ratio test was significant with P < 0.01 in the analysis of the Cercocebus clade, suggesting that positive selection has most likely operated on Cercocebus tetherins (Figure 1f). On the other hand, no significant positive selection was detected for RCM clade, probably because only two highly similar sequences were included in this clade. In addition, the branch-site analysis identified six codons, 10, 14, 39, 93, 103, and 187, to be positively selected sites in the Cercocebus clade (Figure 1f; these sites are also indicated with pink asterisks in Figure 1a), and three out of the six sites, 93, 103, and 187, are located in the extracellular domain. In this regard, it has been recently reported that human tetherin directly binds to immunoglobulin-like transcript 7 (ILT7), a cellular molecule specifically expressed on plasmacytoid dendritic cells, through its extracellular domain and modulates ILT7-mediated signaling leading to the production of type I interferons and proinflammatory cytokines34. Therefore, it is possible that these three sites may associate with the interaction of tetherin and ILT7 in the genus Cercocebus. Taken together, here we firstly demonstrated that Cercocebus tetherins have evolved under stronger positive selection and also identified six codons which may be functionally important.
Antagonism of RCM tetherin by SIVrcm Nef
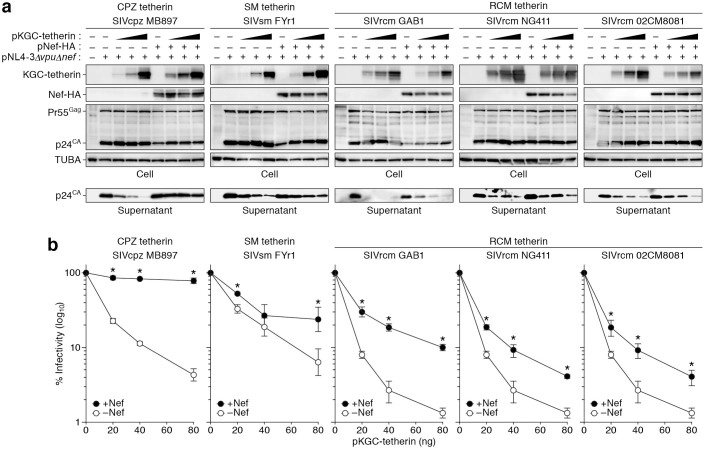
We then assessed the interplay of RCM tetherin and SIVrcm Nef. Western blotting (Figure 2a) and TZM-bl assay (Figure 2b) revealed that increasing concentrations of CPZ, SM and RCM tetherins resulted in a dose-dependent decrease in the release of nascent viral particles. We then investigated whether SIVrcm Nef has the ability to antagonize RCM tetherin. As shown in Figure 2a, the Nefs of SIVcpz, SIVsm, and SIVrcm did not affect the expression levels of tetherins and Gag (Figure 2a), which is consistent with previous reports16,35. Moreover, we revealed that the Nefs of 3 SIVrcm isolates, strains GAB1, NG411, and 02CM8081, enhanced virus release in the presence of RCM tetherin (Figures 2a and 2b). These findings directly demonstrate that SIVrcm Nef is the bona fide antagonist of RCM tetherin, and further support the hypothesis that the anti-tetherin ability of SIVcpz Nef is originated from that of SIVrcm Nef.
Figure 2. Anti-viral activity of RCM tetherin and antagonistic ability of SIVrcm Nefs.
(a) Western blotting. Representative results are shown. Blots have been cropped; full uncropped blots are available as Supplementary Figure 1. (b) TZM-bl assay The data represents the percentage of infectivity compared to the values without tetherin ± SD. The assay was performed in triplicate. The statistic difference (*P < 0.05) is determined by Student's t test.
In the cytoplasmic tail of Cercocebus tetherins, we found a novel positively selected site positioned at 10 (Arginine in RCM and SM tetherins; Figure 1a) with posterior probability greater than 0.95 (Figure 1f). In this regard, previous papers have reported that human tetherin but not OWM tetherins (e.g., RM and AGM tetherins) has the ability to induce NFκB-dependent proinflammatory signaling, and that certain amino acids in the cytoplasmic tail of human tetherin (Figure 1a, with shading in pale blue) are associated with this activity36,37,38. It is particularly noteworthy that the site 10 detected to be under positive selection in Cercocebus tetherin is located in this region and has the same amino acid (arginine) with that in human tetherin (Figure 1a). This raises a hypothesis that Cercocebus tetherin has evolutionarily acquired this NFκB-mediated signaling capacity. To address this possibility, tetherin expression plasmids were cotransfected with an NFκB-luciferase reporter plasmid39 and a vpu-deficient HIV-1-producing plasmid40 into 293T cells. However, although we detected the induction of NFκB-mediated activation by human tetherin, Cercocebus tetherins including RCM and SM tetherins did not elicit this activity (Figure 3b). Although a previous study has shown that CPZ tetherin partially induces this NFκB-mediated signaling36, this effect was not observed in our assay (Figure 3b). This may be due to the addition of KGC tag at the N-terminus of tetherin in our study. These results suggest that an arginine residue at the site 10 is not associated with the gain-of-function to induce NFκB-mediated signaling.
Figure 3. Ability of primate tetherins to induce NFκB-dependent signaling.
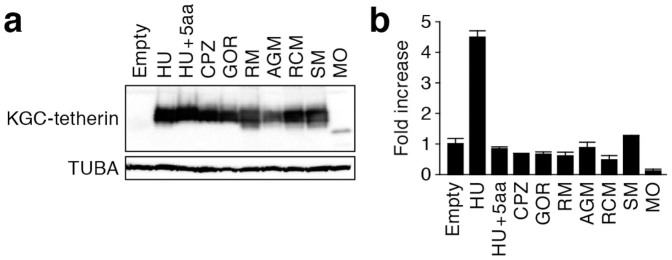
(a) Western blotting. Representative results are shown. Blots have been cropped; full uncropped blots are available as Supplementary Figure 2. (b) Luciferase assay. The data represents the average of fold induction of luciferase activity compared to the empty vector-transfected cells with SD. The assay was performed in triplicate.
Gain-of-function evolution of SIVcpz Nef
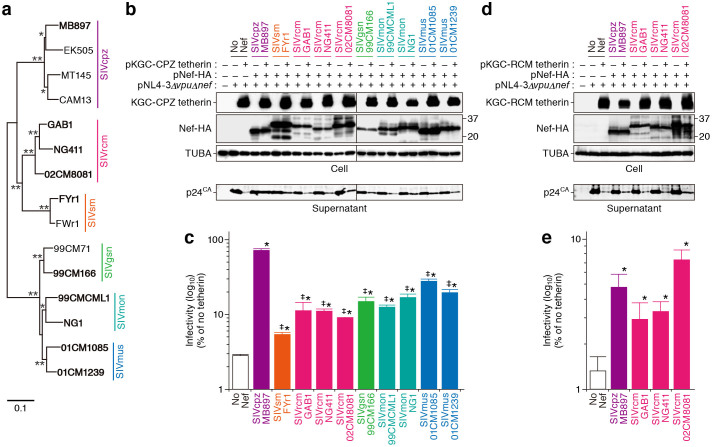
As described above, phylogenetic analyses has assumed that SIVcpz is the recombinant of two OWM SIV linages, SIVgsn/mon/mus and SIVrcm26. Consistent with previous reports5,14, we confirmed that SIVcpz Nef is phylogenetically closer to SIVrcm Nef than SIVgsn, SIVmon, and SIVmus Nefs (Figure 4a). We then directly evaluated the functional evolution process of SIV Nefs in terms of anti-tetherin ability. Although SIVcpz Nef efficiently antagonized CPZ tetherin, the counteraction efficacy of OWM SIV Nefs including SIVsm, SIVrcm and SIVgsn/mon/mus was significantly lower than that of SIVcpz Nef (Figures 4b and 4c). These results suggest that the difference in anti-tetherin activity (Figure 2) is not related to Nef but is due to the differences between RCM and CPZ tetherins. On the other hand, it was of interest that SIVcpz Nef counteracted RCM tetherin in comparable to SIVrcm Nefs (Figures 4d and 4e). Because SIVcpz Nef is able to antagonize RCM tetherin in addition to CPZ tetherin, these findings suggest that SIVcpz Nef has accomplished a gain-of-function during evolution.
Figure 4. Gain-of-function evolution of SIVcpz Nef.
(a) Phylogenic tree of SIV Nef. The strains indicated in bold were used in the experiments shown in panels (b–e). Bootstrap values are shown as follows: *, >50%; **, >80%. (b–e) Anti-viral effect of CPZ and RCM tetherins and the antagonism by SIV Nefs. (b and d) Western blotting. Representative results are shown. The number on the right of blots indicates kilodalton. Blots have been cropped; full uncropped blots are available as Supplementary Figure 3. (c and e) TZM-bl assay. The data represents the percentage of infectivity compared to the values without tetherin with SD. The assay was performed in triplicate. The statistic difference is determined by Student's t test. *, P < 0.05 versus no Nef; ‡, P < 0.05 versus SIVcpz Nef.
Discussion
In this study, we have determined the sequence of RCM tetherin. In addition, we have demonstrated that RCM tetherin has the ability to impair the release of nascent viral particles and that SIVrcm Nef is the bona fide antagonist against RCM tetherin. Although the two concepts: (i) SIVrcm Nef counteracts RCM tetherin; and (ii) this ability is succeeded to SIVcpz Nef, have been proposed elsewhere5,27, no direct evidence has been shown so far. Therefore, to our knowledge, this is the first report directly elucidating the interplay between SIVrcm Nef and RCM tetherin (Figure 2). Moreover, we here firstly performed the phylogenetic analysis of primate tetherins with Cercocebus tetherins including SM and RCM tetherins and revealed that stronger positive selection occurred in the evolution of Cercocebus tetherins (Figure 1).
To better understand the Red Queen dynamics in the co-evolution of PLVs and primates, our findings should be compared with the evolutionary interplay between Vpx and simian SAMHD1. To antagonize the anti-viral ability of simian SAMHD1, certain SIVs including SIVrcm degrade SAMHD1 by a viral protein, Vpx. In this regard, Etienne et al. have recently demonstrated that vpx, which is located at the recombination site of SIVrcm and SIVgsn/mon/mus, was lost during the emergence of SIVcpz and that SIVcpz does not possess any anti-CPZ SAMHD1 factor(s)41. When compared to the evolutionary interplay between SIV Vpx and SAMHD1, our findings on the relationship between SIV Nef and tetherin suggest that anti-tetherin ability was more crucial for PLVs, at least for SIVcpz, to adapt and expand in the new host because SIVcpz Nef has successfully inherited its anti-tetherin ability from SIVrcm Nef. Moreover, SIVcpz Nef has acquired the ability to antagonize CPZ tetherin without the loss of anti-RCM tetherin activity, indicating that SIVcpz Nef has made the way through a gain-of-function evolution (Figure 4). To our knowledge, here we demonstrated that SIVcpz Nef is able to antagonize OWM tetherin for the first time. Our findings can be a milestone to decipher the complicated co-evolutionary process between PLVs and primates.
As described in the beginning of this paper, HIV-2 has resulted from the zoonotic infection of SIVsm from SM25, while SIVrcm in RCM is one of the ancestral viruses of SIVcpz giving rise to HIV-126. These insights imply that the ancestors of these two HIVs have passed through the genus Cercocebus, namely SM or RCM. In this study, we firstly performed molecular phylogenetic analyses of primate tetherins with the genus Cercocebus and revealed that positive selection had operated on Cercocebus tetherins (Figure 1). Since it has been estimated that the genus Cercocebus has evolutionary diversified around 4–4.85 million years ago42,43, the observed positive selection probably occurred after the divergence of the genus Cercocebus. In fact, a recent paper has suggested that PLVs had already existed at least 12 million years ago44. Therefore, it is plausible that a Nef-like protein encoded by ancient PLV(s) can be regarded as a selective pressure against Cercocebus tetherins. Moreover, it is of interest that the 2 types of genetic deletion in CCR5, CCR5Δ2445 and CCR5Δ246, have been observed in the population of genus Cercocebus including RCM and SM, and that the homozygous mutants are resistant to CCR5-tropic SIV infections. To circumvent the restriction at the entry step of infection, some SIVrcm and SIVsm have acquired the ability to use alternative coreceptors for their infections presumably as a result of genetic conflict between viruses and hosts45,46. Although SIVsm is nonpathogenic in SM, it is conceivable that the ancestors of the genus Cercocebus have been subjected to stronger selective pressure from pathogenic PLVs and that the genetic conflict between pathogenic PLVs and the genus Cercocebus, including ancient Nef and Cercocebus tetherins, might have occurred in their evolutionary history.
Methods
Sequencing PCR
Blood was collected from six wild-caught RCMs in Nigeria according to the Guide for the Care and Use of Laboratory Animals47 under a NIAID Animal Care and Use Committee-approved protocol28,29. RCM genomic DNA was isolated from cryopreserved peripheral blood mononuclear cells28,29, and PCR was performed by using PfuUltra High Fidelity DNA polymerase (Agilent Technologies) and the following primers: 5′-CAG CTA GAG GGG AGA TCT GGA TG-3′; 5′-CTC ACT GAC CAG CTT CCT GGG-3′, which were used in a previous study31. The obtained PCR products (approximately 2 kb) were purified by gel extraction and directly sequenced by using BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems) with the two primers described above and the following 4 primers: 5′-GGA CTT CAC CAG ACC CTG AA-3′; 5′-TTC AGG GTC TGG TGA AGT CC-3′; 5′-TCT CTC CTT TGC TCC CAA AA-3′; 5′-TTT TGG GAG CAA AGG AGA GA-3′. The sequencing PCR was performed by using ABI Prism 3130 xl genetic analyzer (Applied Biosystems), and the data was analyzed by Sequencher v5.1 software (Gene Codes Corporation).
Phylogenetic analyses
The two RCM tetherins newly sequenced in this study (note that five out of the six analyzed RCM tetherin sequences were identical) was aligned with 45 primate tetherins (listed in Table 1) by using ClustalW implemented in MEGA548. The resulting alignment was verified manually at amino acid level. Then the phylogenetic tree of 47 primate tetherins was reconstructed using both neighbour-joining (NJ) method49 with MEGA5 and ML method with PhyML50. Since the two methods yield almost identical topology, the unrooted ML tree was used for further analyses.
The detection of positive selection was conducted as follows: first, to detect the positive selection across various primate lineages, two pairs of site models implemented in the PAML package v 4.751 were used to conduct the likelihood ratio tests: M1 (neutral model) versus M2 (selection model) and M7 (neutral model) versus M8 (selection model). The REL model in HyPhy was also employed to this analysis. Second, to calculate the dN/dS ratio of each primate tetherin, a free-ratio branch model in PAML was performed, which assumes an independent dN/dS ratio for each branch of the tree. Third, since we were particularly interested in whether the RCM and Cercocebus clades have evolved under positive selection, the branch-site model in PAML was further applied to our analyses. This model allows dN/dS ratio vary both among sites and branches, which is useful for detecting positive selection along a particular lineage or clade (pre-specified as foreground branches) with positively selected site identified32,52,53,54,55. We conducted two branch-site tests in which RCM and the Cercocebus clades were designated as the foreground branches, respectively.
SIV Nef sequences were aligned by using ClustalW implemented in MEGA548. The phylogenetic tree of SIV Nef was reconstructed using NJ method49 with MEGA5.
Plasmid construction
To construct tetherin expression plasmids tagged with Kusabira green C (KGC) at N-termini, tetherin ORFs were inserted into the KpnI-XhoI site of phmKGC-MC vector (Medical and Biological laboratories, Inc.). The ORFs of HU tetherin (GenBank accession number NM_004335), AGM tetherin (FJ943430), and mouse (Mus Musculus; MO) tetherin (NM_198095) were used in our previous report18. CPZ tetherin (XM_512491), GOR tetherin (GQ925926), RM tetherin (FJ943432), and SM tetherin (FJ864713), and RCM tetherin (AB907706, determined in this study) were generated by overlap extension PCR using artificially synthesized oligonucleotides (Greiner bio-one). The ORF of the 5 amino acid (14GDIWK18)-inserted HU tetherin (designated “HU + 5aa”) was prepared by overlap extension PCR using HU tetherin expression plasmid as the template and the following primers: 5′-GGG GTA CCC CCA TGG CAT CTA CTT CGT ATG-3′; 5′-TTT CTT CCA AAT GTC ATC CAT GGG CAC TCT GCA ATA-3′; 5′-GAT GAC ATT TGG AAG AAA GAC GGG GAT AAG CGC TGT-3′; 5′-CCG CTC GAG CGG ATC TCA CTG CAG CAG AGC GCT GA-3′. To construct SIV Nef expression plasmids tagged with Hemagglutinin (HA) at the C-termini, HA was added at the C-termini of Nef ORFs by PCR and the Nef-HA fragments were then inserted into the XbaI-MluI site of pCGCG-IRES-EGFP vector (kindly provided by Dr. Frank Kirchhoff)56. The ORFs of SIVsm Nef (strain FYr1 [DQ092760]), 2 SIVrcm Nefs (strains NG411 [AF349680] and 02CM8081 [HM803689]), SIVgsn Nef (strain 99CM166 [AF468659]), 2 SIVmon Nefs (strains 99CMCML1 [AY340701] and NG1 [GQ925927]), and 2 SIVmus Nefs (strains 01CM1085 [AY340700] and 01CM1239 [EF070330]) were obtained from GeneArt Gene Synthesis service (Life Technologies). The ORF of SIVrcm Nef (strain GAB1 [AF382829]) was generated by overlap extension PCR using artificially synthesized oligonucleotides (Greiner bio-one). The ORF of SIVcpz Nef (strain MB897 [EF535994]) was prepared by overlap extension PCR using pMB897 (an infectious molecular clone of SIVcpz strain MB897 [JN835461]; kindly provided by Dr. Beatrice Hahn)57 as the template and the following primers: 5′- TTA ATA CCT AGT CTA GAC TAG ATG GGA AAC AAA TGG TCA AAA AGT AGC-3′; 5′- TAG CGA CGC GTC GGC GAG ATC TCT AAG CGT AAT CTG GTA CGT CGT ATG GGT AGA TAT CGC AGT CTT TGT AGT ACT CCG GAT GC-3′. To construct pNL4-3ΔvpuΔnef (an HIV-1-producing plasmid with defects in vpu and nef), pNL43-Udel (a vpu-deficient HIV-1-producing plasmid, based on HIV-1 strain NL4-3 [M19921]; kindly provided by Dr. Klaus Strebel)40 was digested with XhoI, blunted, and then self-ligated. The sequencing PCR was performed by using ABI Prism 3130 xl genetic analyzer (Applied Biosystems), and the data were analyzed by Sequencher v5.1 software (Hitachi).
Cell culture and transfection
293T cells and TZM-bl cells (obtained through NIH AIDS Research and Reference Reagent Program) were maintained in Dulbecco's modified Eagle medium (Sigma) containing FCS and antibiotics. Transfection was performed by using Lipofectamine 2000 (Life Technologies) according to the manufacture's protocol. Various amounts of KGC-tagged tetherin expression plasmids (0, 20, 40, 80 ng) and pNL4-3ΔvpuΔnef (1,500 ng) were cotransfected with or without the expression plasmids of SIV Nefs of their natural hosts (400 ng) into 293T cells. At 48 hours post-transfection, the culture supernatants and transfected cells were harvested and were respectively used for TZM-bl assay and Western blotting as described below.
Viral budding and Western blotting
The culture supernatant harvested at 48 hours post-transfection was centrifuged and then filtered through a 0.45-μm-pore-size filter (Milipore) to produce virus solution. The infectivity of virus solution was measured by TZM-bl assay as previously described18,19,58. Briefly, 10 μl of the virus solution was inoculated into TZM-bl cells in 96-well plate (Nunc), and the β-galactosidase activity was measured by using the Galacto-Star mammalian reporter gene assay system (Roche) and a 2030 ALBO X multilabel counter instrument (PerkinElmer) according to the manufacturers' procedure. Western blotting was performed as previously described18,19,58 by using the following antibodies: anti-p24 polyclonal antibody (ViroStat), anti-KGC antibody (clone 21B10; Medical and Biological Laboratories, Inc.), anti-HA antibody (3F10; Roche), and anti-α-Tubulin (TUBA) antibody (DM1A; Sigma). For the virus solution, 500 μl of the virus solution was ultracentrifuged at 100,000 × g for 1 hour at 4°C using a TL-100 instrument (Beckman). The pellet was lysed with 1 × SDS buffer, and the lysate was used for SDS-PAGE/Western blotting. For the transfected cells, the cells were lysed with 1 × SDS buffer, and the lysate was used for SDS-PAGE/Western blotting.
NFκB reporter assay
One hundred nanogram of either respective tetherin expression plasmid or parental empty vector (phmKGC-MC) was cotransfected with 25 ng of an NFκB-luciferase reporter plasmid (p55A2-Luc; kindly provided by Dr. Takashi Fujita)39, 250 ng of a vpu-deficient HIV-1-producing plasmid (pNL43-Udel; kindly provided by Dr. Klaus Strebel)40, and 125 ng of pUC19 plasmid into 293T cells. At 48 hours post-transfection, the cells were harvested and used for Western blotting and luciferase assay. Luciferase assay was performed as previously described58.
Statistical analyses
The data expressed as average with standard deviation (SD), and significant differences (P < 0.05) were determined by Student's t test.
Author Contributions
T.K., J.S.T., F.R., K.M., K.S., H.T., V.M.H. and Y.Koyanagi wrote the main manuscript text; J.S.T. and F.R. performed molecular phylogenetic analyses and prepared Figure 1; T.K., J.S.T., K.S., Y.Kimura, N.M., R.Y., Y.N. and E.Y. performed the experiments and prepared Figures 2–4; K.M. and V.M.H. contributed to the samples of RCM genomic DNA; K.S. conceived and designed the experiments. All authors reviewed the manuscript.
Supplementary Material
Supprementary Information
Acknowledgments
We would like to thank Drs. Takashi Fujita (Institute for Virus Research, Kyoto University, Japan), Klaus Strebel (National Institute of Allergy and Infectious Diseases, NIH, USA), Frank Kirchhoff (University of Ulm, Germany), and Beatrice Hahn (University of Pennsylvania, USA) for providing p55A2-Luc, pNL43-Udel, pCGCG-IRES-EGFP, and pMB897, respectively. This study was supported in-part by grants from the following: the Aihara Innovative Mathematical Modeling Project, the Japan Society for the Promotion of Science through the “Funding Program for World-Leading Innovative R&D on Science and Technology (FIRST Program),” initiated by the Council for Science and Technology Policy of Japan (to K.S.); Takeda Science Foundation (to K.S.); Sumitomo Foundation Research Grant (to K.S.); Senshin Medical Research Foundation (to K.S.); the intramural research program of NIAID, NIH (to K.M. and V.M.H.); Grants-in-Aid for Scientific Research B24390112 (to Y.Koyanagi) and S22220007 (to Y.Koyanagi) from JSPS; a Grant-in-Aid for Scientific Research on Innovative Areas 24115008 (to Y.Koyanagi) from the Ministry of Education, Culture, Sports, Science and Technology of Japan; and Research on HIV/AIDS (to Y.Koyanagi) from the Ministry of Health, Labor and Welfare of Japan.
References
- Klatt N. R., Silvestri G. & Hirsch V. Nonpathogenic simian immunodeficiency virus infections. Cold Spring Harb. Perspect. Med. 2, a007153 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrosiers R. C. Nonhuman Lentiviruses. Fields Virology (eds Knipe, D. M. & Howley, P. M.) 2215–2243 (Lippincott Williams & Wilkins, Philadelphia, 2007). [Google Scholar]
- Duggal N. K. & Emerman M. Evolutionary conflicts between viruses and restriction factors shape immunity. Nat. Rev. Immunol. 12, 687–695 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford R. J. Viral evolution in deep time: lentiviruses and mammals. Trends Genet. 28, 89–100 (2012). [DOI] [PubMed] [Google Scholar]
- Kirchhoff F. Immune evasion and counteraction of restriction factors by HIV-1 and other primate lentiviruses. Cell Host Microbe 8, 55–67 (2010). [DOI] [PubMed] [Google Scholar]
- Dawkins R. & Krebs J. R. Arms races between and within species. Proc. R. Soc. Lond. B. Biol. Sci. 205, 489–511 (1979). [DOI] [PubMed] [Google Scholar]
- Sheehy A. M., Gaddis N. C., Choi J. D. & Malim M. H. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418, 646–650 (2002). [DOI] [PubMed] [Google Scholar]
- Laguette N. et al. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrecka K. et al. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474, 658–661 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil S. J., Zang T. & Bieniasz P. D. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451, 425–430 (2008). [DOI] [PubMed] [Google Scholar]
- Van Damme N. et al. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3, 245–252 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas J. L. et al. The great escape: viral strategies to counter BST-2/tetherin. PLoS Pathog. 6, e1000913 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia B. et al. Species-specific activity of SIV Nef and HIV-1 Vpu in overcoming restriction by tetherin/BST2. PLoS Pathog. 5, e1000429 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter D. et al. Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe 6, 409–421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra-Moreno R., Zimmermann K., Stern L. J. & Evans D. T. Tetherin/BST-2 antagonism by Nef depends on a direct physical interaction between Nef and tetherin, and on clathrin-mediated endocytosis. PLoS Pathog. 9, e1003487 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F. et al. SIV Nef proteins recruit the AP-2 complex to antagonize Tetherin and facilitate virion release. PLoS Pathog. 7, e1002039 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikovics K. et al. Counteraction of tetherin antiviral activity by two closely related SIVs differing by the presence of a Vpu gene. PLoS One 7, e35411 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T. et al. Identification of amino acids in the human tetherin transmembrane domain responsible for HIV-1 Vpu interaction and susceptibility. J. Virol. 85, 932–945 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K. et al. Comparative study on the effect of human BST-2/Tetherin on HIV-1 release in cells of various species. Retrovirology 6, 53 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M. & Hahn B. H. The evolution of HIV-1 and the origin of AIDS. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 2487–2494 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim J. O. & Worobey M. Dating the age of the SIV lineages that gave rise to HIV-1 and HIV-2. PLoS Comput. Biol. 5, e1000377 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worobey M. et al. Direct evidence of extensive diversity of HIV-1 in Kinshasa by 1960. Nature 455, 661–664 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F. et al. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397, 436–441 (1999). [DOI] [PubMed] [Google Scholar]
- Keele B. F. et al. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science 313, 523–526 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch V. M. et al. An African primate lentivirus (SIVsm) closely related to HIV-2. Nature 339, 389–392 (1989). [DOI] [PubMed] [Google Scholar]
- Bailes E. et al. Hybrid origin of SIV in chimpanzees. Science 300, 1713 (2003). [DOI] [PubMed] [Google Scholar]
- Evans D. T., Serra-Moreno R., Singh R. K. & Guatelli J. C. BST-2/tetherin: a new component of the innate immune response to enveloped viruses. Trends Microbiol. 18, 388–396 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer B. E. et al. Characterization of novel simian immunodeficiency viruses from red-capped mangabeys from Nigeria (SIVrcmNG409 and -NG411). J. Virol. 75, 12014–12027 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinton C. et al. CD4-like immunological function by CD4− T cells in multiple natural hosts of simian immunodeficiency virus. J. Virol. 85, 8702–8708 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R. K. et al. Mutation of a single residue renders human tetherin resistant to HIV-1 Vpu-mediated depletion. PLoS Pathog. 5, e1000443 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim E. S., Malik H. S. & Emerman M. Ancient adaptive evolution of tetherin shaped the functions of Vpu and Nef in human immunodeficiency virus and primate lentiviruses. J. Virol. 84, 7124–7134 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Chen K., Wang J. H. & Zhang C. Molecular evolution of the primate antiviral restriction factor tetherin. PLoS One 5, e11904 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNatt M. W. et al. Species-specific activity of HIV-1 Vpu and positive selection of tetherin transmembrane domain variants. PLoS Pathog. 5, e1000300 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W. et al. Regulation of TLR7/9 responses in plasmacytoid dendritic cells by BST2 and ILT7 receptor interaction. J. Exp. Med. 206, 1603–1614 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F. et al. Nef proteins from simian immunodeficiency viruses are tetherin antagonists. Cell Host Microbe 6, 54–67 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galao R. P. et al. Innate sensing of HIV-1 assembly by Tetherin induces NFκB-dependent proinflammatory responses. Cell Host Microbe 12, 633–644 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokarev A. et al. Stimulation of NF-κB activity by the HIV restriction factor BST2. J. Virol. 87, 2046–2057 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocka L. J. & Bates P. Identification of alternatively translated Tetherin isoforms with differing antiviral and signaling activities. PLoS Pathog. 8, e1002931 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita T. et al. The candidate proto-oncogene bcl-3 encodes a transcriptional coactivator that activates through NF-κ B p50 homodimers. Genes Dev. 7, 1354–1363 (1993). [DOI] [PubMed] [Google Scholar]
- Klimkait T. et al. The human immunodeficiency virus type 1-specific protein vpu is required for efficient virus maturation and release. J. Virol. 64, 621–629 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne L. et al. Gene loss and adaptation to hominids underlie the ancient origin of HIV-1. Cell Host Microbe 14, 85–92 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelman P. et al. A molecular phylogeny of living primates. PLoS Genet. 7, e1001342 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman M. et al. Toward a phylogenetic classification of Primates based on DNA evidence complemented by fossil evidence. Mol. Phylogenet. Evol. 9, 585–598 (1998). [DOI] [PubMed] [Google Scholar]
- Compton A. A. & Emerman M. Convergence and divergence in the evolution of the APOBEC3G-Vif interaction reveal ancient origins of simian immunodeficiency viruses. PLoS Pathog. 9, e1003135 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. et al. Natural infection of a homozygous delta24 CCR5 red-capped mangabey with an R2b-tropic simian immunodeficiency virus. J. Exp. Med. 188, 2057–2065 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddick N. E. et al. A novel CCR5 mutation common in sooty mangabeys reveals SIVsmm infection of CCR5-null natural hosts and efficient alternative coreceptor use in vivo. PLoS Pathog. 6, e1001064 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council. Guide for the care and use of laboratory animals (National Academies Press, Washington, D. C., 1996).
- Tamura K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N. & Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987). [DOI] [PubMed] [Google Scholar]
- Guindon S. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010). [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- Gharib W. H. & Robinson-Rechavi M. The branch-site test of positive selection is surprisingly robust but lacks power under synonymous substitution saturation and variation in GC. Mol. Biol. Evol. 30, 1675–1686 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. & dos Reis M. Statistical properties of the branch-site test of positive selection. Mol. Biol. Evol. 28, 1217–1228 (2011). [DOI] [PubMed] [Google Scholar]
- Zhang J., Nielsen R. & Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 22, 2472–2479 (2005). [DOI] [PubMed] [Google Scholar]
- Yoshida I. et al. Change of positive selection pressure on HIV-1 envelope gene inferred by early and recent samples. PLoS One 6, e18630 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carl S. et al. Modulation of different human immunodeficiency virus type 1 Nef functions during progression to AIDS. J. Virol. 75, 3657–3665 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Heuverswyn F. et al. Genetic diversity and phylogeographic clustering of SIVcpzPtt in wild chimpanzees in Cameroon. Virology 368, 155–171 (2007). [DOI] [PubMed] [Google Scholar]
- Sato K. et al. Modulation of human immunodeficiency virus type 1 infectivity through incorporation of tetraspanin proteins. J. Virol. 82, 1021–1033 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supprementary Information