Abstract
The use of massively parallel sequencing for studying RNA expression has greatly enhanced our understanding of the transcriptome through the myriad ways these data can be characterized. In particular, clinical samples provide important insights about RNA expression in health and disease, yet these studies can be complicated by RNA degradation that results from the use of formalin as a clinical preservative and by the limited amounts of RNA often available from these precious samples. In this study we describe the combined use of RNA sequencing with an exome capture selection step to enhance the yield of on-exon sequencing read data when compared with RNA sequencing alone. In particular, the exome capture step preserves the dynamic range of expression, permitting differential comparisons and validation of expressed mutations from limited and FFPE preserved samples, while reducing the data generation requirement. We conclude that cDNA hybrid capture has the potential to significantly improve transcriptome analysis from low-yield FFPE material.
RNA sequencing (RNA-Seq) approaches are designed to characterize the expressed genome in numerous ways1,2 from defining different types of RNA, such as long noncoding RNAs,3 to comparing RNA expression,4 splice isoforms,5–7 allele-specific expression,8–10 fusions,11–14 RNA editing,15,16 and other complex questions that define RNA. This inquiry has been enriched by the development of massively parallel sequencing applications that permit large data sets to be generated quickly and at relatively low cost. In particular, the characterization of RNA expression as a comparator of diseased versus normal cells from clinical samples extends information gained from DNA-based studies, often revealing insights that would be impossible to ascertain by looking at DNA alone, such as allele-specific or elevated expression levels.10 To date, most of the discovery studies of transcriptome analyses have traditionally been conducted using fresh frozen (FF) tumor samples with stringent criteria applied in terms of cellularity, tumor necrosis, and RNA quality. However, most clinical samples collected are not FF and are complicated by two common characteristics; the use of formalin fixation to preserve protein and cellular structure for pathologic examination of the tissue causes degradation of the RNA over time due to cross-linking and backbone breakage, and clinical samples often are available only in limited amounts that provide an equally limited yield of nucleic acids. Furthermore, the nonuniformity of preservation methods (eg, fixation time, fixative concentration, and tissue size) can negatively affect sample quality. Limited sample material also results when flow sorting or laser capture microdissection is used to purify the cells of interest or when core biopsies or fine needle aspirates are obtained for clinical diagnostic procedures. These low-yield samples obviate the possibility of isolating polyadenylated transcripts in advance of RNA sequencing because this isolation would further decrease the amount of RNA available for library construction, which may introduce sample issues, such as biased transcript representation.
Hence, in the context of pursuing several projects of interest for our cancer genomics research, we attempted to combine RNA-Seq with an intermediate enrichment step of exome capture, which we refer to as cDNA-Capture sequencing, as a means of addressing these challenges. Several studies have set the precedent in describing targeted approaches to RNA sequencing, although they focused on monitoring tens to hundreds of genes using high-quality material.17–19 Our initial application of cDNA-Capture sequencing was from abundant samples for the purposes of identifying transcripts that were mutated and contributed to host immunosurveillance and immunoediting.20 Subsequent studies, described here, have further developed the method of obtaining high-quality RNA-Seq data from samples that have exceptionally low amounts of total RNA or have compromised RNA quality because of the use of formalin fixation. In this article, we present our approach and illustrate the utility of the method for detecting expressed variants from degraded RNA due to formalin-mediated damage and for determining gene expression levels from extremely limited input material. In addition, we demonstrate that the hybrid capture step provides a cost advantage for data generation by concentrating the data yield onto the exome. The resulting data suggest improved validation rates of single-nucleotide variants (SNVs) and detection of gene fusions and splice isoforms while preserving the dynamic range of detection for low-abundance transcripts.
Material and Methods
Transcriptome Sequencing
For the FF tumor RNA samples (LUC4, LUC6, LUC7, LUC13, LUC20) and LNCaP prostate cancer cells, we selected poly(A) mRNA from approximately 950 ng of input total RNA using the Ambion MicroPoly(A)Purist Kit [Thermo Fisher Scientific Inc., Pittsburgh, PA (previously Life Technologies, Carlsbad, CA)] and converted 20 ng of isolated mRNA into cDNA using the Ovation RNA-Seq System version 2 (NuGEN, San Carlos, CA), as previously described.10 All FF samples had an RNA Integrity Number (RIN) value of at least 8.0 except LUC7, which was assessed in duplicate and had RIN values of 6.5 and 7.4 (Supplemental Table S1). Because the LUC7-T FF failed to generate cDNA with poly(A) mRNA, we converted 20 ng of LUC7-T FF total RNA into cDNA. As part of our standard operating procedures, the formalin-fixed, paraffin-embedded (FFPE) LUC6 and LUC7 RNA, 1200 ng and 1120 ng, respectively, was DNase treated and recovered using a 1:1.6 sample to RNAClean XP bead ratio. LUC6 and LUC7 FFPE samples had RIN values of 2.0 and 1.9 and were 4.75 and 5.83 years old, respectively, when RNA was isolated (Supplemental Table S2). FFPE-DNase RNA (150 ng) was used as input into the Ovation RNA-Seq FFPE System (NuGEN) per the manufacturer protocol. Because of the already small fragment size distribution of the NuGEN-generated cDNA, no additional fragmentation was performed. One microgram of each cDNA sample was converted into Illumina-ready libraries as described.
SeqCap EZ Human Exome Library Capture Experiments
The LUC cDNA-converted Illumina libraries were enriched by hybridization to the SeqCap EZ Human Exome Library version 3.0 reagent (Roche NimbleGen, Madison, WI). The targeted genomic regions in this kit cover 63.5 Mb or 2.1% of the human reference genome, including 98.8% of coding regions, 23.1% of untranslated regions (UTRs), and 55.5% of miRNA bases (as annotated by Ensembl version 7321). Each hybridization reaction was incubated at 47°C for 72 hours, and single-stranded capture libraries were recovered and cycle amplified per the manufacturer protocol. The exome capture experimental specifics are listed in Supplemental Table S3, which describes RNA type, library mass used per capture, pooling scheme, and post-capture PCR cycles. Post-capture library sizing used AMpureXP beads to remove residual primer dimers from post-capture PCR amplification, and libraries were diluted to 2 nmol/L for subsequent Illumina sequencing.
cDNA-Capture Dilution Experiment Using Colon Specimens
Because clinically relevant RNA sources may be limiting in quantity, we evaluated the effect of DNase-treated low-input sources by generating a dilution series. Human adult colon RNA and human adult colon adenocarcinoma RNA (Agilent Technologies, Santa Clara, CA) were assessed using Qubit Fluorometric Quantitation and the Quant-iT RNA Assay (Life Technologies, Grand Island, NY). These samples had RIN values of 7.9 and 8.0, respectively. We diluted the normal and adenocarcinoma colon RNA to 5, 1, 0.2, and 0.08 ng/μL in 10 μL of nuclease-free water (Life Technologies, Grand Island, NY). Each dilution was performed in triplicate and corresponded to an RNA mass of 50, 10, 2, and 0.8 ng per sample, respectively. Although our initial experiment, using 60 ng of input RNA, did not undergo a DNase treatment step, we decided to add this step to the lower RNA inputs to mimic our in-house protocol for cellular RNA isolates. We assessed the RIN value for each diluted RNA sample using the Agilent RNA 6000 Pico Assay chip (Agilent Technologies). Next, we treated each 10-μL RNA sample with 2 units of TURBO DNase (Life Technologies), concentrated the DNase-treated RNA samples using a 1:1.8 sample to RNAClean XP bead ratio (Beckman Coulter, Indianapolis, IN), and recovered the RNA in 10 μL of nuclease-free water. Each RIN value was reassessed as above and reported in Supplemental Table S4. These four DNase-treated RNA samples and the 60 ng of non–DNase-treated total RNA were used as input into the Ovation RNA-Seq System version 2 following the manufacturer protocol (NuGEN). The generated cDNA was assessed for concentration using the Quant-iT dsDNA HS Assay (Life Technologies) (Supplemental Table S5). DNA molecular weight distribution analysis used BioAnalyzer 2100 (data not shown) and Agilent DNA 7500 Chip Assay (Agilent Technologies).
We fragmented 100 ng of FF-generated cDNA (for each RNA input, in triplicate) in 1× DNA Terminator End Repair Buffer (Lucigen, Middleton, WI) using the Covaris S2 and microTUBEs (Covaris, Woburn, MA) on the following settings: volume, 50 μL; temperature, 4°C; duty cycle, 5; intensity, 4; cycle burst, 200; and time, 90 seconds. The fragmented ends were converted to blunt ends by adding DNA Terminator End Repair Enzyme following the manufacturer protocol. The blunt-ended DNA was purified using a 1:1.6 sample to AMpure XP bead ratio (Beckman Coulter). Adenylation of the 3′ DNA fragments used 15 units of the Klenow Fragment (3′→5′ exo; New England BioLabs, Ipswich, MA). Each sample was then ligated with 90 nmol/L of an Integrated DNA Technologies (Coralville, IA) synthesized dual same index adapter (oligonucleotide sequences; Illumina, Inc., San Diego, CA). These index adapters are similar to Illumina TruSeq HT adapters but have the same 8 bp index on both strands of the adapter. Binning of multiplexed sample reads requires 100% identity from the forward and reverse index sequencing reaction. For the non–DNase-treated sample (60 ng), the library was generated using the Illumina TruSeq LT single-index adapter. The ligation reactions were accomplished using 5000 units of T4 DNA ligase (New England BioLabs). To purify each ligation reaction and reduce adapter-dimer carryover, we used a 1:1.3 sample to AMpure XP bead ratio. Next, for each library ligation, we performed PCR optimization to prevent overamplification. The PCR optimization procedure used 1 μL of ligated sample into the KAPA SYBRFAST Universal 2× qPCR Master Mix protocol (Kapa Biosystems, Inc., Woburn, MA) and the universal Illumina library primers: forward 5′ P5 primer (5′-AATGATACGGCGACCACCGAGATCTA-3′) and reverse 3′ P7 primer (5′-CAAGCAGAAGACGGCATACGAGAT-3′). PCR amplifications were performed using the Mastercycler ep realplex real-time PCR system (Eppendorf, Hamburg, Germany). Once the optimal PCR cycle number for each sample was determined, we performed eight PCR reactions per sample using the 2× Phusion High-Fidelity PCR Master Mix with HF Buffer (New England BioLabs) and 200 nmol/L P5 and P7 primers. For each sample octet, we combined and purified the PCR reactions using MinElute PCR Purification columns according to manufacturer protocol (Qiagen Inc., Valencia, CA). Each amplified ligation was then assessed for concentration using Quant-iT dsDNA HS Assay and for size using the BioAnalyzer 2100 and the Agilent DNA 1000 Assay (Agilent Technologies).
We used 500 ng of each library for SeqCap EZ Human Exome Library version 3.0 capture. The aliquots were then pooled, totaling 3 μg of pooled library per capture (Supplemental Table S6). Each hybridization reaction was incubated at 47°C for 72 hours, and single-stranded capture hybrid fragments were recovered and cycle amplified per the manufacturer protocol. Capture libraries were subsequently sized to approximately 300 to 500 bp using a 1:0.6 sample to AMpureXP bead ratio to which the supernatant was added to 0.9× volumes of beads. The resulting supernatant was discarded, the beads washed, and size-fractioned capture libraries were eluted and diluted to 2 nmol/L stocks for subsequent Illumina sequencing. These data are available through National Center for Biotechnology Information Sequence Read Archive (http://www.ncbi.nlm.nih.gov/gene; accession number PRJNA228917).
RNA-Seq and cDNA-Capture Analysis
Quality of raw RNA sequence data were assessed by use of FastQC version 0.10.0 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). Paired 2 × 100–bp sequence reads were first trimmed to remove single primer isothermal amplification adapters (ligated during cDNA synthesis) using the read trimmer FAR/Flexbar version 2.17 (http://sourceforge.net/projects/flexbar) with the following parameters set: ‘–adapter CTTTGTGTTTGA –trim-end left –adaptive-overlap yes –format fastq –write-lengthdist yes –nr-threads 4 –min-overlap 7 –max-uncalled 150 –min-read length 25’. After trimming, reads were aligned to a modified version of the human genome reference sequence (National Center for Biotechnology Information build 37) with alternative haplotype sequences omitted. Initial segmented alignments were performed using bowtie version 2.0.0-beta722 followed by spliced alignments with TopHat version 2.0.4.23 During alignment, TopHat was supplied transcript models in gene transfer format (GTF) using the ‘-g’ parameter. Transcript models representing known and predicted human transcripts were obtained from Ensembl version 67.21 The binary sequence alignment files obtained by alignment of RNA-Seq reads with TopHat were summarized by use of SAMStat version 1.08 and SAM tools version 0.1.18 (specifically the idxstats and flagstat utilities).24 Reads aligning to the target region were extracted using samtools view (specifying the BED file of target regions with the ‘–L’ parameter). The percentage of enrichment for the targeted region was calculated as the number of reads with both ends uniquely aligned to the target region divided by the total number of uniquely aligned reads. The quality of alignments was assessed by use of Picard version 1.52 (specifically the RnaSeqMetrics utility; http://picard.sourceforge.net/command-line-overview.shtml). Duplication rates were calculated using Picard MarkDuplicates. After alignment, expression estimates in the form of fragments per kilobase of exon per million bases mapped (FPKM) were calculated by Cufflinks version 2.0.2.25 Transcript models were supplied to Cufflinks using the ‘-g’ option and the same GTF described above. Transcripts corresponding to mitochondrial and ribosomal genes were masked during calculation of transcript expression estimates. Exon-exon junction statistics were obtained by parsing the junctions.bed file produced by TopHat. This file reports the coordinates of all introns observed by splice aware alignment of reads to the genome and the number of reads supporting each. Each observed exon-exon junction was cross-referenced against the known junctions of Ensembl version 67 human transcripts. GC content was calculated as the percentage of GC bases using Ensembl gene annotations. Genes were split into four equal-sized bins based on GC content. Gene expression values were calculated as the mean FPKM across all samples and were subsequently log2 transformed.
Variant allele frequencies (VAFs) were calculated by interrogating binary sequence alignment files with the Bio::DB::Sam BioPerl package at somatic SNV positions detected in whole genome sequence data from tumor and normal DNA samples from the same tumors as those profiled by RNA sequencing. Specifically, the VAF for a variant is the ratio of variant supporting reads to the total number of reads covering the variant position. Somatic variants were detected by a union of VarScan version 2.2.6,26 Somatic Sniper version 1.0.2,27 and Strelka version 0.4.6.2.28 Variants predicted from each of these somatic variant callers were filtered according to the authors' instructions. Variants considered in this analysis were further limited to only Tier 1 variants (ie, those occurring within the protein-coding portion of exons or anywhere within a predicted noncoding RNA).
Gene fusions were detected using ChimeraScan version 0.4.529 with default parameters. Read counts for each fusion were determined by aggregating the encompassing and spanning reads identified by ChimeraScan. Normalized gene fusion read support was calculated as the total number of encompassing and spanning reads per million reads sequenced.
Figures were created in R version 2.15.2 (http://www.r-project.org) using packages ggplot230 and VennDiagram.31
Differential Gene Analysis
Read counts were obtained for the set of Ensembl version 67 transcripts using BEDTools version 2.16.232 for each colon replicate. Transcripts without a corresponding HUGO gene symbol were removed. If a gene had multiple transcripts, only the transcript with the highest overall count across all replicates was kept. For each dilution, lowly expressed genes were removed by requiring at least three samples to have at least 50 read counts. Differentially expressed genes between the three tumor and normal replicates from each dilution were calculated using edgeR version 3.0.833 with a false discovery rate cutoff of 10−5. For each pair of dilutions, Spearman's rank correlation and corresponding P value were calculated between the edgeR log10 P values.
Results
cDNA-Capture on Lung Adenocarcinoma in FF Samples
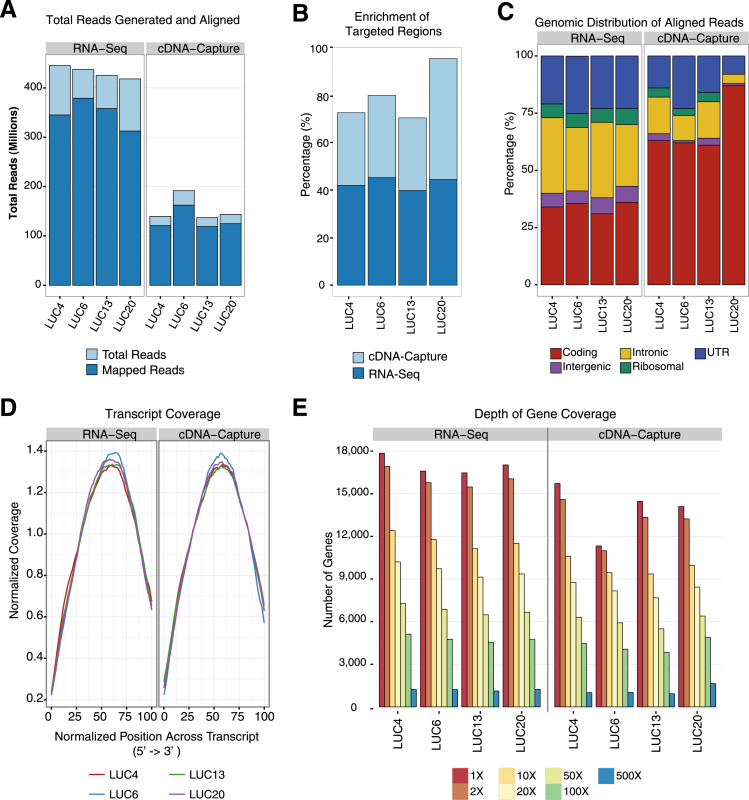
To evaluate the performance of cDNA-Capture using isolated polyA mRNA from FF samples, we first compared data from this approach to previously generated RNA-Seq data from four lung adenocarcinoma (LUC) patients.10 In contrast to the 418 million to 445 million transcriptome reads generated from each RNA-Seq library, only 137 million to 191 million reads were generated from each cDNA-Capture library (Figure 1A). The percentage of reads mapped to the genome was similar for RNA-Seq (74% to 86%) and cDNA-Capture (84% to 86%) (Figure 1A and Supplemental Table S7). However, the distribution of the alignments varied between the two approaches. Hybrid capture led to a >30% increase in the proportion of reads aligning to the targeted regions for each sample (Figure 1B). As a result, relative to RNA-Seq, all of the cDNA-Capture libraries displayed both a decrease in the intronic aligning reads (cDNA-Capture mean, 11.8%; RNA-Seq mean, 30.2%) and an increase in the proportion of reads aligning to coding regions (cDNA-Capture mean, 68.3%; RNA-Seq mean, 34.1%) (Figure 1C). The coverage across transcripts was similar for both cDNA-Capture and RNA-Seq data, with greatest coverage occurring in the middle of transcripts (Figure 1D). We also observed a similar distribution in the depth of gene coverage for RNA-Seq and cDNA-Capture (Figure 1E). Taken together, this finding suggests that cDNA-Capture sequencing using FF specimens achieves similar coverage levels as RNA-Seq, with only one-third the amount of sequencing reads.
Figure 1.
cDNA-Capture sequencing of FF lung adenocarcinomas. A: Total reads generated (light blue) and aligned (dark blue) for both RNA-Seq and cDNA-Capture across four FF lung adenocarcinomas (LUC4, LUC6, LUC13, and LUC20). B: Percentage of reads that aligned uniquely to the target regions for RNA-Seq and cDNA-Capture. C: Distribution of read alignments relative to genomic features, including coding, intergenic, intronic, ribosomal, and UTRs. D: Normalized coverage across transcripts for LUC4, LUC6, LUC13, and LUC20 from 5′ (left) to 3′ (right). E: Frequency of genes expressed at increasing coverage depth (1×, 2×, 10×, 20×, 50×, 100×, and 500×) using RNA-Seq and cDNA-Capture.
Gene Expression Using cDNA-Capture
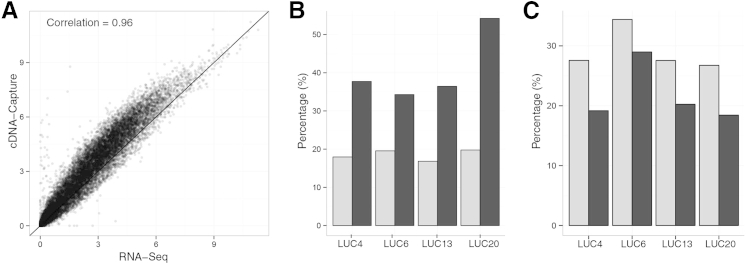
To assess the ability of cDNA-Capture to recapitulate gene expression values observed with RNA-Seq, we measured gene-level expression and compared the two approaches. Of the 19,741 protein-coding genes, 98.8% had corresponding probes in the capture reagent and thus should be enriched by cDNA-Capture. There existed a high concordance of gene expression for the set of all protein-coding genes (Pearson correlation, 0.93 to 0.96; one-sided P < 10−15) across all four lung tumors (Figure 2A and Supplemental Figure S1). More than two-thirds of genes (67.7% to 73.2%) had higher FPKM expression values in cDNA-Capture than RNA-Seq. There was no clear effect of poor probe design on gene expression because even genes that contained several short exons were adequately covered (data not shown). On average, cDNA-Capture was able to rescue high expression levels (FPKM > 1) of 25 genes (range, 18 to 37) that were missed by RNA-Seq (FPKM <0.1). Conversely, for three of the lung tumors, fewer than four genes (range, 3 to 4) displayed high expression in RNA-Seq but were missed by cDNA-Capture, with the exception of LUC20 (65 genes). cDNA-Capture also showed a consistent increase in the percentage of reads spanning exon-exon boundaries, thereby providing higher read depth for alternative splicing analysis (Figure 2B).
Figure 2.
Comparison of cDNA-Capture and RNA-Seq using FF lung tumors. A: Scatterplot of LUC13 gene expression values measured by RNA-Seq and cDNA-Capture. Gene expression is measured as log2(FPKM + 1). B: Percentage of reads aligning to exon-exon junctions from RNA-Seq (gray bars) and cDNA-Capture (black bars). C: Percentage of reads spanning a splice junction that aligned to the highest 1% of expressed genes from RNA-Seq and cDNA-Capture.
One challenge with accurately detecting low-abundance transcripts is that the highest expressing genes consume a significant proportion of the reads generated. cDNA-Capture is designed to increase the representation of the lowest expressed genes in the transcriptome while minimizing the oversequencing of the most highly expressed genes. In all four lung cancer samples, the percentage of reads spanning splice junctions consumed by the top 1% of expressed genes was lower using cDNA-Capture relative to RNA-Seq (Figure 2C). We chose to measure expression using this metric because reads spanning exon junctions are less prone to ambiguous alignments34 and thus may provide a more sensitive and accurate measurement of transcript expression levels. Overall, this suggests that a greater percentage of the reads generated by cDNA-Capture were distributed across genes with lower expression levels. Because increased representation of lower expressed genes commensurately decreases the representation of the highest expressed genes, our next aim was to determine the accuracy of cDNA-Capture expression levels of the most highly expressed genes. We measured the correlation between RNA-Seq and cDNA-Capture for the top 1% (n = 196) of highest expressed genes in RNA-Seq (Supplemental Figure S2). Excluding LUC20, correlations ranged from 0.73 to 0.85, suggesting high accuracy of the expression levels for these genes from cDNA-Capture data. LUC20 had a significantly smaller correlation of 0.36. Interestingly, LUC20 also had a much higher enrichment to the targeted regions (95%) than the other lung tumors (70% to 80%). Therefore, the capture enrichment step may provide a large increase in gene expression values for the lowest expressed genes without sacrificing accuracy of expression levels for the highest expressed genes.
It has previously been demonstrated that GC content can bias RNA-Seq expression.35,36 We chose to investigate whether there is any bias in cDNA-Capture expression due to the GC content of targeted regions. Compared with RNA-Seq data, cDNA-Capture data resulted in increased normalized expression levels across the entire range of GC content, including much larger gains for genes with lower GC content (Supplemental Figure S3A). However, similar to RNA-Seq, cDNA-Capture expression levels had a bias, providing lower expression levels as GC content increases.
Validation of SNVs Using FF Samples
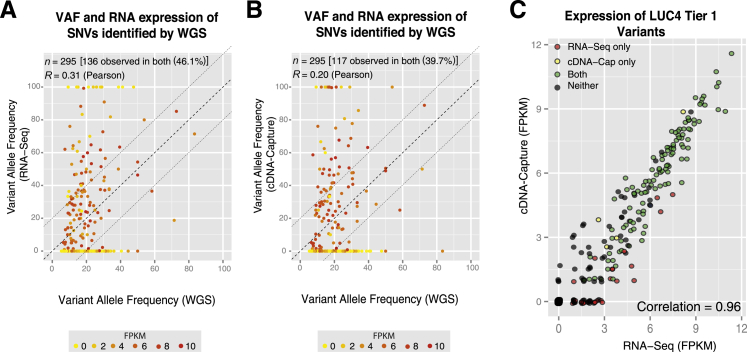
An increasingly common application of RNA-Seq is to validate expressed SNVs identified by whole genome sequencing. For each LUC sample, we previously conducted whole genome analysis to identify SNVs within protein-coding genes or Tier 1 SNVs.10 We compared the ability of RNA-Seq and cDNA-Capture to validate the expression of these SNVs. Because many SNVs reside in genes that are not expressed, or expressed at low levels, we do not expect either RNA-Seq or cDNA-Capture to confirm all SNVs. Of the 295 SNVs detected in one tumor (LUC4), RNA-Seq (Figure 3A) and cDNA-Capture (Figure 3B) had similar validation rates of 46.1% and 39.7%, respectively. These percentages are fairly consistent across the remaining samples as cDNA-Capture validated 31.7% to 42.0% of Tier 1 SNVs compared with 37.9% to 45.7% validated by RNA-Seq (Supplemental Figure S4). The SNVs that were not confirmed by either cDNA-Capture or RNA-Seq commonly resided in genes with negligible expression (0 FPKM). Most SNVs confirmed by RNA-Seq or cDNA-Capture had >3 FPKM, whereas SNVs missed by both RNA-Seq and cDNA-Capture commonly resided in genes with low expression (Figure 3C). In addition, as expected based on the gene expression analysis, FPKM expression of genes harboring SNVs were highly correlated between the two approaches (Pearson correlation, 0.93 to 0.97; one-sided P < 10−15) (Figure 3C and Supplemental Figure S4). Overall, RNA-Seq and cDNA-Capture had similar SNV validation rates despite having three times more sequence data generated from RNA-Seq.
Figure 3.
Validation of expressed SNVs in LUC4 FF tissue using RNA-Seq and cDNA-Capture. Scatterplots highlight the VAF of Tier 1 SNVs discovered by whole genome sequencing relative to VAF supported by RNA-Seq (A) and cDNA-Capture (B) in LUC4 using FF tissue. Each protein-coding gene harboring an SNV is colored based on its normalized expression level [0 FPKM (yellow) to 10 + FPKM (red)]. C: Correlation of expressed SNVs detected by cDNA-Capture or RNA-Seq and their corresponding normalized expression values. SNVs are color-coded based on whether they are found by both approaches (green), neither approach (black), cDNA-Capture only (yellow), or RNA-Seq only (red).
Gene Fusion Detection Using cDNA-Capture
Because none of the lung tumor samples harbored any experimentally validated gene fusions, we chose to compare RNA-Seq and cDNA-Capture on the well-characterized LNCaP prostate cancer cell line, which contains eight validated fusions.12 We generated 355 million RNA-Seq and 192 million cDNA-Capture reads. ChimeraScan29 was used to identify gene fusions and rediscovered all eight experimentally validated gene fusions in both RNA-Seq and cDNA-Capture. cDNA-Capture provided approximately 10 times more reads supporting the fusion between MIPOL1 and DGBK, which has been reported to result in the activation of the adjacent gene ETV1, an oncogenic transcription factor commonly up-regulated in prostate cancer patients through gene fusions (Supplemental Figure S5A).11 Because we generated almost twice as many sequence reads using RNA-Seq, we developed a normalized fusion score representing the total number of fusions supporting reads per million reads generated. All of the fusions had a higher cDNA-Capture normalized fusion score compared with RNA-Seq (Supplemental Figure S5B).
cDNA-Capture Using FFPE Material
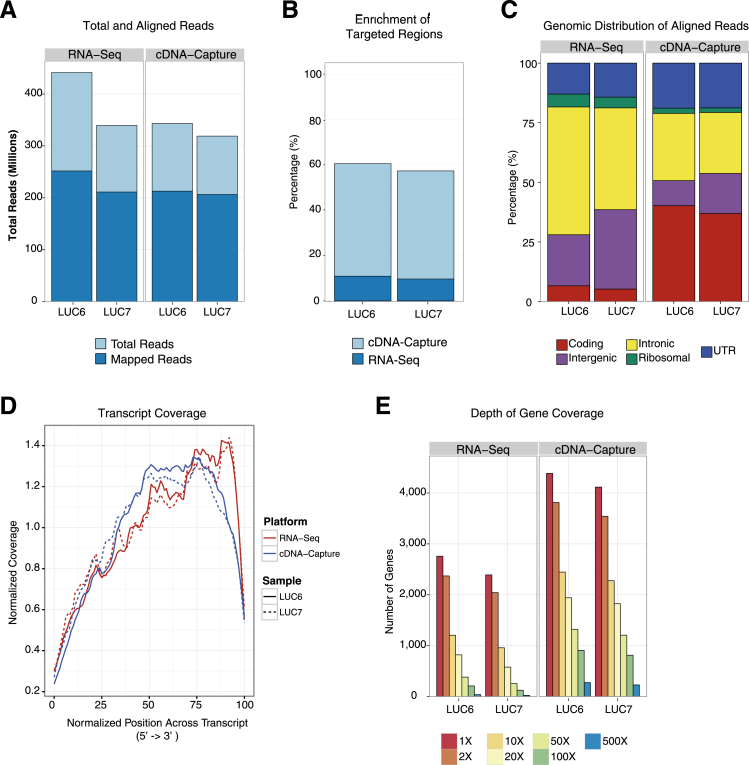
We next compared RNA-Seq and cDNA-Capture using FFPE material from two lung adenocarcinomas, LUC6 and LUC7. In total, we generated 441 million and 339 million RNA-Seq reads and 343 million and 318 million cDNA-Capture reads for LUC6 and LUC7, respectively (Supplemental Table S7). The percentage of reads aligned to the genome was nearly equivalent for RNA-Seq (57% to 62%) and cDNA-Capture (62% to 64%) (Figure 4A). Despite having similar alignment percentages, the genomic distribution of aligned reads for the FFPE material exhibited a shift between cDNA-Capture and RNA-Seq. Namely, cDNA-Capture exhibited a sixfold increase in the proportion of aligned reads that mapped to a targeted region (Figure 4B). Using cDNA-Capture, the percentage of reads aligning to coding regions increased by 33.6% and 31.7% for LUC6 and LUC7, respectively, compared with RNA-Seq (Figure 4C). There also was a slight increase in the alignment percentages to the UTRs (mean, 5.2%). These increases coincide with a corresponding decrease in reads aligning to the ribosomal (mean, 2.9%), intronic (mean, 21.2%), and intergenic regions (mean, 13.7%). We also observed a bias in coverage across transcripts toward the 3′ end (Figure 4D). However, use of cDNA-Capture resulted in a shift upstream from the 3′ end, thereby improving coverage across transcripts. In addition, the number of highly covered genes increased when using cDNA-Capture relative to RNA-Seq (Figure 4E). For instance, cDNA-Capture detected a mean of 6744 genes with splice junctions having at least 10× coverage compared with only 2310 genes detected at this coverage level with RNA-Seq. This was also accompanied by an increase in the proportion of reads aligning to splice junctions (Supplemental Figure S6).
Figure 4.
Comparison of cDNA-Capture and RNA-Seq using archived material. A: Total reads generated and aligned from RNA-Seq and cDNA-Capture on FFPE material in lung adenocarcinomas LUC6 and LUC7. B: Percentage of reads that aligned uniquely to the target regions for RNA-Seq and cDNA-Capture. C: Distribution of read alignments relative to genomic features, including coding, intergenic, intronic, ribosomal, and UTRs. D: Normalized coverage across transcripts. E: Dynamic range of gene coverage at varying depths.
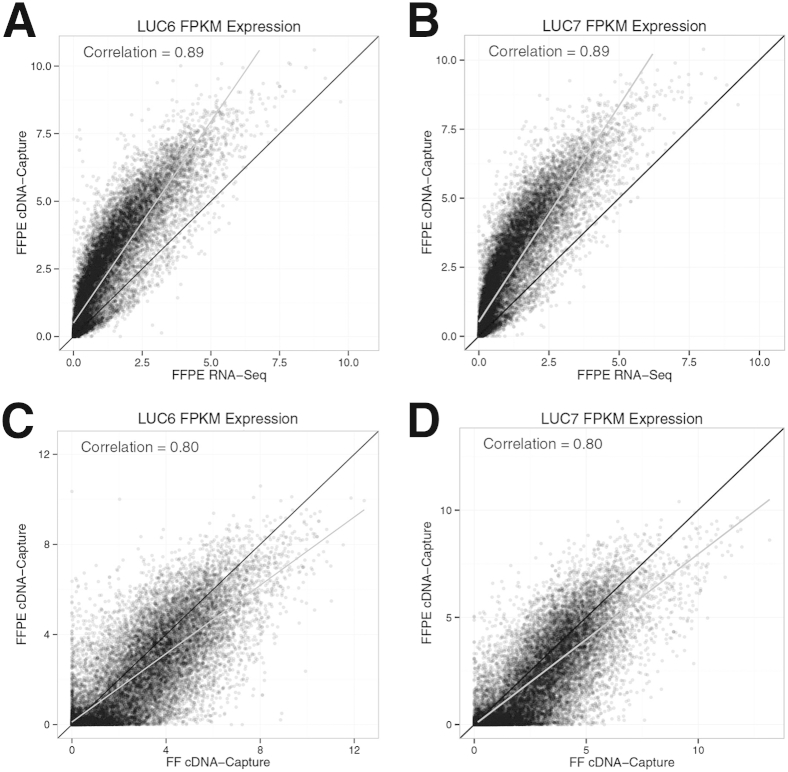
A comparison of the cDNA-Capture and RNA-Seq gene expression values using FFPE revealed significant correlations in both LUC6 (correlation, 0.89; one-sided P < 10−15) (Figure 5A) and LUC7 (correlation, 0.89; P < 10−15) (Figure 5B). Furthermore, genes tended to have higher expression levels in cDNA-Capture, indicated by the least-squares regression line deviating above what is expected if the expression levels were identical (the 45° line). This is likely the byproduct of using an enrichment step to increase the depth of coverage. Although cDNA-Capture appears to offer an improvement relative to RNA-Seq when using FFPE material, we wanted to confirm that it accurately recapitulates the biology of the tumor. Therefore, we compared gene expression between cDNA-Capture from FFPE and FF material and found significant correlations for LUC6 (correlation, 0.80; one-sided P < 10−15) (Figure 5C) and LUC7 (correlation, 0.80, P < 10−15) (Figure 5D). A similar GC bias was observed for FFPE compared with FF material (Supplemental Figure S3B). However, cDNA-Capture from FFPE provided increased expression levels across the entire range of GC content, including much larger gains for genes with lower GC content when compared with RNA-Seq from FFPE.
Figure 5.
Comparison of cDNA-Capture and RNA-Seq using FF and archived material. Scatterplots comparing LUC6 (A) and LUC7 (B) gene expression values calculated from FFPE material using cDNA-Capture and RNA-Seq. The least-squares regression line is shown in gray and the 45° line in black. Gene expression is measured as log2(FPKM + 1). Correlation of LUC6 (C) and LUC7 (D) gene expression values measured from FFPE and FF material using cDNA-Capture.
Validation of SNVs Using FFPE
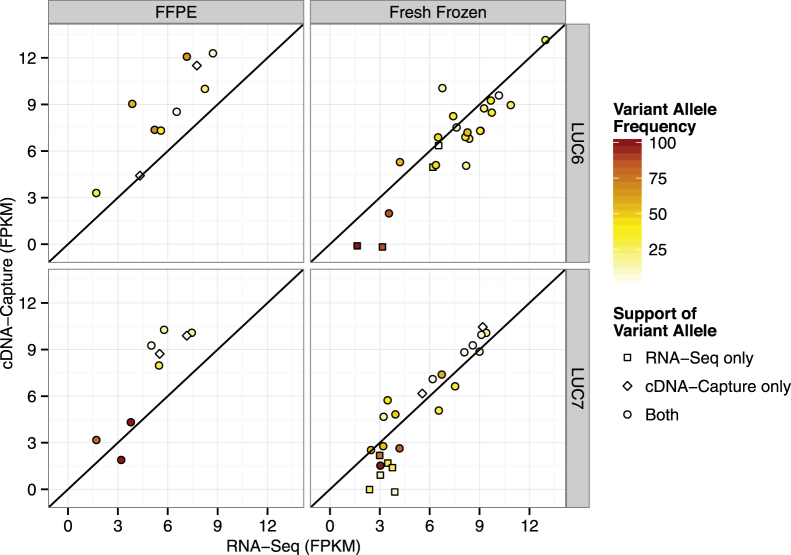
We further examined the detection of expressed Tier 1 SNVs in LUC6 and LUC7, comparing RNA-Seq and cDNA-Capture from FFPE material. Although expressed SNVs were detected from FFPE specimens, not surprisingly both LUC6 and LUC7 had a greater number of expressed SNVs detected by both RNA-Seq and cDNA-Capture from FF material (Figure 6). Of the SNVs validated from FFPE material, 80.0% and 77.7% were common to both RNA-Seq and cDNA-Capture in LUC6 and LUC7, respectively. Although the SNVs detected only by cDNA-Capture when using FFPE material had low VAFs, RNA-Seq failed to validate any SNVs missed by cDNA-Capture. Furthermore, the genes harboring validated Tier 1 SNVs appeared to have a slight increase in the normalized expression values (FPKM) in cDNA-Capture data relative to RNA-Seq data.
Figure 6.
Validation of expressed Tier 1 SNVs using FFPE material. Scatterplots highlight concordance between expressed Tier 1 SNVs using cDNA-Capture and RNA-Seq on FFPE and FF material. The x and y axes indicate the normalized expression level (FPKM) of the genes harboring the SNVs. Only Tier 1 SNVs with at least one read supporting the variant are displayed. Circles indicate SNVs supported by both RNA-Seq and cDNA-Capture, squares for SNVs supported by only RNA-Seq, and diamonds for SNVs supported only by cDNA-Capture. For SNVs detected by both RNA-Seq and cDNA-Capture, the color indicates the mean VAF between cDNA-Capture and RNA-Seq. Otherwise, the color indicates the VAF of the approach validating the SNV.
Comparison between FF and FFPE cDNA-Capture
We have already demonstrated a high correlation between cDNA-Capture FF and FFPE gene expression values and a larger number of expressed SNVs detected in FF tissue–derived RNA than FFPE. We next compared additional metrics between LUC6 and LUC7 FF and FFPE to determine the amount of potential information lost when sequencing FFPE material. Across both RNA-Seq and cDNA-Capture, a much higher percentage of reads aligned to the genome when using FF than FFPE (84% to 87% versus 57% to 65%) (Supplemental Table S7). In addition, the FF samples had a larger proportion of reads aligning to the target region than FFPE (70% to 95% versus 57% to 61%). However, when comparing cDNA-Capture reads to RNA-Seq reads, FFPE material had a much larger gain in target enrichment than FF material (sixfold versus twofold increase). For both RNA-Seq and cDNA-Capture, a larger percentage of mapped reads from FF spanned an exon-exon junction than from FFPE (Supplemental Table S7). Interestingly, cDNA-Capture using FFPE had as many or more mapped reads span a junction than FF RNA-Seq (LUC6: 19.71% versus 19.55%; LUC7: 17.74% versus 8.32%). This same pattern is observed for the percentage of mapped reads that aligned to coding regions: FF cDNA-Capture had the largest percentage (mean, 56%), followed by FFPE cDNA-Capture (mean, 32%), FF RNA-Seq (mean, 25%), and FFPE RNA-Seq (mean, 6%). Additional comparisons are complicated by the large discrepancy in the number of reads generated among the four experiments. These results demonstrate that, although more sequencing may be required for FFPE-derived tissues due to decreased mapping efficiency, FFPE cDNA-Capture appears to have similar performance to FF RNA-Seq.
cDNA-Capture Using Lower-Input Libraries
To assess the consequence of lower-input material on the quality of sequencing results, we applied our cDNA-Capture strategy using varying quantities of RNA input (60, 50, 10, 2, and 0.8 ng), in triplicate, from a colorectal tumor and adjacent normal tissue. Consistent with our normal protocol, the 50 ng and lesser inputs underwent DNase treatment. However, for the 60 ng input amount, this step was skipped. There was a positive correlation between the quantity of starting material and total number of reads generated (Supplemental Figure S7A and Supplemental Table S8). There was also a slight decrease in the percentage of reads aligning to the genome for lower-input libraries. Furthermore, the sequence duplication rate increased as the quantity of starting material decreased (Supplemental Figure S7B). Of the reads that aligned, their genomic distribution was fairly consistent across the varying input levels (Supplemental Figure S7C). However, despite having a similar distribution, the higher duplication rates in the lower-input libraries resulted in less coverage per gene (Supplemental Figure S7D).
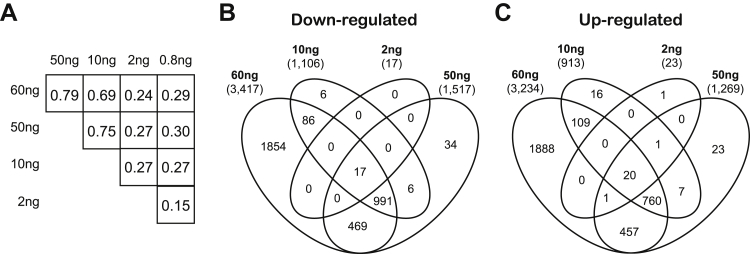
One of the primary uses of RNA-Seq from limited material is to detect genes with altered expression. Therefore, differentially expressed genes were calculated using edgeR33 for each library and compared to assess the degree of association between the ranked gene lists using Spearman's correlation (Figure 7A and Supplemental Figure S8). A significant positive correlation was found between the different RNA inputs (range, 0.15 to 0.79; one-sided P < 10−15 for each correlation). The most notable decline in correlation between any two RNA inputs occurred between 2 and 10 ng (decreasing from 0.75 between 50 and 10 ng to 0.27 between 10 and 2 ng). This finding suggests that the level of reliable differential gene expression analysis currently diminishes to <10 ng of RNA input.
Figure 7.
Differential gene expression analysis using cDNA-Capture on low-input libraries. A: Spearman rank correlation of edgeR P values between varying levels of RNA input. B and C: Venn diagrams show the overlap between the down- (B) and up-regulated (C) genes among the 60, 50, 10, and 2 ng input amounts. The total number of differentially expressed genes is shown under the RNA input. These plots suggest that the reliability of discovering differentially expressed genes diminishes for RNA inputs <10 ng.
Just as the gene coverage decreased as the RNA input level decreased, the number of differentially expressed genes identified also decreased (Figure 7, B and C, and Supplemental Table S9; 0.8 ng libraries not shown). In total, we observed 6651 differentially expressed genes between the tumor and normal 60 ng libraries, whereas there were only 40 differentially expressed genes from the 2 ng libraries. However, the specific genes that were differentially expressed in the lower-input libraries typically represent a subset of the differentially expressed genes identified at the highest input, 60 ng. The lack of library-specific differentially expressed genes suggests that lower-input libraries are capturing a subset of the expected altered genes without introducing any additional false-positive results. The largest percent decrease in the number of differentially expressed genes occurred between 2 and 10 ng (2019 genes for 10 ng and 40 genes for 2 ng, a 98% decrease). This decrease is despite the fact that the 2 ng input had a greater number of sequenced reads than the 10 ng input. This finding further suggests that the reliability of discovering differentially expressed genes currently diminishes for RNA inputs <10 ng.
Discussion
The clinical utility of monitoring gene expression can be exemplified by previous efforts using microarrays and RT-PCR for biomarker discovery37 and patient stratification.38,39 Transcriptome sequencing has further enabled our ability to reveal functionally relevant events (ie, overexpressed oncogenes, gene fusions, alternative splicing variants, or expressed deleterious SNVs), many of which simply cannot be detected from DNA-based assays. However, conventional RNA-Seq using low-input and archived material typically results in suboptimal performance. cDNA-Capture may offer improved results over RNA-Seq at low input by enriching for coding regions, hence rescuing the gene expression signals masked by noise from RNA degradation. Our results suggest that the enrichment is sufficient to maintain the biological interpretation observed in FF material, such as gene expression signatures, while requiring only one-third the amount of sequencing data. Even with the additional cost of the exome capture kit, cDNA-Capture costs approximately 50% less per sample than RNA-Seq when considering the increase in usable read yield provided by the capture step (Supplemental Table S10). Although cDNA-Capture may slightly decrease the accuracy of quantitated gene expression of the most highly expressed genes, it results in providing more even and comprehensive coverage across all expressed genes. This is a significant advance for generating sufficient transcript coverage from low-input and archived specimens in a cost-effective manner and ultimately makes it possible to maximize the wealth of information offered by monitoring the transcriptome in these precious clinical samples.
Despite the improved gene coverage using cDNA-Capture relative to RNA-Seq, the FFPE material lacked the same conformity as the FF material. This in turn may have contributed to the reduced ability to fully recapitulate results from FF samples as exemplified by the lower quantity of expressed SNVs validated via RNA-Seq and cDNA-Capture when using FFPE. Although most of the SNVs were detected by both RNA-Seq and cDNA-Capture, the only SNVs validated by a single approach were SNVs with low VAFs detected by cDNA-Capture. Despite the improved SNV validation rate achieved by cDNA-Capture in FFPE specimens, transcriptome analysis of archived material may require a greater depth of coverage to recapitulate results that would have been obtained with higher-quality FF specimens.
Gene fusion detection is one of the most important features of transcriptome sequencing. Using a well-characterized prostate cancer cell line, we were able to identify validated gene fusions using cDNA-Capture. In addition, we demonstrated that after normalizing by the total number of reads generated, cDNA-Capture provided more reads supporting every fusion than RNA-Seq. Unfortunately, neither of the FFPE samples studied contained a validated gene fusion. Therefore, future work is needed to fully elucidate what limitations may exist when detecting fusions in FFPE material using cDNA-Capture.
For the FFPE material, we used 150 ng input into the Ovation FFPE protocol. Thus, future experiments will evaluate improvement of transcript representation by increasing FFPE RNA input with the single primer isothermal amplification-based Ovation RNA-Seq FFPE System and the newly designed Ovation Human FFPE RNA-Seq system (NuGEN, San Carlos, CA). In cases where FFPE material is limiting, we will assess methods that first fragment RNA before the cDNA synthesis. In addition, although we have evaluated cDNA-Capture using an exome reagent, the probe design could be customized to cover any specific subset of the genome, thereby minimizing the cost and maximizing the coverage for a given experiment. Ultimately, cDNA-Capture will enable a cost-effective approach to achieve higher depths of coverage, which can sometimes be beneficial when using archived specimens.
Another critical hurdle toward conducting transcriptome analysis of clinically meaningful samples is the ability to sequence the limited quantities of material extracted from biopsy specimens. By assaying various levels of RNA input, we were able to demonstrate a reasonable threshold, of approximately 10 ng input, for using cDNA-Capture while reliably recapitulating results compared with higher-input amounts. Although we observed some diminishing returns corresponding to lower inputs (ie, fewer differentially expressed genes), the signal we were able to detect from as low as 10 ng appeared to be an accurate representation of gene expression for the genes detected. Because it is difficult to obtain high RNA yields from FFPE sections, a similar dilution experiment that involved assaying various low levels of FFPE could provide additional insights. However, because of limited amounts of available material, we were unable to perform this experiment and leave it as an open research question.
In summary, we have found that cDNA-Capture, the combination of exome capture and RNA-Seq, provides an efficient and cost-effective means to monitor expression and mutational status within a targeted subset of genomic regions using low-input and archived specimens. Although our results highlight the potential of cDNA-Capture, further experimentation in a broader range of patients and cancer types will determine the utility of this technique for routine clinical use.
Acknowledgments
We thank Gue Su Chang (The Genome Institute, Washington University) for help with characterizing the SeqCap EZ Human Exome Library version 3.0 capture regions.
Footnotes
Supported by an NIH Pathway to Independence Award (grant R00 CA149182), LUNGevity Career Development Award, and American Lung Association Biomedical Research Grant (C.A.M.). Computing and sequencing infrastructure at The Genome Institute was supported by the National Human Genome Research Institute (grant U54 HG003079; PI R.K. Wilson).
C.R.C. and V.M. contributed equally to this work.
Disclosures: None declared.
Supplemental Data
Comparison of cDNA-Capture and RNA-Seq using fresh frozen lung tumors. Scatterplots of LUC4 (A), LUC6 (B), and LUC20 (C) gene expression values measured by RNA-Seq and cDNA-Capture. Gene expression is measured as log2(FPKM + 1). The least-squares regression line is shown in red and the 45-degree line in black.
Comparison of cDNA-Capture and RNA-Seq in highly expressed genes. Scatterplots comparing log2(FPKM + 1) gene expression values for the top 1% highly expressed RNA-Seq genes in LUC4 (A), LUC6 (B), LUC13 (C), and LUC20 (D). The least-squares regression line is shown in red and the 45-degree line in black.
Gene expression versus GC-content. Distribution of gene-level expression values after binning by GC-content for RNA-Seq and cDNA-Capture using fresh frozen (A) and FFPE (B) material. Genes were split into four bins based on the GC-content, where bin 1 corresponds to the 25% of genes with the lowest percentage of GC bases and bin 4 corresponds to the 25% of genes with the highest percentage of GC bases. Gene expression values were calculated as the mean FPKM across all samples, and were subsequently log2 transformed. cDNA-Capture shows an increase in expression levels across all bins, although there is a much larger increase for genes with lower GC-content. Similar to RNA-Seq, cDNA-Capture shows a bias for lower expression levels as GC-content increases.
Validation of expressed SNVs using RNA-Seq and cDNA-Capture on fresh frozen tissue. Scatterplots highlight the variant allele frequency (VAF) of Tier 1 SNVs discovered by whole genome sequencing relative to VAF supported by RNA-Seq and cDNA-Capture using fresh frozen tissue in LUC6 (A), LUC13 (B), and LUC20 (C). Each protein-coding gene harboring an SNV is colored by its normalized expression level [0 FPKM (yellow) to 10+ FPKM (red)]. Correlation of expressed SNVs, and corresponding normalized expression values, detected by cDNA-Capture or RNA-Seq are shown on the right.
Comparison of RNA-Seq and cDNA-Capture for gene fusion detection in LNCaP prostate cancer cells. A: Schematic of the MIPOL1 and DGKB gene fusion in LNCaP cells with a representation of the total reads supporting the fusion junction in log-scale. B: A line plot of the normalized read support for all experimentally validated gene fusions in LNCaP cells for RNA-Seq and cDNA-Capture.
Comparison of canonical splice site reads between cDNA-Capture and RNA-Seq using FFPE material. Percentage of reads aligning to exon-exon junctions from RNA-Seq and cDNA-Capture.
cDNA-Capture using colorectal tumor and normal low-input libraries. A: Total and aligned reads grouped by RNA input material. B: Duplication rates across samples grouped by RNA input material and color-coded based on the normal or tumor sample. C: Distribution of read alignments relative to genomic features including coding, intergenic, intronic, ribosomal, and untranslated regions (UTR). D: Dynamic range of gene coverage at varying depths.
Differential gene expression analysis using cDNA-Capture on low-input libraries. Scatterplots showing the association of edgeR P values (log10 transformed) between each pair of RNA input amounts. Each plot also reports the Spearman rank correlation between input amounts. The correlations greatly decrease for the 2 and 0.8 ng input amounts, suggesting that 10 ng is a reasonable threshold for employing cDNA-Capture while reliably recapitulating results compared to higher input amounts.
References
- 1.Wang Z., Gerstein M., Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ozsolak F., Milos P.M. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet. 2011;12:87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prensner J.R., Iyer M.K., Balbin O.A., Dhanasekaran S.M., Cao Q., Brenner J.C., Laxman B., Asangani I.A., Grasso C.S., Kominsky H.D., Cao X., Jing X., Wang X., Siddiqui J., Wei J.T., Robinson D., Iyer H.K., Palanisamy N., Maher C.A., Chinnaiyan A.M. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 5.Sultan M., Schulz M.H., Richard H., Magen A., Klingenhoff A., Scherf M., Seifert M., Borodina T., Soldatov A., Parkhomchuk D., Schmidt D., O'Keeffe S., Haas S., Vingron M., Lehrach H., Yaspo M.-L. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 6.Pan Q., Shai O., Lee L.J., Frey B.J., Blencowe B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 7.Wang E.T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S.F., Schroth G.P., Burge C.B. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rozowsky J., Abyzov A., Wang J., Alves P., Raha D., Harmanci A., Leng J., Bjornson R., Kong Y., Kitabayashi N., Bhardwaj N., Rubin M., Snyder M., Gerstein M. AlleleSeq: analysis of allele-specific expression and binding in a network framework. Mol Syst Biol. 2011;7:522. doi: 10.1038/msb.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skelly D.A., Johansson M., Madeoy J., Wakefield J., Akey J.M. A powerful and flexible statistical framework for testing hypotheses of allele-specific gene expression from RNA-seq data. Genome Res. 2011;21:1728–1737. doi: 10.1101/gr.119784.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Govindan R., Ding L., Griffith M., Subramanian J., Dees N.D., Kanchi K.L., Maher C.A., Fulton R., Fulton L., Wallis J., Chen K., Walker J., McDonald S., Bose R., Ornitz D., Xiong D., You M., Dooling D.J., Watson M., Mardis E.R., Wilson R.K. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. 2012;150:1121–1134. doi: 10.1016/j.cell.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maher C.A., Kumar-Sinha C., Cao X., Kalyana-Sundaram S., Han B., Jing X., Sam L., Barrette T., Palanisamy N., Chinnaiyan A.M. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458:97–101. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maher C.A., Palanisamy N., Brenner J.C., Cao X., Kalyana-Sundaram S., Luo S., Khrebtukova I., Barrette T.R., Grasso C., Yu J., Lonigro R.J., Schroth G., Kumar-Sinha C., Chinnaiyan A.M. Chimeric transcript discovery by paired-end transcriptome sequencing. Proc Natl Acad Sci U S A. 2009;106:12353–12358. doi: 10.1073/pnas.0904720106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palanisamy N., Ateeq B., Kalyana-Sundaram S., Pflueger D., Ramnarayanan K., Shankar S., Han B., Cao Q., Cao X., Suleman K., Kumar-Sinha C., Dhanasekaran S.M., Chen Y., Esgueva R., Banerjee S., LaFargue C.J., Siddiqui J., Demichelis F., Moeller P., Bismar T.A., Kuefer R., Fullen D.R., Johnson T.M., Greenson J.K., Giordano T.J., Tan P., Tomlins S.A., Varambally S., Rubin M.A., Maher C.A., Chinnaiyan A.M. Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer and melanoma. Nat Med. 2010;16:793–798. doi: 10.1038/nm.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson D.R., Kalyana-Sundaram S., Wu Y.-M., Shankar S., Cao X., Ateeq B., Asangani I.A., Iyer M., Maher C.A., Grasso C.S., Lonigro R.J., Quist M., Siddiqui J., Mehra R., Jing X., Giordano T.J., Sabel M.S., Kleer C.G., Palanisamy N., Natrajan R., Lambros M.B., Reis-Filho J.S., Kumar-Sinha C., Chinnaiyan A.M. Functionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancer. Nat Med. 2011;17:1646–1651. doi: 10.1038/nm.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park E., Williams B., Wold B.J., Mortazavi A. RNA editing in the human ENCODE RNA-seq data. Genome Res. 2012;22:1626–1633. doi: 10.1101/gr.134957.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramaswami G., Zhang R., Piskol R., Keegan L.P., Deng P., O'Connell M.A., Li J.B. Identifying RNA editing sites using RNA sequencing data alone. Nat Methods. 2013;10:128–132. doi: 10.1038/nmeth.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levin J.Z., Berger M.F., Adiconis X., Rogov P., Melnikov A., Fennell T., Nusbaum C., Garraway L.A., Gnirke A. Targeted next-generation sequencing of a cancer transcriptome enhances detection of sequence variants and novel fusion transcripts. Genome Biol. 2009;10:R115. doi: 10.1186/gb-2009-10-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ueno T., Yamashita Y., Soda M., Fukumura K., Ando M., Yamato A., Kawazu M., Choi Y.L., Mano H. High-throughput resequencing of target-captured cDNA in cancer cells. Cancer Sci. 2012;103:131–135. doi: 10.1111/j.1349-7006.2011.02105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mercer T.R., Gerhardt D.J., Dinger M.E., Crawford J., Trapnell C., Jeddeloh J.A., Mattick J.S., Rinn J.L. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat Biotechnol. 2012;30:99–104. doi: 10.1038/nbt.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsushita H., Vesely M.D., Koboldt D.C., Rickert C.G., Uppaluri R., Magrini V.J., Arthur C.D., White J.M., Chen Y.-S., Shea L.K., Hundal J., Wendl M.C., Demeter R., Wylie T., Allison J.P., Smyth M.J., Old L.J., Mardis E.R., Schreiber R.D. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482:400–404. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flicek P., Ahmed I., Amode M.R., Barrell D., Beal K., Brent S. Ensembl 2013. Nucleic Acids Res. 2012;41:D48–D55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koboldt D.C., Zhang Q., Larson D.E., Shen D., McLellan M.D., Lin L., Miller C.A., Mardis E.R., Ding L., Wilson R.K. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larson D.E., Harris C.C., Chen K., Koboldt D.C., Abbott T.E., Dooling D.J., Ley T.J., Mardis E.R., Wilson R.K., Ding L. SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics. 2012;28:311–317. doi: 10.1093/bioinformatics/btr665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saunders C.T., Wong W.S.W., Swamy S., Becq J., Murray L.J., Cheetham R.K. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28:1811–1817. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- 29.Iyer M.K., Chinnaiyan A.M., Maher C.A. ChimeraScan: a tool for identifying chimeric transcription in sequencing data. Bioinformatics. 2011;27:2903–2904. doi: 10.1093/bioinformatics/btr467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wickham H. Springer; New York: 2009. ggplot2: elegant graphics for data analysis. [Google Scholar]
- 31.Chen H., Boutros P.C. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics. 2011;12:35. doi: 10.1186/1471-2105-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cabanski C.R., Wilkerson M.D., Soloway M., Parker J.S., Liu J., Prins J.F., Marron J.S., Perou C.M., Hayes D.N. BlackOPs: increasing confidence in variant detection through mappability filtering. Nucleic Acids Res. 2013;41:e178. doi: 10.1093/nar/gkt692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pickrell J.K., Marioni J.C., Pai A.A., Degner J.F., Engelhardt B.E., Nkadori E., Veyrieras J.-B., Stephens M., Gilad Y., Pritchard J.K. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Risso D., Schwartz K., Sherlock G., Dudoit S. GC-content normalization for RNA-Seq data. BMC Bioinformatics. 2011;12:480. doi: 10.1186/1471-2105-12-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewis F., Maughan N.J., Smith V., Hillan K., Quirke P. Unlocking the archive–gene expression in paraffin-embedded tissue. J Pathol. 2001;195:66–71. doi: 10.1002/1096-9896(200109)195:1<66::AID-PATH921>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 38.Paik S., Shak S., Tang G., Kim C., Baker J., Cronin M., Baehner F.L., Walker M.G., Watson D., Park T., Hiller W., Fisher E.R., Wickerham D.L., Bryant J., Wolmark N. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 39.Gianni L., Zambetti M., Clark K., Baker J., Cronin M., Wu J., Mariani G., Rodriguez J., Carcangiu M., Watson D., Valagussa P., Rouzier R., Symmans W.F., Ross J.S., Hortobagyi G.N., Pusztai L., Shak S. Gene expression profiles in paraffin-embedded core biopsy tissue predict response to chemotherapy in women with locally advanced breast cancer. J Clin Oncol. 2005;23:7265–7277. doi: 10.1200/JCO.2005.02.0818. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of cDNA-Capture and RNA-Seq using fresh frozen lung tumors. Scatterplots of LUC4 (A), LUC6 (B), and LUC20 (C) gene expression values measured by RNA-Seq and cDNA-Capture. Gene expression is measured as log2(FPKM + 1). The least-squares regression line is shown in red and the 45-degree line in black.
Comparison of cDNA-Capture and RNA-Seq in highly expressed genes. Scatterplots comparing log2(FPKM + 1) gene expression values for the top 1% highly expressed RNA-Seq genes in LUC4 (A), LUC6 (B), LUC13 (C), and LUC20 (D). The least-squares regression line is shown in red and the 45-degree line in black.
Gene expression versus GC-content. Distribution of gene-level expression values after binning by GC-content for RNA-Seq and cDNA-Capture using fresh frozen (A) and FFPE (B) material. Genes were split into four bins based on the GC-content, where bin 1 corresponds to the 25% of genes with the lowest percentage of GC bases and bin 4 corresponds to the 25% of genes with the highest percentage of GC bases. Gene expression values were calculated as the mean FPKM across all samples, and were subsequently log2 transformed. cDNA-Capture shows an increase in expression levels across all bins, although there is a much larger increase for genes with lower GC-content. Similar to RNA-Seq, cDNA-Capture shows a bias for lower expression levels as GC-content increases.
Validation of expressed SNVs using RNA-Seq and cDNA-Capture on fresh frozen tissue. Scatterplots highlight the variant allele frequency (VAF) of Tier 1 SNVs discovered by whole genome sequencing relative to VAF supported by RNA-Seq and cDNA-Capture using fresh frozen tissue in LUC6 (A), LUC13 (B), and LUC20 (C). Each protein-coding gene harboring an SNV is colored by its normalized expression level [0 FPKM (yellow) to 10+ FPKM (red)]. Correlation of expressed SNVs, and corresponding normalized expression values, detected by cDNA-Capture or RNA-Seq are shown on the right.
Comparison of RNA-Seq and cDNA-Capture for gene fusion detection in LNCaP prostate cancer cells. A: Schematic of the MIPOL1 and DGKB gene fusion in LNCaP cells with a representation of the total reads supporting the fusion junction in log-scale. B: A line plot of the normalized read support for all experimentally validated gene fusions in LNCaP cells for RNA-Seq and cDNA-Capture.
Comparison of canonical splice site reads between cDNA-Capture and RNA-Seq using FFPE material. Percentage of reads aligning to exon-exon junctions from RNA-Seq and cDNA-Capture.
cDNA-Capture using colorectal tumor and normal low-input libraries. A: Total and aligned reads grouped by RNA input material. B: Duplication rates across samples grouped by RNA input material and color-coded based on the normal or tumor sample. C: Distribution of read alignments relative to genomic features including coding, intergenic, intronic, ribosomal, and untranslated regions (UTR). D: Dynamic range of gene coverage at varying depths.
Differential gene expression analysis using cDNA-Capture on low-input libraries. Scatterplots showing the association of edgeR P values (log10 transformed) between each pair of RNA input amounts. Each plot also reports the Spearman rank correlation between input amounts. The correlations greatly decrease for the 2 and 0.8 ng input amounts, suggesting that 10 ng is a reasonable threshold for employing cDNA-Capture while reliably recapitulating results compared to higher input amounts.