Abstract
Somatic mutations are present in various proportions in numerous developmental pathologies. Somatic activating missense mutations of the GNAS gene encoding the Gsα protein have previously been shown to be the cause of fibrous dysplasia of bone (FD)/McCune-Albright syndrome (MAS). Because in MAS patients, tissues as diverse as melanocytes, gonads and bone are affected, it is generally accepted that the GNAS mutation in this disease must have occurred early in development. Interestingly, it has been shown that the development of an active FD lesion may require both normal and mutant cells. Studies of the somatic mosaic states of FD/MAS and many other somatic diseases need an accurate method to determine the ratio of mutant to normal cells in a given tissue. A new method for quantification of the mutant:normal ratio of cells using a PNA hybridization probe-based FRET technique was developed. This novel technique, with a linear sensitivity of 2.5% mutant alleles, was used to detect the percentage mutant cells in a number of tissue and cell culture samples derived from FD/MAS lesions and could easily be adapted for the quantification of mutations in a large spectrum of diseases including cancer.
INTRODUCTION
Many diseases such as McCune–Albright syndrome (MAS), Alzheimer’s, Duchenne muscular dystrophy and a number of tumor pathologies are characterized by point mutations in the sequence of particular genes. These mutations may lead to a specific pathology when the number of cells expressing the mutation reach a critical level. For example, the total number of cells containing specific point mutations in tissues could be important in the switch from benign to malignant forms or from primary to metastatic forms of tumors. A variety of individual point mutations in mitochondrial DNA with various clinical phenotypes have been reported to be associated with aging (1), degenerative diseases such as Alzheimer’s disease (2), Parkinson disease (3), cardiomyopathy and encephalomyopathy (4), MELAS (myopathy, encephalopathy, lactic acidosis and stroke-like episodes) syndrome (5), MIDD (maternally inherited diabetes and deafness) syndrome (6) and progressive kidney disease (7). Quantification of the precise mutational load is also critical in the discrimination of a mosaic from a full heterozygotic carrier (8).
MAS is characterized by the clinical triad of polyostotic fibrous dysplasia (FD), café au lait skin hyperpigmentation lesions, and endocrine hyperfunction (9–11). Several different activating missense mutations in the GNAS gene, which encodes the α subunit of the stimulatory G protein (Gsα), have been identified in all patients studied with MAS and FD. The vast majority of the patients, however, have been shown to have affected cells/tissues with R201H or R201C mutations [where Arg(CGT) at position 201 is replaced with either His(CAT) or Cys(TGT), respectively] (12 ). Although rare, R201G, R201S and R201L mutations have also been reported for the GNAS R201codon (13–15).
The causative mutations occur post-zygotically, leading to a somatic mosaic state. It is commonly believed that variations in the number and locations of the affected sites within the organism reflect the time of mutation, and determine the extent and severity of the mutation-associated disease. An important consequence of mosaicism is that different tissues or even different areas within a tissue may have anywhere from a few to a predominance of mutant cells. Indeed, it has been shown that the fibrous dysplastic bone lesion itself may require both normal and mutant cells (16), raising the important point of degree of mosaicism as a potential determinant of their natural history as well as their severity. Variable representation of the disease genotype in a single tissue needs methods of analysis that can determine the ratio of mutant to normal cells. Addressing these issues require not so much an extremely sensitive method but rather an accurate and precise quantitative method for detecting the disease genotype within a wide range of mutational loads.
Such a quantitative method for the determination of the percentage of mutant cells in numerous pathologies can be applied to a variety of known point mutations. Smith et al. (1999) developed a quantitative fluorescent multiplex PCR assay for the quantification of somatic mosaicism for a point mutation in Duchenne muscular dystrophy with 5% sensitivity (8). The method relies on changes in the size of the amplification product for wild-type and mutant DNA, and therefore may not be convenient for quantification of point mutations in other pathologies. Furthermore, a number of techniques have been reported for the quantification of heteroplasmy in mitochondrial DNA (mtDNA). Heteroplasmy is a phenomenon in which tissues harbor a mixture of mutant and wild-type mtDNA molecules. The A3243G mutation of mtDNA is associated with MELAS syndrome and diabetes mellitus. Hybridization-based methods, allele-specific oligonucleotide (ASO) hybridization and allele-specific PCR are limited in quantification of low frequency mutant alleles (17). Restriction fragment length polymorphism (RFLP) and Southern blot analysis have been applied for the quantification of the A3243G mutation; however, partial digestion by ApaI which is created by the mutation, and variations in efficiency of transfer and hybridization reduce the accuracy of quantification. Although the combination of RFLP analysis with PCR improves the sensitivity, the risk of partial digestion remains problem (18). The inclusion of a single fluorescence-based primary extension step to PCR-RFLP eliminated heteroduplex formation and the risk of partial digestion; however, the method is limited to a specific subset of mutations (19). Similarly, PCR-single strand conformation polymorphism (PCR-SSCP) has also been used to determine the mutation load. The method is semi-quantitative, with a sensitivity of 5–10% (20). Differential termination of fluorescence-based primary extension assay requires an electrophoretic step and relies on fluorescent band intensities associated with wild-type and mutant DNA-derived extension products (21). PCR ligation detection reaction (PCR-LDR) has been applied for the detection and quantification of point mutations and is based on the alternative ligation of one of two upstream oligonucleotides to a common downstream oligonucleotide at the 3′ end, the site of the mutation. However, the sensitivity is ∼10% of total DNA as reported for the A3243G mutation and the technique may not be applicable to all mutations (22,23). Recently, a novel approach has been reported for heteroplasmy quantification of A8344G tRNALys mutation in tissue samples that uses two differently labeled molecular beacon probes (a novel class of nucleic acid probes that become fluorescent when bound to a complementary sequence) for real-time fluorescence monitoring of PCR (24). The sensitivity of the assay has been shown to be 5% and offers some promise, but may not work for all mutations. Finally, techniques based on multiple clonal analysis (25), solid-phase minisequenceing (26) and direct peak comparison in a sequence chromatogram (27) have been used; however, they differ in the speed, accuracy, sensitivity and ease of application. Although all of these techniques can provide some information on the quantification of mutations, the applicability to new diseases often remains a concern. We have developed an assay that would appear to be applicable to quantify a wide variety of mutations due to an increased sensitivity of distinguishing perfect from single base pair mismatches.
Peptide nucleic acid (PNA) is a synthetic DNA analog (made on a DNA synthesizer) in which the normal phosphodiester backbone is replaced with a 2-aminoethylglycine chain to which nucleobases have been chemically attached via a methylene carbonyl linker for the interaction of the bases in the normal A–T and G–C geometry (28,29). Perfectly matched PNA–DNA hybrids are actually more stable than are those for DNA–DNA of the same length. PNA–DNA hybrids that contain a single mismatch, however, typically melt at a temperature 10–18°C lower than the same pair lacking the mismatch, a much larger difference than that seen for normal DNA–DNA interactions with single mismatches (30). The fluorescence resonance energy transfer (FRET) concept is characterized by the excitation of a donor fluorophore with an emission spectrum that overlaps the excitation spectrum of an acceptor fluorophore in very close proximity (31). A signal is generated only when both probes are bound to the template DNA.
We describe here the development of a quantitative PCR method relying on a PNA hybridization probe-based FRET principle, which allows for the direct and rapid detection and quantification of activating GNAS mutations with a sensitivity of ∼2.5% of mutant alleles (corresponding to 5% of mutant cells) from different tissues and cell culture samples derived from FD of MAS patients’ lesions. We report that samples from various FD lesions contain different ratios of mutant to normal cells.
MATERIALS AND METHODS
Patient samples
Biopsy specimens were obtained for this study from nine different patients with FD/MAS under an IRB-approved protocol (NIH, 98-D-0145). Nine fresh biopsy specimens from eight patients with FD/MAS were used for DNA extraction. From one patient, biopsies were obtained from both the FD-affected iliac crest, and from the contralateral iliac crest that was clinically and radiographically free of disease. Samples from six fresh specimens were used for establishing both non-clonal (multicolony-derived, from two patients) and clonal (single colony-derived, from four patients) marrow stromal cell cultures as previously described (16). Biopsy specimens from six normal donors obtained under an IRB-approved protocol (NIH, 94-D-0188) were used to establish multicolony strains. Genomic DNA from either fresh tissue samples or cell cultures was extracted and purified as described previously (16).
Fresh tissue taken from the femur (surgical waste from a corrective surgery) obtained from one patient was used to assess the potential effects of paraffin embedding on the sensitivity/accuracy of mutation detection in comparison with fresh, unfixed and unprocessed tissue. In this case, three samples were excised from the specimen using a 2.3 mm trepan, and then each core was divided in half to create paired samples for both histological processing (fixation, decalcification and paraffin embedding) as described previously (32) and direct DNA extraction while fresh. Genomic DNA from paraffin-embedded tissues was extracted using the Puregene DNA Isolation Kit (Gentra systems, Minneapolis, MN) as described previously (33).
Synthesis and labeling of oligonucleotide primers and DNA probes
Synthesis supports, reagents and monomers were purchased from Applied Biosystems, Foster City, Glen Research (Sterling, VA) and Roche Molecular Biochemicals (Indianapolis, IN). All oligonucleotide primers and fluorescently labeled probes were synthesized on Applied Biosystems Model 392/394 DNA synthesizers, using the manufacturer’s pre-programmed synthesis cycles and phosphoramidite chemistry with a β-cyanoethyl protecting group on the 3′ phosphorus moiety. The donor fluorescent DNA probe used with the LightCycler was synthesized using a CPG-fluorescein synthesis column. The acceptor fluorescent DNA probe used with the LightCycler was phosphorylated at the 3′ end. Following synthesis, the support-bound acceptor probe was labeled at the 5′ end with LightCycler Red 705 phosphoramidite (Roche Molecular Biochemicals). Cleavage and final deprotection of the Red 705-labeled probe was performed by incubation in 28% ammonia for 1 h, at 55°C. Following deprotection, the fluorescently labeled probes were resuspended in deionized water prior to a single purification by RP-HPLC using a 0.1 M triethylammonium acetate buffer gradient and dual monitoring at 260 and 685 nm (for the Red 705-labeled probe) and 494 nm (for the fluorescein-labeled probe). The donor probe was purified on Delta-Pak C18, 5 µm 300 Å, 7.8 × 300 mm (Waters Corporation, Milford, USA) and the acceptor probe was purified on a POROS OligoR3, 4.6 × 50 mm column. HPLC fractions containing the fluorescent products were evaporated to remove acetonitrile prior to lyophilization. The purified labeled probes were stored at –20°C. Sequences of primers and of the DNA and PNA hybridization probes are described in Table 1.
Table 1. Sequences of primers, and DNA and PNA hybridization probes.
| Sequence | |
|---|---|
| Primers | |
| F1 | 5′-GAGCCTGACCTTGTAGAGAGAC-3′ |
| R1 | 5′-GGGTTCTTCTCTATAAACAGTGC-3′ |
| R2 | 5′-GGTGAATGTCAAGAAACCATGA-3′ |
| Hybridization probes | |
| FDP R201 DNA | 5′-CCGTGTCCTGACTTCTGGAATC-3′ |
| FDP R201H DNA | 5′-CCATGTCCTGACTTCTGGAATC-3′ |
| FDP R201C DNA | 5′-CTGTGTCCTGACTTCTGGAATC-3′ |
| RAP705 DNA | 5′-TTGAGACCAAGTTCCAGGTGGACAAA-3′ |
| FDP DNA | 5′-GGTGAGATCCATTGACCTCAATTTTGTTTCAGGACCTGCT-3′ |
| RAP640 R201 PNA | 5′-CGCTGCCGTGTCCT-3′ |
| RAP640 R201H PNA | 5′-CGCTGCCATGTCCT-3′ |
| RAP640 R201C PNA | 5′-CGCTGCTGTGTCCT-3′ |
The underlined letters represent single nucleotide mutations. F1 = forward primer, R1 = reverse primer, R2 = reverse, more 5′ primer, FDP = fluorescein donor probe, RAP = Red acceptor probe.
Synthesis and labeling of PNA oligomers with Red 640
PNA synthesis columns, PNA monomers and reagents were purchased from Applied Biosystems (Foster City, CA) and Roche Molecular Biochemicals. PNAs were synthesized on a PerSeptive Biosystem ExpediteTM 8900 Nucleic Acid Synthesis System, using the manufacturer’s pre-programmed synthesis cycles and the 9-fluorenylmethoxycarbonyl (Fmoc) chemistry mediated by HATU [O-(7-azabenzo-triazol-1-yl)-1,1,3,3-tetramethylluronium hexafluorophosphate]. Following synthesis, the support-bound PNAs were labeled at the N-terminal end (equivalent to 5′-end of a DNA oligo) with LightCycler Red 640-NHS (Roche Molecular Biochemicals) using the manufacturer’s protocol except that the fluorescent label was attached to the PNA via linkers (8-amino-3,6-dioxaoctanoid acid). The labeled PNAs were cleaved from their supports with a cocktail of trifluoroacetic acid (TFA):m-cresol (4:1, v:v) for 2 h at room temperature. Following cleavage, the PNA products were precipitated and washed with methyl tert-butyl ether (MTBE). The PNA pellets were resuspended in 0.1% TFA/water prior to a single-step purification on RP-HPLC using Delta-Pak C18, 5 µm 300 Å, 7.8 × 300 mm (Waters Corporation) with a gradient of 0.1% TFA/water and 0.1% TFA/acetonitrile, and dual monitoring at 260 and 620 nm. HPLC fractions containing the PNA products were evaporated to remove acetonitrile prior to lyophilization. The purified labeled PNAs were characterized by matrix-assisted laser desorption ionization mass spectrometry analysis (MALDI-TOF) and were stored at –20°C.
PCR protocol for the preparation of standards and samples
Template DNAs for R201 wild type and R201H mutant DNA were prepared from a biopsy sample taken from a patient with MAS. An 801 bp DNA fragment containing the R201 site was amplified by routine PCR and subcloned into pBluescript SK (Stratagene, La Jolla, CA). Two separate clones, R201 wild type and R201H mutant, were isolated, verified by sequencing, and used for the preparation of DNA standards. The R201C mutant DNA was made from the wild-type clone above by in situ mutagenesis using the appropriate mutant oligonucleotides, Pfu DNA polymerase and the QuikChange Site-Directed Mutagenesis kit (Stratagene). The full-length sequence for this mutant clone standard was verified by sequencing.
For the preparation of standards, the 801 bp DNA fragment of the wild type and the two Gsα mutations were separately amplified in a standard PCR using the corresponding plasmids as templates. The PCR was carried out in the presence of 1× PCR buffer (20 mM Tris–HCl, pH 8.4 and 50 mM KCl), 2.5 mM MgCl2, 2.5 U of platinum Taq DNA polymerase (all from Invitrogen Life Technologies, Carlsbad, CA), 0.25 mM of each deoxynucleotide triphosphate (New England Biolabs, Beverly, CA), 0.5 µg of each primer (F1 and R1), and 10 ng of either wild-type or mutant template DNA in a final volume of 100 µl. Amplification reactions were performed under the following cycle conditions: 95°C denaturation and enzyme activation step for 1 min followed by 25 cycles of 95°C denaturation for 30 s, 55°C annealing for 30 s and 72°C extension for 45 s. The reaction was completed after 72°C termination for 7 min. The amplified products were purified using a PCR purification kit (Qiagen, Valencia, CA), eluted in 50 µl of ddH2O, and then used for the preparation of single-stranded DNA (ssDNA).
For the preparation of ssDNA standards, 400 bp antisense ssDNA strands, containing the codons for R201, R201H or R201C, were separately amplified using the above double-stranded DNA (dsDNA) as templates. The PCR was carried out in the same reaction conditions as above except that only the more 5′ antisense primer (R2), and a 1:20 dilution of either wild-type or mutant template dsDNA (as template) were used in separate 100 µl reactions. Amplification reactions were the same as above except that the 72°C extension was for only 30 s. The ssDNAs from these standards were then analyzed by FRET.
Similarly, an 801 bp fragment of the GNAS centered on the R201 region was amplified by PCR using the genomic DNA isolated from various patient samples. Samples were heated at 95°C for 15 min before amplification by PCR for 40 cycles with the identical conditions described above for standards. For each sample, the corresponding ssDNA was then amplified under the identical conditions as described above. For each analysis, 400 bp single-stranded standards and experimental samples were then directly used for analysis by FRET. Standard curves using pre-measured combinations of mutant and wild-type ssDNA were prepared for quantifying the mutant allele (range: 0–50% mutant/wild-type allele).
FRET with DNA hybridization probes
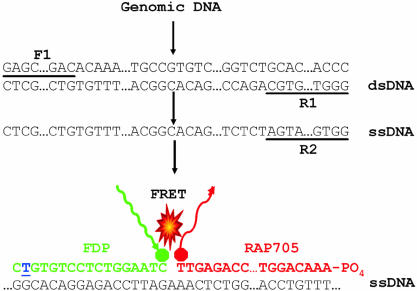
The ssDNA was detected with two specially designed DNA oligonucleotide probes. The donor probes containing either R201H or R201C were 3′-labeled with fluorescein. The acceptor probe was 5′-labeled with Red 705 (Fig. 1). The sequences of the oligonucleotides were selected such that the donor–acceptor pairs hybridize to the amplified ssDNA fragment in a head to tail arrangement with the two fluorochromes juxtaposed. The donor dye was excited at 470 nm and entered a higher energy state (Fig. 1). FRET occurred only when the two dyes were in close proximity and the transferred energy excited the acceptor dye, Red 705, attached to the second hybridization probe. The Red 705 subsequently emitted a red fluorescent light at 710 nm and the intensity of the light emitted was measured. The amount of measured fluorescence is directly proportional to the amount of bound donor and acceptor pairs. Those fluorochromes free in solution do not fluoresce at 710 nm to any significant degree.
Figure 1.
Diagram showing the PCR amplification of a target sequence and detection using FRET-based DNA hybridization probes with R201C mutation. The double-stranded cDNA of the target sequence (dsDNA) was amplified with forward (F1) and reverse (R1) primers. Subsequently, a shorter single strand was amplified (ssDNA) using only one, more 5′ reverse primer (R2). DNA hybridization probes (DNA-HP) were designed in such a way that donor and acceptor probes localize in a head to tail arrangement when hybridized to ssDNA. The R201C mutation site is indicated by an underlined blue nucleotide. When the fluorescent donor probe (FDP) was excited at 470 nm, the FDP transferred its energy to the acceptor probe (RAP705) that then emitted light of 705 nm for DNA-HP. The intensity of this signal was proportional to the amount of target DNA molecule bound. Both wild-type and mutant sequences were detected by FRET in the single tube, with the mutant probe melting off at a higher temperature than the mismatched wild-type probe. Similar approaches were also taken for R201H mutation analysis. The FRET-based PNA hybridization probes with R201C and R201H mutations were also designed with similar approaches where DNA was substituted by PNA.
FRET with PNA hybridization probes
The FRET with PNA hybridization probe procedure was similar to that described for the DNA hybridization probes except that the acceptor probe was a PNA hybridization probe (containing either the R201H or R201C mutation) labeled with Red 640 dye at the position equivalent to a corresponding DNA oligonucleotide’s 5′ end. The intensity of the light emitted by the Red 640 (640 nm) was directly proportional to the amount of target DNA present.
Use of LightCycler for FRET analysis
Analysis was performed by taking 13 µl of ssDNA from the PCR described above, bringing it to a 20 µl volume, including a final concentration of: 4 mM MgCl2, 0.2 µM fluorescent DNA donor probe and 0.3 µM Red dye 705 DNA acceptor probe. The sample was prepared in a capillary tube and analyzed in the LightCycler (Roche Applied Science) by the melting curve technique. The melting step for DNA hybridization probes consisted of an initial denaturation at 95°C (ramp rate 20°C/s), followed by cooling to 45°C (ramp rate 20°C/s, 5 min hold), then heating to 80°C at 0.1°C/s during which fluorescence was continuously monitored at 710 nm. Similarly, for the PNA hybridization probe analysis, 20 µl of reaction mixture consisting of ssDNA template (13 µl), MgCl2 (4 mM), fluorescent DNA donor probe (0.2 µM) and Red dye 640 PNA acceptor probe (0.3 µM) was prepared. The melting steps for PNA hybridization probes were identical to those for DNA–FRET except that fluorescence was monitored continuously at 640 nm. The fluorescence signal from the acceptor fluorophore was plotted as the negative first derivative of the fluorescence with respect to temperature versus temperature (–dF/dT). A separate standard curve was produced for both R201H and R201C by diluting the respective mutant cDNA standards to 50, 40, 30, 20, 10, 5 and 2.5% using wild-type cDNA standard as diluent. The relative amount of mutant allele was determined by comparing the peak heights of the high temperature (mutant) and low temperature (wild-type) melting curves and plotting the ratio against the percentage of mutant DNA present in the standards. The percentage of mutant cells in each unknown sample was determined by interpolation.
Statistical analyses
The relationship between the ratio of mutant to normal and standards was calculated by simple linear regression and the strength of the relationship was assessed by correlation analysis using the WinStat statistical package embedded in Microsoft Excel.
RESULTS
Standard curve generation by DNA hybridization probes
In order to quantify the percentage of mutant cells in a variety of biological samples, we first developed standards for R201H and R201C mutations. Pure 801 bp PCR DNA containing either the wild-type allele R201, or the mutant alleles R201H or R201C were carefully adjusted to have equal concentrations (as determined by SYBR green fluorescence). Subsequently, a second PCR was carried out for the production of the 401 bp single-stranded, antisense materials needed for analysis. A linear standard curve was produced by mixing the single-stranded standards into the proper ratios. The LightCycler was used first to allow the binding of the labeled probes to the antisense strands and then to detect the loss of the fluorescence signal as the samples were slowly heated until the acceptor oligo with a single-mismatch melted off the wild-type ssDNA. With the continued increase in temperature, the signal was totally lost as the perfect match, acceptor mutant oligonucleotide dissociated from the target ssDNA. Converting this information into the negative first derivative (–dF/dT) resulted in ‘melting peaks’ whose relative heights were a very consistent measure of the relative abundance of the two sequences in the sample. The results were plotted by taking the ratio of height of perfect match (mutant allele) and mismatch (wild-type allele) peaks. Using a DNA–DNA FRET hybridization probe set, the difference between the Tm for the 100% ssDNA R201H mutation (perfect match) and the 100% wild-type ssDNA R201 (single mismatch) allele yielded two melting peaks separated by ∼5°C using the LightCycler in the temperature-controlled fluorometer mode (Fig. 2A, left). Linear regression (r2 = 0.998) provided an excellent fitting curve (Fig. 2A, right). The results obtained from biological samples were then compared against the standard curve to determine the percentage of mutant allele present. The R201H mutation could easily be quantified using this approach. However, using the DNA–DNA FRET oligo pairs, the results of standards for R201C mutations show that the difference between the Tm of the wild-type and mutant allele was only ∼1°C (Fig. 2B). Thus, it was not possible to easily distinguish two peaks and to accurately acquire a standard curve for R201C mutations with DNA hybridization probes.
Figure 2.
The quantification of R201H (A) and R201C (B) mutations by using FRET-based DNA hybridization probes. The left panels show the negative first derivative of fluorescence versus temperature of standards (0–50% mutant alleles corresponding to 0–100% mutant cells). (A) Left: the two melting peaks, clearly separated by 5°C, correspond to the perfect match between mutant sequence and mutant fluorescent donor probe (FDP) (melting temperature of ∼65°C) and the single mismatch between wild-type sequence and mutant FDP (melting temperature of ∼60.5°C). There are two peaks because mutant cells have both mutant and wild-type alleles. The one peak in the case of the wild-type sequence was exclusively the lower melting, single mismatch peak. Right: a standard curve was plotted by using pre-mixed ssDNA standards in the 0–50% range of R201H, with the remainder being wild type. (B) Two melting peaks are separated by only ∼1°C (as shown by red and blue bars); thus, they are not easily quantified for the analysis. Standard curves could not be plotted by using DNA standards.
Standards curve generation by PNA hybridization probes
Since it was not possible to quantify the mutation load in the samples from patients with an R201C mutation with DNA–DNA FRET hybridization probes, PNA–DNA FRET hybridization probes were developed. In this case, the difference between the Tm for R201C mutation and the wild-type R201 allele was ∼ 8°C, clearly yielding two well separated melting peaks (Fig. 3A, left). A linear standard curve was plotted as defined above with an r2 = 0.995 value (Fig. 3A, right). Furthermore, the results of standards for R201H mutations showed that the difference between the Tm of the wild-type and mutant allele improved to ∼14°C (Fig. 3B, left). The standard curve for the R201H experiments resulted in a linear correlation of r2 = 0.998 (Fig. 3B, right). In order to determine the intra-assay variability for each, mutation type (three separate experiments with three independent observations) was investigated and the variability was <5% for both types (data not shown). Thus, the PNA–DNA FRET probe sets were selected to quantify both types of mutant alleles in a variety of biological samples from patients with FD/MAS. PNA–DNA FRET analysis over a range of total genomic DNA amounts (15–100 ng) obtained from the samples resulted in the same final ratios of mutant to normal cells (data not shown).
Figure 3.
The quantification of R201C (A) and R201H (B) mutations by using FRET-based PNA hybridization probes. The left panels show the negative first derivative of fluorescence versus temperature of standards (0–50% mutant alleles corresponding to 0–100% mutant cells) and the right panels show standard curve plots by using pre-mixed ssDNA standards in the 0–50% range to determine the percentage mutant DNA present in the samples from the mosaic subjects. In the left panel of (A), two melting peaks are clearly separated by ∼8°C. Two peaks observed in mutant cell standards were an equal balance between the peaks due to a perfect match (melting temperature of ∼72°C) between mutant sequence and mutant Red 640 acceptor probe (RAP640), and a peak due to a mismatch (melting temperature of ∼64°C) between wild-type sequence and mutant RAP640. The one peak in the case of the wild-type sequence is exclusively the lower temperature mismatch peak. In the right panel of (A), a standard curve was plotted by using pre-mixed ssDNA standards in the 0–50% range to determine the percentage of mutant DNA cells present in R201C samples from the mosaic subjects. The left panel of (B) shows that two melting peaks were separated by ∼14°C (melting temperatures were ∼72°C for a perfect match and ∼58°C for a mismatch) in R201H mutations. Similarly, in the right panel of (B), a standard curve was plotted by using pre-mixed ssDNA standards in the 0–50% range to determine the percentage mutant DNA present in R201H samples from the mosaic subjects.
When using the linear portion of the standard curves, it was found that the assay does not distinguish the difference between 0% (i.e. completely wild type) and 2% mutant alleles. Similarly, the linear portion of the assay does not distinguish between 48 and 50% (i.e. completely mutant) mutant alleles. Such data are therefore reported as <2.5% mutant alleles (i.e. <5% mutant cells) or >47.5% mutant alleles (i.e. >95% mutant cells). Therefore, DNA from completely normal cells is reported as <2.5% mutant alleles, and even pure mutant cells would be reported as >47.5% mutant alleles.
Quantification of mutational load in samples from patients with FD/MAS
Genomic DNA samples routinely isolated from fresh biopsies, cell cultures and paraffin blocks from either normal individuals or patients with FD/MAS were quantified by comparison with their corresponding mutation-specific standard curve. The results are summarized in Table 2. Less than 5% mutant cells (i.e. not distinguished from wild-type) were detected in a biopsy sample taken from the clinically uninvolved iliac crest of a MAS patient with an R201C mutation (Table 2, Cys01). Lack of involvement was confirmed by histological analysis, which demonstrated normal bone and bone marrow tissues (data not shown). In contrast, 72% mutant cells were detected in the specimen obtained from the contralateral, FD-involved iliac crest in the same patient (Table 2, Cys02), in which typical histopathological features of FD were observed (data not shown). Analysis of data from several R201C mutation samples revealed a mutation rate from 49 to 70% (Table 2, Cys03-07). Histopathological observations were correlated with the severity of the mutation rates (data not shown). Also analyzed were biopsy samples from two patients with the R201H mutation. In one case, 16% mutant cells were detected (Table 2, His01), and histology demonstrated an admixture of FD areas with areas of normal bone and marrow (data not shown). In the second case, 77% mutant cells were detected (Table 2, His02), and histology demonstrated typical FD histopathological features throughout (data not shown). Because the R201H mutation generates a novel NlaIII restriction site, the relative amount of this allele can be estimated by digesting the double-stranded PCR products with NlaIII using the restriction analysis technique described by Collins et al. (34). The patient DNA for the same two R201H patients described above gave digestion patterns suggesting a relatively greater amount of mutation in the His02 sample (∼59% by band quantification) compared with the His01 sample (∼12%; data not shown), thereby grossly confirming the FRET data presented above. The R201C mutation, like many mutations, does not directly generate or delete a useful restriction site, so the restriction digestion and band quantification approach is very often not as practical as making a PNA–FRET pair for quantitative analysis.
Table 2. R201 mutations quantified in bone biopsy samples from patients with FD/MAS, cell cultures generated from normal individuals or FD patients’ biopsy samples, and fresh biopsies and paraffin blocks obtained from the same sample.
| Sample ID | DNA source | Mutation | % allele | % cells |
|---|---|---|---|---|
| Cys01 | Biopsy | R201C | <2.5 | <5 |
| Cys02 | Biopsy | R201C | 36 | 72 |
| Cys03 | Biopsy | R201C | 26 | 52 |
| Cys04 | Biopsy | R201C | 33.5 | 67 |
| Cys05 | Biopsy | R201C | 24.5 | 49 |
| Cys06 | Biopsy | R201C | 35 | 70 |
| Cys07 | Biopsy | R201C | 33 | 66 |
| His01 | Biopsy | R201H | 8 | 16 |
| His02 | Biopsy | R201H | 38.5 | 77 |
| Control01 | Cell culture | R201 | <2.5 | <5 |
| Control02 | Cell culture | R201 | <2.5 | <5 |
| Control03 | Cell culture | R201 | <2.5 | <5 |
| Control04 | Cell culture | R201 | <2.5 | <5 |
| Control05 | Cell culture | R201 | <2.5 | <5 |
| Control06 | Cell culture | R201 | <2.5 | <5 |
| CC01 | Cell culture | R201C | 17 | 34 |
| CC02 | Cell culture | R201C | <2.5 | <5 |
| CC03 | Cell culture | R201C | >47.5 | >95 |
| CC04 | Cell culture | R201H | 35 | 70 |
| CC05 | Cell culture | R201H | <2.5 | <5 |
| CC06 | Cell culture | R201H | >47.5 | >95 |
| P01 | Paraffin block | R201C | 29 | 58 |
| P02 | Paraffin block | R201C | 40.5 | 81 |
| P03 | Paraffin block | R201C | 43 | 86 |
| F01 | Fresh biopsy | R201C | 27 | 54 |
| F02 | Fresh biopsy | R201C | 42.5 | 85 |
| F03 | Fresh biopsy | R201C | 44 | 88 |
One biopsy sample was divided in half, with one piece prepared for demineralization and subsequent paraffin embedding and the other half used as fresh sample. DNA was isolated from both samples and the R201C mutation rate was quantified (P01–P03 are the paraffin-embedded samples corresponding to the fresh samples, F01–F03). Results are shown as a percentage of mutant alleles and cells.
The relative amounts of normal and mutant cells in two multicolony cell culture populations obtained from both R201H and R201C FD lesions (Table 2, CC01 and CC04) were also quantified. A different proportion of mutant cells was observed in the two cultures, consistent with the expected, variable frequency of mutant and wild-type cells in non-clonal cultures established from FD lesions. As expected, analysis of four cloned strains revealed proportions of mutant cells <5% in two cases (Table 2, CC02 and CC05), and >95% in two cases (Table 2, CC03 and CC06), although the current assay does not possess the sensitivity to definitively declare that a clone is absolutely pure in nature. While the use of PNA clamping (13) could prove the purity of wild-type cells, the PNA–FRET assay is the best current method to determine if clones are at least >95% pure mutant cells. Also, as expected, six multicolony-derived strains from normal donors did not reveal either R201H or R201C mutations within the limit of sensitivity of the assay (Table 2, Control01–Control06).
In order to determine whether decalcification and paraffin embedding of biopsy samples would affect the results of quantitative mutation analysis, three samples were analyzed. Paired paraffin-embedded and fresh samples were prepared from two halves of single trepan cores from different areas of a surgical waste specimen from a patient with the R201C mutation. Similar frequencies of mutant cells were detected in both paired halves of the three samples. The observed frequencies of mutant cells in paraffin blocks versus matching fresh tissues were 58 versus 54% (P01 versus F01), 81 versus 85% (P02 versus F02) and 86 versus 88% (P03 versus F03), respectively (Table 2).
DISCUSSION
Post-zygotic somatic activating mutations of the GNAS gene, which encodes the α-stimulatory G-protein, are now recognized as the underlying cause of isolated endocrinopathies, FD, and the even more rare MAS. (The MAS phenotype includes skin hyperpigmentation and endocrinopathies in addition to FD.) As a consequence of the post-zygotic nature of these mutations, these patients represent a broad spectrum of somatic mosaicism. From the descriptions of diseases caused by somatic mutations, it has long been hypothesized that tissues and even different regions within an individual tissue may contain different numbers of cells harboring the mutation. While it has been shown in a transplantation model that both mutant and normal cells are required to generate an FD lesion (16), no study has quantitatively shown the range of mutational loads that exist within naturally occurring lesions and histologically normal tissues.
PNAs have inspired the development of a variety of hybridization-based methods for detection, quantification, purification and characterization of nucleic acids. Inhibition of a PCR amplification of a specific target by PCR clamping has been used successfully in the detection of single base pair gene mutations (35). We have previously developed a PNA-based PCR clamping technique for the detection of R201H and R201C mutations in FD/MAS (33). In that report, it was shown that suppression of the amplification of the wild-type allele by PNA in a single PCR led to a subsequent sequencing reaction that was dominated by the mutant sequence, even when the percentage of the mutant cells was as low as 0.1%. This technique is useful in the detection of the mutations in samples with low mutation frequency including peripheral blood cells in which mutations were detected in four out of nine MAS patients’ blood samples (36). Although this technique permitted the determination of the identity of the mutation for each patient, it could not provide quantification of the mutational load. A robust method of quantifying the number of mutant cells in a biopsy, paraffin block or dish of cells grown from an affected tissue has been lacking, and such a method is reported here. The assay is based on the concept that when bound to its target DNA, a fluorescently labeled PNA will obtain energy from a paired fluorescein-labeled donor oligonucleotide and emit a signal at a longer wavelength. The strength of the signal is proportional to the number of bound PNA molecules. In the assay, a portion of the PNA–FRET signal is lost upon heating as the subpopulation of PNAs containing a single mismatch (representing the wild-type allele) melts off the target ssDNA. The remaining signal is lost at a higher temperature as the perfectly matched PNA–target DNA pair (mutant allele) subsequently melts off. Analysis of these ‘melting peaks’ yields a very high correlation to the ratio of mutant to normal alleles in the standards.
Using this novel technique, it was observed that fresh biopsy specimens that appeared to be clinically and histologically unaffected in FD patients harbored <5% mutant cells, the lowest limit of detection using the linear portion of the standard curves. A range of mutation load (16–88%) was detected in specimens from clinically affected areas of bone. Importantly, it was shown that paraffin embedding, at least under the conditions used, did not appear to affect the outcome of the ratio of mutant to normal cells. It is also of interest to note that a relatively low portion of mutant cells (16%) in some areas proved to be a mixture of normal and pathological cells upon histological examination. It was also shown that the variability in the number of mutant cells in the parent lesion is reflected in non-clonal cell cultures established from those lesions, whereas cultures from single cell-derived colonies were shown to be either mutant (>95% mutant cells) or wild-type (<5% mutant cells) within the detection limit of the assay, with no variability. This is of utmost importance in view of the need for cell-based studies to determine the downstream effects of activating GNAS mutations. Variation in mutational load undoubtedly impacts upon outcomes of such studies, and reliable identification of non-mutant and mutant clonal strains would greatly facilitate changes in cell function as a direct effect of mutation.
Our data support the notion that MAS and related pathologies such as FD and non-skeletal isolated endocrine lesions associated with GNAS mutations represent a spectrum of somatic mosaicism of the same basic disorder (37). However, further investigation is needed to determine the impact of specific levels of mosaicism on the function of cells and tissues that are affected. We predict that our technique should prove effective in accurately determining the ratio of mutant to normal cells in a wide variety of clinical samples.
The assay can be modified into a multiplex format, somewhat like that reported for K-ras mutations in primary colon tumors (23), to quantify different R201 mutations in various samples by using different wavelength FRET pairs for each site to be probed. Our LightCycler has the capacity to mutiplex two or possibly three different FRET pairs, but other systems such as the Mx4000® Multiplex Quantitative PCR System (Stratagene, La Jolla, CA), with more wavelengths available, may be able to mutiplex even more. This would depend on the availability of the specific fluorescent molecules that can be added to the PNA and oligonucleotide probes. We used PNA hybridization probes specifically designed for R201H and R201C mutations in order to obtain higher precision. However, theoretically, a wild-type PNA hybridization probe could result in a melting temperature difference with any type of R201 mutation (R201C, R201H, R201G, R201L, R201S and R201P) present in the sample, allowing use of the technique in screening large number of samples for the presence/absence of an R201 mutation without multiplex format. PNA–FRET analysis of any known point mutation site or single nucleotide polymorphism (SNP) could similarly be screened using properly developed probes. Because all of the steps up to the melting analysis of the FRET probes are standard DNA purification, PCR, etc., the assay would seem to be easily adapted to high throughput. Even generating the melting curves (which we could do at a theoretical rate of ∼60/h with our LightCycler) could be further adapted to high throughput by using appropriately designed real-time PCR machines for the 96-well format.
Data presented here show that this novel technique could lead to more advanced investigations of pathological lesions, at the cellular and molecular level, not only in FD/MAS, but also in other applications. For example, the detection and quantification of SNPs observed in aging and a number of tumor pathologies, neurodegenerative disorders and inherited diseases including sickle cell anemia (38), several diseases of skeletal muscles (39) and familial hypertrophic cardiomyopathy (40) have been reported to have both mutant and wild-type alleles, and the wide spectrum of clinical symptoms may be due in part to the mutational load. The study of many of these diseases may benefit as much as FD/MAS does from the use of the PNA–FRET quantitative analysis of point mutational loads as long as the location of the mutation thought to cause the disorder is within a known 10–15 bp region, the size of a typical PNA. We used a combination of a nucleotide probe (anchor probe) and a PNA partner, but the ability to use PNAs for both probes in the FRET conjunction could effectively double the size of the area probed.
Our technique, by combining the FRET principle and a PNA hybridization probe, provides a mutant allele detection approach that is sensitive to 2.5% mutant alleles. This assay could probably be made more sensitive if necessary with additional purification of the PNA probes (allowing for sharper melting curves) and attention to non-linear portions of the standard curves. The emphasis of this study, however, is on obtaining accurate ratios of two sequences differing by one base pair and not on identifying low abundance somatic mutations per se. Furthermore, it is likely that, unlike many of the techniques that are sequence specific (e.g. use of restriction enzymes), the PNA–FRET approach will probably work for most mutations and SNPs. While other oligonucleotide-based assays such as molecular beacons and TaqMan probes can be inherently sensitive, accurately obtaining the ratio of one sequence to another within a single sample (e.g. wild-type to mutant) will sometimes suffer from the same dependency of the local sequences that we saw when using DNA-based FRET probes with the R201C mutation of GNAS. Because of the inherent properties of PNAs, a successful PNA-based FRET pair can probably be found for nearly all short genomic sequences. With the ability to isolate DNA from paraffin sections, this technique should also enable us to answer questions of mutational load even within microscopic areas as long as sufficient amounts of genomic DNA can be recovered and amplified by PCR.
In conclusion, it was shown that DNA obtained from fresh tissue, paraffin-embedded tissue and cell cultures of MAS patient lesions contains a range of mutation loads while normal areas and control patient samples had no detectable mutational load. Thus the mutational load may determine the phenotype or severity of the lesion. Furthermore, this novel technique could easily be adapted for the quantification of mutations related to a wide spectrum of diseases.
Acknowledgments
ACKNOWLEDGEMENT
The support of Telethon Fondazione Onlus grant E1029 (to P.B.) is gratefully acknowledged.
REFERENCES
- 1.Michikawa Y., Mazzucchelli,F., Bresolin,N., Scarlato,G. and Attardi,G. (1999) Aging-dependent large accumulation of point mutations in the human mtDNA control region for replication. Science, 286, 774–779. [DOI] [PubMed] [Google Scholar]
- 2.Davis R.E., Miller,S., Herrnstadt,C., Ghosh,S.S., Fahy,E., Shinobu,L.A., Galasko,D., Thal,L.J., Beal,M.F., Howell,N. et al. (1997) Mutations in mitochondrial cytochrome c oxidase genes segregate with late-onset Alzheimer disease. Proc. Natl Acad. Sci. USA, 94, 4526–4531. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3.Tanaka M., Kovalenko,S.A., Gong,J.S., Borgeld,H.J., Katsumata,K., Hayakawa,M., Yoneda,M. and Ozawa,T. (1996) Accumulation of deletions and point mutations in mitochondrial genome in degenerative diseases. Ann. NY Acad. Sci., 786, 102–111. [DOI] [PubMed] [Google Scholar]
- 4.Kovalenko S.A., Tanaka,M., Yoneda,M., Iakovlev,A.F. and Ozawa,T. (1996) Accumulation of somatic nucleotide substitutions in mitochondrial DNA associated with the 3243 A-to-G tRNA(leu)(UUR) mutation in encephalomyopathy and cardiomyopathy. Biochem. Biophys. Res. Commun., 222, 201–207. [DOI] [PubMed] [Google Scholar]
- 5.Goto Y., Nonaka,I. and Horai,S. (1990) A mutation in the tRNA(Leu)(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature, 348, 651–653. [DOI] [PubMed] [Google Scholar]
- 6.vandenOuweland J.M., Lemkes,H.H., Ruitenbeek,W., Sandkuijl,L.A., de Vijlder,M.F., Struyvenberg,P.A., van de Kamp,J.J. and Maassen,J.A. (1992) Mutation in mitochondrial tRNA(Leu)(UUR) gene in a large pedigree with maternally transmitted type II diabetes mellitus and deafness. Nature Genet., 1, 368–371. [DOI] [PubMed] [Google Scholar]
- 7.Jansen J.J., Maassen,J.A., van der Woude,F.J., Lemmink,H.A., van den Ouweland,J.M., t’ Hart,L.M., Smeets,H.J., Bruijn,J.A. and Lemkes,H.H. (1997) Mutation in mitochondrial tRNA(Leu(UUR)) gene associated with progressive kidney disease. J. Am. Soc. Nephrol., 8, 1118–1124. [DOI] [PubMed] [Google Scholar]
- 8.Smith T.A., Yau,S.C., Bobrow,M. and Abbs,S.J. (1999) Identification and quantification of somatic mosaicism for a point mutation in a Duchenne muscular dystrophy family. J. Med. Genet., 36, 313–315. [PMC free article] [PubMed] [Google Scholar]
- 9.McCune D. (1937) Osteodystrophia fibrosa. Am. J. Disabl. Child., 54, 806–848. [Google Scholar]
- 10.Albright F. (1937) Syndrome characterized by osteitis fibrosa disseminata, areas of pigmentation and endocrine dysfunction, with precocious puberty in females. N. Engl. J. Med., 216, 727–741. [Google Scholar]
- 11.Shenker A., Weinstein,L.S., Sweet,D.E. and Spiegel,A.M. (1994) An activating Gs alpha mutation is present in fibrous dysplasia of bone in the McCune–Albright syndrome. J. Clin. Endocrinol. Metab., 79, 750–755. [DOI] [PubMed] [Google Scholar]
- 12.Schwindinger W.F., Francomano,C.A. and Levine,M.A. (1992) Identification of a mutation in the gene encoding the alpha subunit of the stimulatory G protein of adenylyl cyclase in McCune–Albright syndrome. Proc. Natl Acad. Sci. USA, 89, 5152–5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Candeliere G.A., Roughley,P.J. and Glorieux,F.H. (1997) Polymerase chain reaction-based technique for the selective enrichment and analysis of mosaic arg201 mutations in G alpha s from patients with fibrous dysplasia of bone. Bone, 21, 201–206. [DOI] [PubMed] [Google Scholar]
- 14.Riminucci M., Fisher,L.W., Majolagbe,A., Corsi,A., Lala,R., De Sanctis,C., Robey,P.G. and Bianco,P. (1999) A novel GNAS1 mutation, R201G, in McCune–Albright syndrome. J. Bone Miner. Res., 14, 1987–1989. [DOI] [PubMed] [Google Scholar]
- 15.Mockgridge K.A.L.M., Reed,L.A., Post,E., Kaplan,F.S., Jan de Beur,S.M., Deng,Z., Ding,C., Howard,C. and Schnur,R,E. (1999) Polyendocrinopathy, skeletal dysplasia, organomegaly and distinctive facies associated with a novel, widely expressed Gsα mutation. Am. J. Hum. Genet., 65 (Suppl.), A426. [Google Scholar]
- 16.Bianco P., Kuznetsov,S.A., Riminucci,M., Fisher,L.W., Spiegel,A.M. and Robey,P.G. (1998) Reproduction of human fibrous dysplasia of bone in immunocompromised mice by transplanted mosaics of normal and Gsalpha-mutated skeletal progenitor cells. J. Clin. Invest., 101, 1737–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong L.J. and Senadheera,D. (1997) Direct detection of multiple point mutations in mitochondrial DNA. Clin. Chem., 43, 1857–1861. [PubMed] [Google Scholar]
- 18.Zhong S., Ng,M.C., Lo,Y.M., Chan,J.C. and Johnson,P.J. (2000) Presence of mitochondrial tRNA(Leu(UUR)) A to G 3243 mutation in DNA extracted from serum and plasma of patients with type 2 diabetes mellitus. J. Clin. Pathol., 53, 466–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobi F.K., Meyer,J., Pusch,C.M. and Wissinger,B. (2001) Quantitation of heteroplasmy in mitochondrial DNA mutations by primer extension using Vent(R)(exo-) DNA polymerase and RFLP analysis. Mutat. Res., 478, 141–151. [DOI] [PubMed] [Google Scholar]
- 20.Mashima Y., Saga,M., Hiida,Y., Oguchi,Y., Wakakura,M., Kudoh,J. and Shimizu,N. (1995) Quantitative determination of heteroplasmy in Leber’s hereditary optic neuropathy by single-strand conformation polymorphism. Invest. Ophthalmol. Vis. Sci., 36, 1714–1720. [PubMed] [Google Scholar]
- 21.Fahy E., Nazarbaghi,R., Zomorrodi,M., Herrnstadt,C., Parker,W.D., Davis,R.E. and Ghosh,S.S. (1997) Multiplex fluorescence-based primer extension method for quantitative mutation analysis of mitochondrial DNA and its diagnostic application for Alzheimer’s disease. Nucleic Acids Res., 25, 3102–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nigou M., Parfait,B., Clauser,E. and Olivier,J.L. (1998) Detection and quantification of the A3243G mutation of mitochondrial DNA by ligation detection reaction. Mol. Cell Probes, 12, 273–282. [DOI] [PubMed] [Google Scholar]
- 23.Khanna M., Park,P., Zirvi,M., Cao,W., Picon,A., Day,J., Paty,P. and Barany,F. (1999) Multiplex PCR/LDR for detection of K-ras mutations in primary colon tumors. Oncogene, 18, 27–38. [DOI] [PubMed] [Google Scholar]
- 24.Szuhai K., Ouweland,J., Dirks,R., Lemaitre,M., Truffert,J., Janssen,G., Tanke,H., Holme,E., Maassen,J. and Raap,A. (2001) Simultaneous A8344G heteroplasmy and mitochondrial DNA copy number quantification in myoclonus epilepsy and ragged-red fibers (MERRF) syndrome by a multiplex molecular beacon based real-time fluorescence PCR. Nucleic Acids Res., 29, e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Howell N., Xu,M., Halvorson,S., Bodis-Wollner,I. and Sherman,J. (1994) A heteroplasmic LHON family: tissue distribution and transmission of the 11778 mutation. Am. J. Hum. Genet., 55, 203–206. [PMC free article] [PubMed] [Google Scholar]
- 26.Juvonen V., Huoponen,K., Syvanen,A.C., Nikoskelainen,E. and Savontaus,M.L. (1994) Quantification of point mutations associated with Leber hereditary optic neuroretinopathy by solid-phase minisequencing. Hum. Genet., 93, 16–20. [DOI] [PubMed] [Google Scholar]
- 27.Olsson C., Zethelius,B., Lagerstrom-Fermer,M., Asplund,J., Berne,C. and Landegren,U. (1998) Level of heteroplasmy for the mitochondrial mutation A3243G correlates with age at onset of diabetes and deafness. Hum. Mutat., 12, 52–58. [DOI] [PubMed] [Google Scholar]
- 28.Nielsen P.E., Egholm,M., Berg,R.H. and Buchardt,O. (1991) Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science, 254, 1497–1500. [DOI] [PubMed] [Google Scholar]
- 29.Hanvey J.C., Peffer,N.J., Bisi,J.E., Thomson,S.A., Cadilla,R., Josey,J.A., Ricca,D.J., Hassman,C.F., Bonham,M.A., Au,K.G. et al. (1992) Antisense and antigene properties of peptide nucleic acids. Science, 258, 1481–1485. [DOI] [PubMed] [Google Scholar]
- 30.Kyger E.M., Krevolin,M.D. and Powell,M.J. (1998) Detection of the hereditary hemochromatosis gene mutation by real-time fluorescence polymerase chain reaction and peptide nucleic acid clamping. Anal. Biochem., 260, 142–148. [DOI] [PubMed] [Google Scholar]
- 31.Szollosi J., Damjanovich,S. and Matyus,L. (1998) Application of fluorescence resonance energy transfer in the clinical laboratory: routine and research. Cytometry, 34, 159–179. [PubMed] [Google Scholar]
- 32.Corsi A., Collins,M.T., Riminucci,M., Howell,P.G., Boyde,A., Robey,P.G. and Bianco,P. (2003) Osteomalacic and hyperparathyroid changes in fibrous dysplasia of bone: core biopsy studies and clinical correlations. J. Bone Miner. Res., 18, 1235–1246. [DOI] [PubMed] [Google Scholar]
- 33.Bianco P., Riminucci,M., Majolagbe,A., Kuznetsov,S.A., Collins,M.T., Mankani,M.H., Corsi,A., Bone,H.G., Wientroub,S., Spiegel,A.M. et al. (2000) Mutations of the GNAS1 gene, stromal cell dysfunction and osteomalacic changes in non-McCune–Albright fibrous dysplasia of bone. J. Bone Miner. Res., 15, 120–128. [DOI] [PubMed] [Google Scholar]
- 34.Collins M.T., Sarlis,N.J., Merino,M.J., Monroe,J., Crawford,S.E., Krakoff,J.A., Guthrie,L.C., Bonat,S., Robey,P.G. and Shenker,A. (2003) Thyroid carcinoma in the McCune-Albright syndrome: contributory role of activating Gs alpha mutations. J. Clin. Endocrinol. Metab., 88, 4413–4417. [DOI] [PubMed] [Google Scholar]
- 35.Orum H., Nielsen,P.E., Egholm,M., Berg,R.H., Buchardt,O. and Stanley,C. (1993) Single base pair mutation analysis by PNA directed PCR clamping. Nucleic Acids Res., 21, 5332–5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hannon T.S., Noonan,K., Steinmetz,R., Eugster,E.A., Levine,M.A. and Pescovitz,O.H. (2003) Is McCune–Albright syndrome overlooked in subjects with fibrous dysplasia of bone? J. Pediatr., 142, 532–538. [DOI] [PubMed] [Google Scholar]
- 37.Ringel M.D., Schwindinger,W.F. and Levine,M.A. (1996) Clinical implications of genetic defects in G proteins. The molecular basis of McCune–Albright syndrome and Albright hereditary osteodystrophy. Medicine, 75, 171–184. [DOI] [PubMed] [Google Scholar]
- 38.Ingram V. (1957) Gene mutations in human haemoglobin: the chemical difference between normal and sickle cell haemoglobin. Nature, 180, 326–328. [DOI] [PubMed] [Google Scholar]
- 39.Hoffman E.P., Lehmann-Horn,F. and Rudel,R. (1995) Overexcited or inactive: ion channels in muscle disease. Cell, 80, 681–686. [DOI] [PubMed] [Google Scholar]
- 40.Nier V., Schultz,I., Brenner,B., Forssmann,W. and Raida,M. (1999) Variability in the ratio of mutant to wildtype myosin heavy chain present in the soleus muscle of patients with familial hypertrophic cardiomyopathy. A new approach for the quantification of mutant to wildtype protein. FEBS Lett., 461, 246–252. [DOI] [PubMed] [Google Scholar]