Abstract
The pteridine nucleoside analog 3-methyl isoxanthopterin (3-MI) is highly fluorescent, with a quantum yield of 0.88, and it can be synthesized as a phosphoramidite and incorporated into oligonucleotides through a deoxyribose linkage. Within an oligonucleotide, 3-MI is intimately associated with native bases and its fluorescence is variably quenched in a sequence-dependent manner. Bend ing, annealing, binding, digestion or cleavage of fluorophore-containing oligonucleotides can be detected by monitoring changes in fluorescence properties. We developed a single step method for detecting annealing of complementary DNA sequences using 3-MI-containing oligonucleotides as hybridization probes. One of the complementary strands contains the fluorophore as an insertion and when annealing occurs, the fluorophore bulges out from the double strand, resulting in increased fluorescence intensity. We have examined the sequence dependency, optimal strand length and impact of multiple fluorophores per strand in terms of brightness and impact on the annealing process. We describe the application of this technique to the detection of positive PCR products using an HIV-1 detection system. This sequence-dependent hybridization technique can result in fluorescence intensity increases of up to 27-fold. Fluorescence intensity increases are only seen upon specific binding to bulge-generating complements, removing issues of high background from non-specific binding.
INTRODUCTION
The pteridine nucleoside analog 3-methyl isoxanthopterin (3-MI) can be synthesized in a phosphoramidite form and incorporated into oligonucleotides through a deoxyribose linkage identical to that of endogenous bases in native DNA, using a conventional DNA synthesizer (1). The fluorescence properties (intensity, lifetime, spectral shifts, anisotropy and energy transfer) of the incorporated pteridine nucleoside analog are affected by interactions with the neighboring purine or pyrimidine bases (2–8). The fluorescence of 3-MI is variably quenched by incorporation into an oligonucleotide (depending on neighboring base sequence) and events that structurally alter the fluorophore-containing DNA can be monitored in real time by observing changes in fluorescence. For example, we previously described an HIV-1 integrase assay in which cleavage of the terminal dinucleotide from the 3-MI-containing oligonucleotide serving as the integrase substrate resulted in a measurable increase in fluorescence intensity (1).
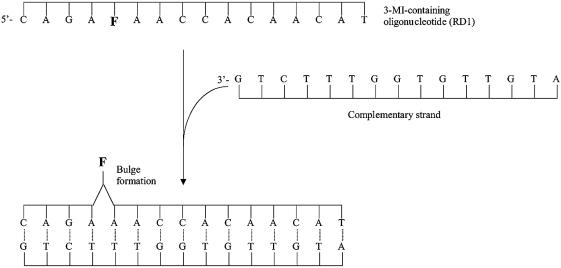
In many cases annealing of a probe-containing oligonucleotide with its complement results in a measurable loss in fluorescence intensity (3,8), however, in order to enhance detection of the annealing process, we have devised another approach. In this technique a bulge composed of the fluorophore is formed on annealing to a complementary strand that does not contain a base pairing partner for the fluorophore (Fig. 1). Pushing the fluorophore out of close proximity to neighboring bases can reduce quenching and increase fluorescence intensity, depending on the sequence. This bulge-forming hybridization technique would allow a fluorophore-containing oligonucleotide to serve as a specific hybridization probe that is quenched in the single-strand state and fluoresces when it anneals to its complementary strand, obviating the need for separation of a mixture of DNA sequences. We applied this method to the detection of PCR products using a 3-MI-containing PCR probe.
Figure 1.
Formation of a single-base bulge as a result of annealing a 3-MI-containing oligonucleotide to its complementary strand, which does not contain a base pairing partner for 3-MI. Displacement of 3-MI from the base stacked configuration of the single strand, which quenches the fluorescence of the fluorophore, results in an increase in fluorescence intensity with hybridization.
MATERIALS AND METHODS
Synthesis and purification of oligonucleotides
3-Methyl-8-(2-deoxy-5-O-dimethoxytrityl-β-d-ribofuranosyl) isoxanthopterin-3′-O-(β-cyanoethyl, N-diisopropyl) phosphoramidite was synthesized as previously described (1). DNA oligonucleotides were synthesized on an automated DNA synthesizer (Applied Biosystems, Foster City, CA) using the standard instructions provided by the manufacturer with 3-MI phosphoramidite in the bottle 5 position. 3-MI-containing strands were de-blocked in the standard manner and purified using 20% denaturing PAGE. Bands were detected in the gel by UV shadowing, excised and extracted using an electro-elution device (Elutrap; Schleicher & Schuell, Keene, NH) followed by ethanol precipitation.
Spectroscopic analysis
Fluorescence measurements were performed on a Photon Technology International modular fluorometer (Brunswick, NJ) with a double excitation monochrometer, water-cooled photomultiplier and a 150 W xenon arc lamp. The sample chamber was equipped with a Peltier temperature controller. All samples were measured at 25°C. Fluorescence intensity was derived by integration of the emission scan from 360 to 500 nm after excitation at 350 nm. Emission polarizers were set at magic angle positions to reduce Raman scatter in dilute samples.
Absorbance was measured with a Hewlett Packard Model 8452a spectrophotometer (Palo Alto, CA) equipped with a Peltier temperature controller. Oligonucleotide melting curves were generated in the presence of 10 mM NaCl at optical densities between 0.2 and 0.8 absorbance units using Hewlett Packard software.
Hybridization with bulge formation
Seventeen 3-MI-containing oligonucleotides and their complementary strands were examined (Table 1) to assess the effect of forced bulge formation upon hybridization on the fluorescence intensity of the fluorophore. Complementary strands did not contain a base pairing partner for 3-MI, such that when the strands annealed, 3-MI would be forced out of its position in the strand (Fig. 1). The effect of the sequence of bases on the increase in fluorescence intensity upon annealing was assessed with these oligonucleotides.
Table 1. Sequences of 3-MI-containing oligonucleotides used to test the effect of bulge formation on fluorescence intensity.
| Oligonucleotide sequence (5′→3′) | Increase | |
|---|---|---|
| HP6 | cctctaagaggtgtaaFaatgtggagaatctcc | 27 |
| RD9 | cttgagcccccgaFaactctgactcgg | 21 |
| RD11 | agcagttgctaaagaaFaattgaacacgctcggacttgc | 21 |
| HP4 | cctctaagaggtgtaaFagtgtggagaatctcc | 14 |
| RD4 | tagacgcttctcaaFaactggaca | 12 |
| PCR2 | gcaagatggaggaaacaaFggctggagccaa | 11 |
| SK19 | atcctgggattaaataaFaatagtaagaatgtatagccctac | 10 |
| JS-6 | taaataaFaatagtaggtagggctatacattct | 9 |
| RD7 | cctgagcgtgagaFaagctggac | 8 |
| JS-5 | tggagaaFgagctacacctggccgtcaggcagc | 7 |
| PCR4 | attccacaaFgccgtgtca | 6 |
| PCR1 | ggctttcgagagFaacccactaccca | 5 |
| RD5 | agaagaagataaFagagcgaggcgtcccca | 4 |
| A1Sf | ctgcagFaatgggatagagtgcatccagtg | 3 |
| RD1 | cagaFaaccacaacat | 2 |
| HP5 | cctctaagaggtgtccFcctgtggagaatctcc | 0.3 |
| HPR | cctctaagaggtgtacFagtgtggagaatctcc | –1.5 |
F represents the position of 3-MI in the oligonucleotide sequence. The fold increase (or decrease represented by a negative number) in fluorescence resulting from annealing and bulge formation over fluorescence intensity of the single-stranded oligonucleotide is shown in the far right column.
3-MI-containing oligonucleotide strands were combined with a slight molar excess (10%) of their complementary strand in 10 mM Tris pH 7.5, and 10 mM NaCl. The control for each experiment contained the identical concentration of the 3-MI-containing oligonucleotide and NaCl with an equivalent volume of buffer added in place of the complementary strand. Annealing was performed by heating the mixture to 90°C for 2–3 min and then allowing the mixture to gradually cool to room temperature. The change in fluorescence intensity for each 3-MI-containing oligonucleotide shown in Table 1 was determined by comparing the integrals of the fluorescence emission spectra of the double strand with bulge formation to the identical single strand (each measured at 25°C). The fold increase is the ratio of the integral of the double strand to the integral of the single strand. A range of concentrations of complementary oligonucleotide pairs was tested in each case to identify at which concentration the fold increase in fluorescence intensity from bulge formation was being negated by the higher background fluorescence intensity from the single strand. Each strand was tested in a concentration series at least three times.
Effect of strand length
Sequences shown in Table 2 were each analyzed in the manner described above to define the most effective length of strand for annealing.
Table 2. Sequences of 3-MI-containing oligonucleotides (5′→3′) used to determine the minimum length of strand for an optimal change in fluorescence intensity upon bulge formation.
| Name | Oligonucleotide sequence (5′→3′) | Fold fluorescence increase |
|---|---|---|
| HIVPCR-11 | aataaFaatag | 1.1 |
| HIVPCR-13 | aaataaFaatagt | 1.2 |
| HIVPCR-15 | taaataaFaatagta | 1.2 |
| HIVPCR-18 | taaataaFaatagtaAga | 7.9 |
| HIVPCR-21 | taaataaFaatagtaAgaatg | 13.6 |
| HIVPCR-24 | taaataaFaatagtaAgaatgtat | 13.6 |
Each of the 3-MI-containing strands was paired to complementary strands in which 3-MI did not have a base pairing partner.
Multiple probes per strand
The effect of having multiple probes in a strand was evaluated using the sequences shown in Table 3. Annealing conditions were as described above. Melting temperatures were determined to assess the extent of disruption of base pairing caused by the insertions (probes).
Table 3. Sequences of 3-MI-containing oligonucleotides used to assess the optimal number of fluorophores per oligonucleotide.
| Name | Oligonucleotide sequence (5′–3′) | Increase | Tm |
|---|---|---|---|
| 1 | gcaagatggaggaaacaaFggctggagccaa | 8 | 70.4 |
| 2 | gcaagatggaggFaaacaaFggctggagccaa | 8 | 68.6 |
| 3 | gcaaFgatggaggFaaacaaFggctggagccaa | 10 | 66.6 |
| 4 | gcaaFgatggaggFaaacaaFggctggFagccaa | 10 | 63.6 |
| Control | gcaagatggaggaaacaaggctggagccaa | 71.4 | |
| Complement | ttggctccagccttgtttcctccatcttgc |
F represents the position of 3-MI. The fold increase in fluorescence and the melting temperatures (Tm, in °C) of the double-stranded oligonucleotides are shown in the right hand column.
Multiple base bulges
Sequences identical to 5′-cct cta aga ggt gtg t{}g tgt gga gaa tct cc-3′ containing one {F}, two {tF} or three {tFg} base insertions that would form a bulge upon annealing were examined. Annealing conditions were as described above.
PCR conditions
HIV-1 gag primers SK38 and SK39 were used to develop the PCR product detection technique (9). The fluorescent hybridization probe (5′-taa ata aFaa tag taaF gaa tgt ata gcc cta cca-3′, where F represents 3-MI) contains two 3-MI insertions and is complementary to a 33 base region between the primers in the 115 bp PCR product. Reactions contained varying amounts of template from the plasmid pSum9 generated from a full-length HIV molecular clone (10).
PCR reactions contained 10 mM Tris pH 8.3, 50 mM KCl, 2 mM MgCl2, 200 µM dNTP mixture, 2.5 U Amplitaq polymerase (Perkin Elmer, Foster City, CA), 0.2 µM primer pair (SK38, 5′-ata atc cac cta tcc cag tag gag aaa t-3′; SK39, 5′-ttt ggt cct tgt ctt atg tcc aga atg c-3′) and between 6.4 and 290 nM 3-MI-containing hybridization probe (5′-taa ata aFaa tag taaF gaa tgt ata gcc cta cc-3′) in a total volume of 100 µl. All components except for the template (or the variable being tested) were combined in a master mix and aliquots were dispensed prior to adding the final component in order to overcome some of the chronic problems associated with quantitation of amplified products (11). Each test of a particular concentration of reactant (such as template) was done in duplicate with experiments repeated at least three times. Reaction mixtures were initially heated to 94°C for 2 min and in each subsequent cycle heated to 94°C for 0.5 min, cooled to 60°C for 0.5 min and heated to 72°C for 0.5 min. This was repeated for 30 cycles using a Perkin Elmer thermocycler. The products already contained the hybridization probe and no additional annealing conditions were pursued.
After amplification, samples were held at 72°C for 10 min and then brought to 4°C until analysis, when they were equilibrated at 25°C for 5 min before fluorescence scanning. Annealing of the 3-MI-containing hybridization probe to the amplified PCR product results in bulge formation at the sites of 3-MI, leading to an increase in fluorescence intensity. For each experiment, a scan of a blank containing all components except the template was subtracted from the template-containing scan. Products were also analyzed using 2% agarose gel electrophoresis and visualized with ethidium bromide staining. A negative control consisting of a 3-MI-containing oligonucleotide (5′-gaa gaa gat aaFa gag cga ggc gtc ccc a-3′) not complementary to the PCR product was also tested.
RESULTS
Bulge formation
Annealing of the 3-MI-containing strands to their bulge-inducing complementary strands resulted in up to a 27-fold increase in fluorescence intensity (Table 1). The magnitude of the increase in fluorescence intensity upon annealing appears to be influenced by the identity of the bases surrounding the fluorophore. The greatest increases were observed when the fluorophore was flanked by two purines on each side and with adenosines immediately flanking the fluorophore (Fig. 2). The fluorescence from 3-MI is more heavily quenched in the single-stranded form with these sequences and the baseline fluorescence intensity (before annealing) is lower than what would be expected with sequences in which 3-MI is surrounded by pyrimidines, as reported previously (1).
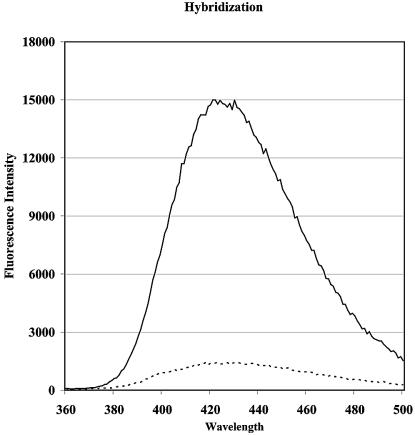
Figure 2.
Fluorescence emission scans of the single-stranded 3-MI-containing oligonucleotide, SK19 from Table 1 (dashed line) and SK19 annealed to its complementary strand, which does not contain a base pairing partner for 3-MI (solid line). Samples were measured at 25°C.
Although a modest increase in fluorescence intensity was observed with the formation of a two or three base bulge, the greatest increases were seen when a single base was forced out of the base stacked configuration upon annealing (Fig. 1). Oligonucleotides with a sequence identical to 5′-cct cta aga ggt gtg t{}g tgt gga gaa tct cc-3′ containing one {F}, two {tF} or three {tFg} base insertions (where F represents 3-MI) that would form a bulge upon annealing to complementary strands that lacked base pairing partners for F, tF or tFg were analyzed. The fluorescence intensity increased by 7-, 1.6- and 1.6-fold for 1, 2 and 3 base bulges, respectively.
A series of 3-MI-containing oligonucleotides ranging from 11 to 24 bases in length with identical sequences were annealed to appropriate complementary strands to determine the optimal probe length (based on the fold increase in fluorescence intensity) (Table 2). For this sequence the data reveal that the greatest increases in fluorescence intensity occur with an oligonucleotide of at least 21 bases in length.
Oligonucleotides (30mer) with the identical sequence of native bases and from one to four 3-MI insertions were annealed to a complementary strand which did not contain base pairing partners for 3-MI (Table 3). The actual counts per second for numbers 1 and 2 in Table 2, were 900 and 1850, respectively, a doubling of the signal for the bulged form. Each 3-MI insertion reduced the melting temperature by 1–2°C. The additional fluorophores resulted in only a modest increase in the fold change in fluorescence intensity upon annealing, primarily because of the increase in baseline fluorescence intensity of the single-stranded oligonucleotides containing multiple copies of 3-MI.
Detection of PCR products
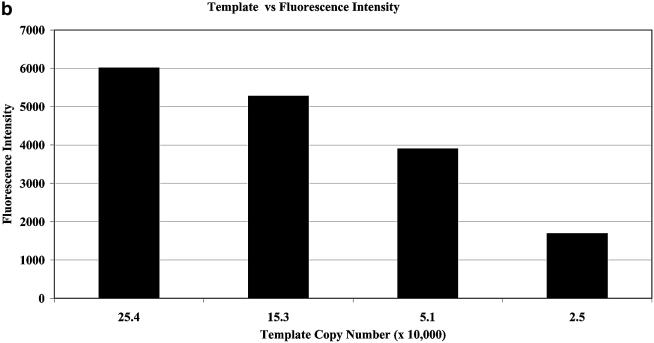
Fluorescence emission scans from a representative positive PCR reaction and an equivalent blank (containing no template) are shown in Figure 3a. The fluorescence intensity of the reaction mixture is increased when the probe anneals to the complementary section of PCR product and bulges form at the sites of the two 3-MI insertions. Figure 3b shows the relationship between template copy number and fold increase in fluorescence intensity. A negative control containing the same probe concentration as the positive samples cancels out any fluorescence-based contribution from the presence of unannealed probe. Detection using the 3-MI-containing PCR probe correlated well with results from analysis of PCR products separated using an agarose gel and visualized with ethidium bromide staining. In a series of experiments examining various concentrations of probe, we found that multiple products (as seen with gel analysis) were formed at concentrations of probe >200 nM. In control experiments a 3-MI-containing oligonucleotide that was not complementary to the PCR product did not result in an increase in fluorescence intensity after amplification. PCR reactions were repeated multiple times (in excess of three) in duplicate for each concentration of template.
Figure 3.
(a) Fluorescence emission scans from a typical PCR reaction containing 2.5 × 106 copies of the template and 25.6 nM 3-MI containing PCR probe (solid line) and from a negative control containing no copies of the template (dashed line), representing the background fluorescence of the single-stranded 3-MI-containing PCR probe. Samples were measured at 25°C. (b) Results of increasing template copy number as reflected by increases in fluorescence intensity in the post-amplification PCR reaction mixture containing the 3-MI-containing probe at a concentration of 0.2 µM. Negative controls containing all components except for template were subtracted from each point to correct for fluorescence from unannealed probe. Data shown are from averaged values of a duplicate set.
DISCUSSION
Bulge formation
Theoretically, the degree to which the absolute fluorescence intensity could potentially change in a given sequence as a result of bulge formation is directly related to the quantum yields of the 3-MI monomer (0.88 relative to quinine sulfate) (1) and the 3-MI-containing single strand. In sequences with 3-MI in a purine environment (that of most high yield bulge probes), the relative quantum yields have been found to range between 0.03 and 0.05 (3). So in a bulge-forming duplex, if most of the quenching associated with neighboring bases is removed, then a realistic increase in fluorescence would be ∼20-fold. There may be sequences in which the initial quenching is greater, which can yield a larger fold increase. A relatively modest increase in fluorescence intensity is depicted in Figure 2, in which the counts per second increase from 1500 to 15 000 for that particular sequence. In most of the 3-MI-containing oligonucleotide sequences shown in Table 1, an increase in fluorescence intensity is observed when 3-MI is forced into a bulge by annealing to a complementary strand that does not contain a base pairing partner for 3-MI. However, the degree to which the fluorescence intensity increases with bulge formation can vary greatly among oligonucleotides with similar sequences. For example, sequences, HP6, RD11, RD4, SK19 and JS-6 (Table 1) are similar in having adenosines on both sides of the probe, but the increases in fluorescence intensity on bulge formation varied from 9- to 27-fold. Studies on the impact of unpaired nucleotides on duplex formation by Morden et al. (12) have confirmed that duplex structure is determined by a delicate balance of many forces, including the nature of the base that is in the unpaired site. Conditions for promoting annealing of complementary strands are complex and in the final analysis a sequence should be tested under the conditions of the experiment. The hybridization conditions chosen for this study include low NaCl concentrations (10 mM) in order to minimize the addition of excess salt to the duplex. Often these probes are used in the presence of enzymes and biological systems in which high NaCl could be a problem. For this reason it was important to characterize the technique under low salt conditions.
In general, however, a 3-MI probe that is bordered by two consecutive adenosines on each side has the potential to yield a substantial signal increase upon annealing. Conversely, sequence HPR, in which the sequence around the probe is 5′-acFag-3′, displays a decrease in fluorescence intensity upon annealing. In the shorter sequences, RD1 and PCR4, the results could be affected by the shorter strand length. Variations in bulge formation and subsequent increases in fluorescence may offer some insights into the sequence-dependent flexibility of DNA.
Oligonucleotide sequences designed to create bulges that contain two or three bases resulted in more modest increases in fluorescence intensity than an identical sequence with only a single base (3-MI) in the bulge. These data suggest that some base stacking interactions may be maintained within a two or three base bulge. The structural conformations of one, two and three base bulges have been studied extensively (13–16), with the finding that the base stacking interactions occurring within these bulges vary considerably depending on the bases involved. The effect that 3-MI will have on these interactions is not yet known.
The dependence of bulge formation and fluorescence intensity changes on the length of oligonucleotide sequences is probably directly related to the general conditions for annealing. Factors known to affect hybridization, such as temperature, reaction buffer, overall sequence (GC content) and salt concentration, will influence the optimal length of strand that will yield the greatest increase in fluorescence intensity. This experiment was based on a single specific sequence and it is possible that other sequences may display different results. The distance between the probe and the end of a sequence will likely also contribute to the result. The optimum conditions for a particular sequence should be determined experimentally.
The presence of 3-MI as a bulge in a double strand appears to have minimal effects on the stability of the double strand. Each 3-MI bulge depresses the Tm by between 1 and 2°C per 3-MI insertion in the sequences tested (Table 2). We reported previously that the effect of 3-MI on Tm in double strands where it is paired to cytidine is approximately equivalent to a single base pair mismatch (∼10°C depression) (1). So the presence of 3-MI as a bulge in a double strand is quite a bit less disruptive than the presence of 3-MI as a complementary base in a double strand.
Detection of PCR products
For PCR application, we selected a primer pair (SK38 and SK39) that is commonly used for HIV-1 detection, targeting a highly conserved region of the HIV-1 gag gene (9). Within the 115 bp product defined by these primers several likely target sequences were identified and tested for fluorescence yield when annealed to 3-MI bulge-forming sequences. The strand that yielded the greatest fluorescence increase contained two 3-MI molecules in a sequence 35 bases long (see Materials and Methods). Using this probe, we detected up to 4-fold increases in fluorescence intensity in the positive PCR samples as compared to negative controls containing no template. Figure 3a displays results from a typical PCR reaction. The blank represented by the dotted line reveals a shoulder at ∼400 nm that represents scattered excitation, an interference often seen in dilute samples, referred to as ‘Raman scatter’. At concentrations of probe exceeding 0.20 µM, the concentration used for the primers, the fluorescence intensity increase was less and multiple products were generated, as shown by agarose gel analysis. We assume that this is due to the probe-containing strand competing with the primers and functioning as a primer itself. The sensitivity of the fluorophore to increasing concentrations of template is demonstrated in Figure 3b. The selectivity of this technique is an improvement over analysis of products based on their migration through gels. The most sensitive and accurate way to quantify PCR amplification is through monitoring the reaction rates in real time through changes in fluorescence, as is done with molecular beacons (17). Using molecular beacons, as few as 10 copies of target have been detected in PCR analyses. Conditions used to achieve this result included much higher concentrations of probe (0.14–0.50 µM as compared to the 6.4–290 nM concentrations that we used). Also at the limits of detection, these authors amplified the samples for between 35 and 40 cycles while we have used only 30 cycles (18). We were unable to monitor samples in real time because hardware designed to scan PCR samples during amplification is not (as yet) equipped with the UV light source required for excitation at 350 nm (19). There are also factors inherent to the PCR amplification process making it difficult to quantitate products. These issues stem from the fact that amplification is an exponential process and consequently even small differences in any of the variables can significantly change the products. We have controlled for this as much as possible by using a master mix containing all components of the reaction except for template, however, even small differences in temperature within the heat block can lead to variation (11). We have not attempted to improve on the sensitivity of the 3-MI bulge hybridization (shown to be 25 000 copies in Fig. 3b), but it is likely that by increasing concentrations of probe, amplifying for more cycles and monitoring in real time using a laser equipped instrument, the sensitivity would be significantly improved.
The PCR application is only one example of how these bulge hybridization probes might be used. Other uses might explore the kinetics of hybridization where the hairpins used with molecular beacons would likely cause significant decreases in probe affinity and hybridization kinetics (20).
SUMMARY
A sequence with the potential to yield the best signal change (and a low background fluorescence) would contain one or two single base bulges of 3-MI surrounded by two adenosines on each side, in a sequence that is at least 21 bases long. The bulge-forming application is very specific, concentration dependent and should be useful for identification and quantification of sequences in a mixture. Unbound sequences are relatively ‘silent’ because the probe is quenched until it is bulged out of the sequence in the annealing process. The fact that only one probe is needed to achieve this result on annealing and that the highly stable pteridine probe, 3-MI, can be incorporated site specifically into oligonucleotides through automated DNA synthesis make this technique very attractive.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to express our appreciation to Dr Hiraoki Mitsuya for the plasmid pSUM9 and to Dr Shizuko Sei for assistance with the PCR conditions.
REFERENCES
- 1.Hawkins M.E., Pfleiderer,W., Mazumder,A., Pommier,Y.G. and Balis,F.M. (1995) Incorporation of a fluorescent guanosine analog into oligonucleotides and its application to a real time assay for the HIV-1 integrase 3′-processing reaction. Nucleic Acids Res., 23, 2872–2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wojtuszewski K., Hawkins,M., Cole,J.L. and Mukerji,I. (2001) HU binding to DNA: evidence for multiple complex formation and DNA bending. Biochemistry, 40, 2588–2598. [DOI] [PubMed] [Google Scholar]
- 3.Hawkins M.E., Pfleiderer,W., Balis,F.M., Porter,D. and Knutson,J.R. (1997) Fluorescence properties of pteridine nucleoside analogs as monomers and incorporated into oligonucleotides. Anal. Biochem., 244, 86–95. [DOI] [PubMed] [Google Scholar]
- 4.Hawkins M.E. (2003) Fluorescent nucleoside analogues as DNA probes. In Lakowicz,J.R. (ed.), DNA Technology. Kluwer Academic/Plenum Publishers, New York, NY, Vol. 7, pp. 151–175. [Google Scholar]
- 5.Rachofsky E.L., Sowers,L.C., Hawkins,M.E., Balis,F.M., Laws,W.R. and Ross,J.B.A. (1998) Emission kinetics of fluorescent nucleoside analogs. SPIE, 3256, 68–75. [Google Scholar]
- 6.Seibert E., Chin,A.S., Pfleiderer,W., Hawkins,M.E., Laws,W.R., Osman,R. and Ross,J.B.A. (2003) pH-dependent spectroscopy and electronic structure of the guanine analogue 6,8-dimethylisoxanthopterin. J. Phys. Chem. A, 107, 178–185. [Google Scholar]
- 7.Deprez E., Tauc,P., Leh,H., Mouscadet,J.-F., Auclair,C., Hawkins,M.E. and Brochon,J.-C. (2001) DNA binding induces dissociation of the multimeric form of HIV-1 integrase: a time-resolved fluorescence anisotropy study. Proc. Natl Acad. Sci. USA, 98, 10090–10095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Driscoll S.L., Hawkins,M.E., Balis,F.M., Pfleiderer,W. and Laws,W.R. (1997) Fluorescence properties of a new guanosine analog incorporated into small oligonucleotides. Biophys. J., 73, 3277–3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ou C.Y., Kwok,S., Mitchell,S.W., Mack,D.H., Sninsky,J.J., Krebs,J.W., Feorino,P., Warfield,D. and Schochetman,G. (1988) DNA amplification for direct detection of HIV-1 in DNA of peripheral blood mononuclear cells. Science, 239, 295–297. [DOI] [PubMed] [Google Scholar]
- 10.Tanaka M., Srinivas,R.V., Ueno,T., Kavlick,M.F., Hui,F.K., Fridland,A., Driscoll,J.S. and Mitsuya,H. (1997) In vitro induction of human immunodeficiency virus type 1 variants resistant to 2′-beta-fluoro-2′,3′-dideoxyadenosine. Antimicrob. Agents Chemother., 41, 1313–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilliland G., Perrin,S. and Bunn,H.F. (1990) Competitive PCR for quantitation of mRNA. In Innis,M., Gelfand,D., Sninsky,J. and White,T. (eds), PCR Protocols: A Guide to Methods and Applications. Academic Press, San Diego, CA, pp. 60–61. [Google Scholar]
- 12.Morden K.M., Gunn,B.M. and Maskos,K. (1990) NMR studies of a deoxyribodecanucleotide containing an extrahelical thymidine surrounded by an oligo(dA)-oligo(dT) tract. Biochemistry, 29, 8835–8845. [DOI] [PubMed] [Google Scholar]
- 13.Rosen M.A., Live,D. and Patel,D.J. (1992) Comparative NMR study of An-bulge loops in DNA duplexes: intrahelical stacking of A, A-A and A-A-A bulge loops. Biochemistry, 31, 4004–4014. [DOI] [PubMed] [Google Scholar]
- 14.Rosen M.A., Shapiro,L. and Patel,D.J. (1992) Solution structure of a trinucleotide A-T-A lbulge loop within a DNA duplex. Biochemistry, 31, 4015–4026. [DOI] [PubMed] [Google Scholar]
- 15.Kalnik M.W., Norman,D.G., Zagorski,M.G., Swann,P.F. and Patel,D.J. (1989) Conformational transitions in cytidine bulge-containing deoxytridecanucleotide duplexes: extra cytidine equilibrates between looped out (low temperature) and stacked (elevated temperature) conformations in solution. Biochemistry, 28, 294–303. [DOI] [PubMed] [Google Scholar]
- 16.Kalnik M.W., Norman,D.G., Li,B.F., Swann,P.F. and Patel,D.J. (1990) Conformational transitions in thymidine bulge-containing deoxytridecanucleotide duplexes. J. Biol. Chem., 265, 636–647. [PubMed] [Google Scholar]
- 17.Tyagi S. and Kramer,R.R. (1996) Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol., 14, 303–308. [DOI] [PubMed] [Google Scholar]
- 18.Vet J., Majithia,A.R., Marras,S., Tyagi,S., Dube,S., Poiesz,B.J. and Kramer,F.R. (1999) Multiplex detection of four pathogenic retroviruses using molecular beacons. Proc. Natl Acad. Sci. USA, 96, 6394–6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeSilva D., Reiser,A., Herrmann,M., Tabiti,K. and Wittwer,C. (1998) Rapid genotyping and quantification on the LightCycler with hybridization probes. Biochemica, 2, 12–15. [Google Scholar]
- 20.Kushon S.A., Jordan,J.P., Seifert,J.L., Nielsen,H., Nielsen,P.E. and Armitage,B.A. (2001) Effect of secondary structure on the thermodynamics and kinetics of PNA hybridization to DNA hairpins. J. Am. Chem. Soc., 123, 10805–10813. [DOI] [PubMed] [Google Scholar]