Abstract
EGFR targeted monoclonal antibodies are effective in a subset of metastatic colorectal tumors (mCRC). Inevitably, all patients develop resistance, which occurs through emergence of KRAS mutations in approximately 50% of the cases. We show that amplification of the MET proto-oncogene is associated with acquired resistance in patients who do not develop KRAS mutations during anti-EGFR therapy. Amplification of the MET locus was present in circulating tumor DNA before relapse was clinically evident. Functional studies demonstrate that MET activation confers resistance to anti-EGFR therapy both in vitro and in vivo. Notably, in patient-derived CRC xenografts, MET amplification correlated with resistance to EGFR blockade which could be overcome by MET kinase inhibitors. These results highlight the role of MET in mediating primary and secondary resistance to anti-EGFR therapies in CRC and encourage the use of MET inhibitors in patients displaying resistance as a result of MET amplification.
Keywords: EGFR, MET, cetuximab, Acquired resistance, Anti-EGFR therapy, Met tyrosine kinase receptor, Colorectal cancer
Introduction
Drugs targeting the Epidermal Growth Factor Receptor (EGFR) — antibodies binding the extracellular domain and small-molecule tyrosine kinase inhibitors — have expanded treatment options for several solid tumors (1). The EGFR targeted monoclonal antibodies cetuximab and panitumumab have been extensively studied in metastatic colorectal cancer (mCRC), whereas tyrosine kinase inhibitors have thus far shown weak or no activity in this setting (2–4). Cetuximab or panitumumab appear to have similar therapeutic efficacy, achieving fairly modest but clinically meaningful objective ~10% response rates when used as monotherapy in genetically unselected patients with chemotherapy-refractory, EGFR-expressing mCRC (5, 6).
It has been clearly established that KRAS mutational status is the key predictor of tumor suitability for anti-EGFR therapy (7, 8). As KRAS is a downstream component of the EGFR signaling pathway, cells with mutant KRAS do not respond to anti-EGFR therapies. BRAF mutations, which are mutually exclusive with KRAS, have also been associated to lack of response to cetuximab and panitumumab (9). Deregulation of other effectors of the EGFR signaling cascade (PIK3CA, PTEN, NRAS) or of EGFR modulators (HER2, EGFR ligands) is also thought to affect primary response to EGFR blockade (10–12). Altogether, these primary mechanisms of resistance account for 70–80% of the cases unresponsive to anti-EGFR therapies, suggesting that there might be additional, yet undiscovered, biomarkers of resistance to these agents.
Importantly, the clinical efficacy of EGFR targeted antibodies is limited by the development of acquired (secondary) resistance, which typically occurs within 3–12 months since starting of therapy (5, 6). Further therapeutic options for these patients are very limited. Understanding the molecular bases of relapse to EGFR blockade in CRC is therefore clearly relevant to develop novel therapeutic strategies.
Multiple mechanisms of secondary resistance to anti-EGFR antibodies have been reported, such as expression of EGFR ligands, HER2 amplification and deregulation of the EGFR recycling process (12–16). We recently discovered that secondary KRAS mutations arise and are responsible for acquired resistance in approximately 50% of the patients who initially respond to cetuximab or panitumumab (17, 18). KRAS mutant alleles can be detected in patients’ blood using highly sensitive circulating tumor DNA analysis methods before disease progression is clinically manifest (17, 18). In the present work, we have studied the molecular bases of relapse in those patients who do not develop KRAS mutations during the course of anti-EGFR therapy.
Results
MET amplification is associated to acquired resistance to cetuximab or panitumumab in mCRC patients
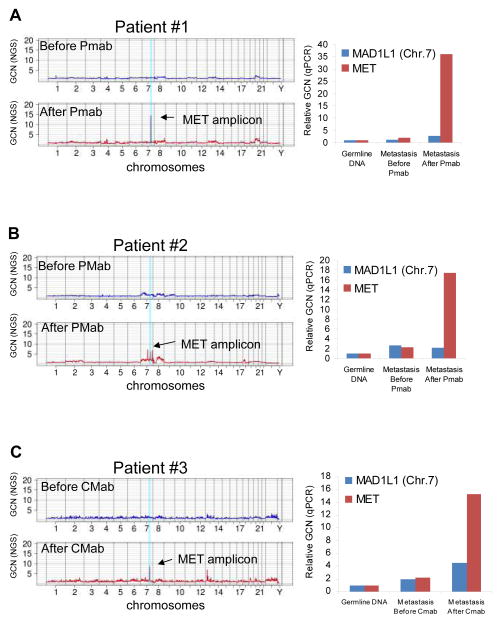
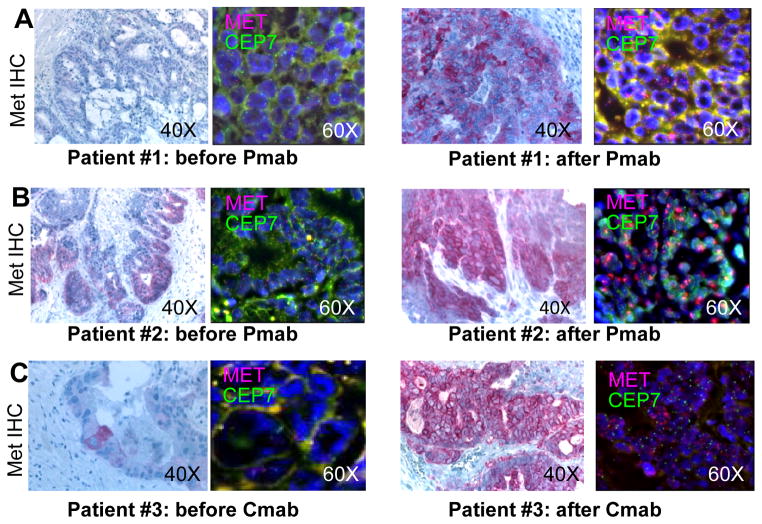
We analyzed seven CRC patients who initially responded to panitumumab or cetuximab-based treatment and then relapsed (Table 1). Of these, four did not display KRAS mutations in plasma samples analyzed by the highly sensitive BEAMing technique (18). For three of these patients (#1, #2, #3, Table 1) tumor tissue – pre and post anti-EGFR therapy- was available through surgical or bioptic procedures. Genomic DNA extracted from these cases was subjected to exome sequencing and next-generation Digital Karyotyping analyses with the aim of identifying sequence and copy number alterations present only in the post-relapse tissue. In all three cases, in the tissue obtained after anti-EGFR treatment, we detected amplification of a genomic fragment encompassing the MET gene, encoding the tyrosine kinase receptor for Hepatocyte Growth Factor. Quantitative PCR analysis confirmed the presence of MET amplification in the post-therapy samples but not in the matched pre-treatment tissues (Fig. 1). The absence of KRAS mutations was verified in both pre and post tissues, thus confirming the analyses performed in blood (data not shown). Mutations in other genes known to be involved in EGFR signaling (such BRAF, NRAS and PIK3CA) were also excluded by next-generation sequencing (data not shown). FISH analysis confirmed MET amplification (see methods for details) in the samples of patients #1, #2 and #3 obtained at relapse (Fig. 2). FISH analysis showed that MET was not amplified in the tumor tissue obtained before anti-EGFR treatment for patients #1 and #2 (Figs. 2A, 2B); however, it revealed the presence of rare MET amplified cells in the sample from patient #3 obtained before treatment with cetuximab (Fig. 2C). At least in this instance, we can therefore hypothesize that EGFR targeted therapies acted as a selective pressure to expand a pre-existing minor subclonal population of cancer cells carrying MET amplification. Immunohistochemistry (IHC) was then employed to assess whether MET amplification translated into overexpression of the MET receptor. Stronger MET immunostaining was present in the post relapse compared to the pre-relapse tissue (Fig. 2). In an additional patient (#4), where exome analyses could not be performed due to the low amount of material retrieved by the bioptic procedure upon relapse, we were able to exclude the presence of genetic alterations in genes previously implicated with primary resistance to anti-EGFR therapies (KRAS, BRAF, NRAS, HRAS, PIK3CA, EGFR, HER2; data not shown). Since in patient #4 FISH and IHC ruled out MET amplification or overexpression (data not shown), the mechanisms of acquired resistance to anti EGFR therapy remains to be elucidated. Finally, IHC showed that the levels of MET expression were low or undetectable in the post relapse tissue samples of patients #5, #6 and #7 that displayed KRAS mutations (Supplementary Fig. S1).
Table 1.
Patients’ clinical characteristics.
| Patient ID | Treatment | Duration of treatment | Pre-EGFR therapy sample | Post-EGFR therapy sample | BEAMing on Post-EGFR cfDNA (# mutant alleles/total) |
|---|---|---|---|---|---|
| #1 | Panitumumab | 12 months (08-2010 to 08-2011) | Para-aortic lymphnodes: KRAS wt, MET wt | Liver metastasis: MET ampl, KRAS wt | Plasma ctDNA: KRAS wt (0/16900 events) |
| #2 | Panitumumab | 13 months (03-2010 to 04-2011) | Liver metastasis: KRAS wt, MET wt | Liver metastasis: MET ampl, KRAS wt | Plasma ctDNA: KRAS wt (0/126700 events) |
| #3 | Cetuximab+ irinotecan | 12 months (02-2011 to 02-2012†) | Liver metastasis: KRAS wt, MET wt* | Lung metastasis: MET ampl, KRAS wt | Plasma ctDNA: KRAS wt (0/114600 events) |
| #4 | Cetuximab+ irinotecan | 6 months (03-2011 to 09-2011) | Abdominal lymphnode: KRAS wt, MET n.a. | Liver metastasis: KRAS wt, MET wt | Plasma ctDNA not available§ |
| #5 | Cetuximab+ irinotecan | 22 months (05-2009 to 03-2011) | Colon: KRAS wt, MET wt | Liver metastasis: KRAS p.G13D, MET wt | Plasma ctDNA: KRAS p.G13D (1355/33300 events) |
| #6 | Panitumumab | 6 months (12-2009 to 06-2010) | Colon: KRAS wt, MET wt | Liver metastasis: KRAS p.G12V, MET wt | Plasma ctDNA KRAS p.G12V (37/5500 events) |
| #7 | Panitumumab | 8 months (01-2010 to 09-2010) | Colon: KRAS wt, MET wt | Liver metastasis: KRAS p.G13D, MET wt | Plasma ctDNA KRAS p.G13D (27/9400 events) |
Patient #3 received cetuximab monotherapy from May 2011 to February 2012 (after receiving cetuximab+irinotecan from February to May 2011).
The pre-existence of single and rare MET amplified cells was observed by FISH analysis in the pre-cetuximab sample from Patient #3.
KRAS was found to be wild-type in the DNA from the liver metastatic biopsy obtained after anti-EGFR treatment (0/165000 events).
Figure 1. Whole exome analysis reveals increased MET copy number in CRC samples from patients who developed resistance to anti-EGFR treatment.
A–C left side. Whole exome gene copy number analysis of colorectal tumor samples from three patients taken before (in blue) and after (in red) therapy with the EGFR targeted monoclonal antibodies panitumumab (Pmab, patients 1 and 2) or cetuximab (Cmab, patient 3). Individual chromosomes are indicated on the x-axis. The lines indicate the sequencing depth as copy number values relative to a diploid exome (y-axis) over windows of 500,000 base pairs. A–C right side MET amplification was confirmed in the paired tumor samples by quantitative PCR gene copy number analysis. GCN= Gene Copy Number; MAD1L1: reference gene on Chr.7; NGS= next generation sequencing.
Figure 2. MET amplification is selected for in KRAS wild-type CRC samples from patients who developed resistance to anti-EGFR treatment.
FISH analysis showing amplification of the MET gene in formalin-fixed paraffin embedded tissue sections from three CRC patients (#1, #2 and #3, A–B–C) who developed resistance to therapy with the anti-EGFR monoclonal antibodies panitumumab or cetuximab. MET (7q31) specific probe is labeled with Texas red, while chromosome 7 centromeric probe D7Z1 (7p11.1-q11.1) is marked in green. Immunohistochemical staining for MET is shown in the left side of each panel. FISH, Original magnification 60x. IHC, original magnification 40x. Pmab= Panitumumab; Cmab= Cetuximab.
Non-invasive monitoring of MET amplification in blood samples
We reasoned that a genetic-based strategy to specifically detect MET gene amplification would allow us to assess whether this event was already present in a subset of the tumor cells before anti-EGFR therapy was initiated. In principle, such a biomarker could also be used to non-invasively monitor the emergence of MET-driven secondary resistance to anti-EGFR therapy in circulating tumor DNA from blood samples.
We initially used real time PCR to detect increased gene copy number in DNA extracted from plasma of patients with or without amplification of the MET gene. Although this approach could readily detect MET gene amplification in DNA extracted from cancer cell lines or tissues (Fig. 1), it was not successful using plasma-derived DNA. We hypothesized that this was due to the limited specificity and sensitivity of this approach when applied to circulating cell-free DNA, which is composed of a mixture of tumor and normal nucleic acids. Indeed, while somatic point mutations (such as those in KRAS) are tumor-specific, increased dosage of wild type loci is highly affected by the concomitant presence of the circulating normal DNA. We reasoned that genetic events occurring concomitantly with intra or extra chromosomal DNA amplification (19, 20) might be exploited as unique (tumor specific) genetic identifiers. As these molecular events often occur within non coding regions, we extended the exome analysis by performing whole genome sequencing on the post-relapse tumor.
As a test case, we analyzed patient #2 for whom tumor tissue obtained before and after anti-EGFR therapy as well as a longitudinal collection of blood samples obtained while on therapy and at relapse were available. Using an algorithm designed to identify amplification-associated chromosomal rearrangements (20–22) we retrieved sequencing reads encompassing two non-contiguous loci (surrounding the amplified region, see Supplementary Fig. S2).
Next, PCR primers were designed to detect the presence of this rearrangement (which represents the genomic breakpoint associated with the amplification of the MET gene) as an 89bp tumor-specific PCR product. We also designed a control assay that encompassed the same locus generating a 124bp product that was not affected by the presence of MET amplification (Supplementary Fig S2).
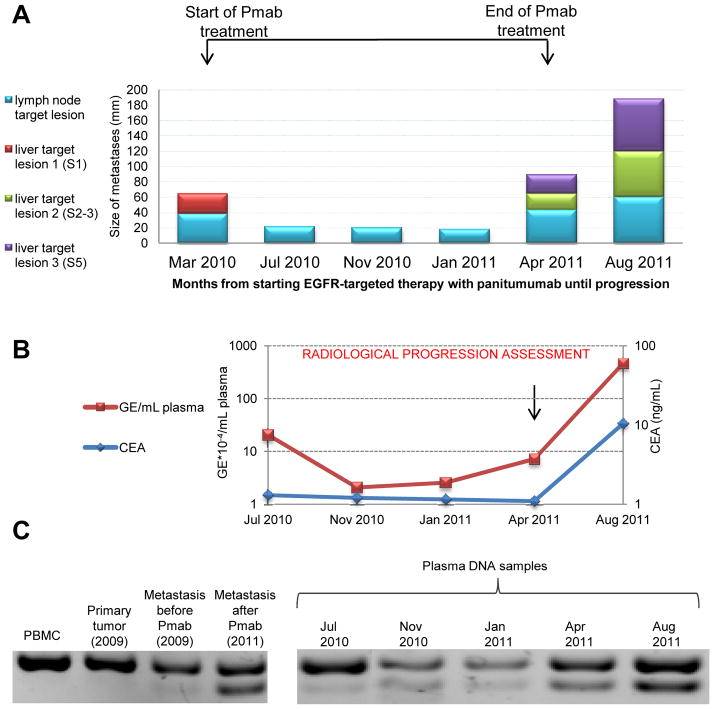
The 89bp product specific to the MET amplified rearrangement was not present in control DNAs obtained from a CRC cell line and in the germline DNA (PBMC derived) of patient #2, but it was detected at high levels in the post relapse tissue (Figure 3). Interestingly, very low levels of the MET amplification-associated chromosomal rearrangement were already present in the surgical specimen obtained before initiation of the anti-EGFR therapy (Figure 3). Using the same approach, we analyzed DNA extracted from plasma samples obtained at 3-month intervals from the initiation of EGFR therapy. The MET amplification-specific product was evident in the blood of this patient before relapse as determined by CT scan, suggesting that detection of MET amplification may provide a highly sensitive method for monitoring molecular resistance and recurrence in this setting. The same approach was then applied to patient #2. Also in this case a genomic breakpoint associated with the amplification of the MET gene could be identified by whole genome analysis of post relapse tissue (Figure S3). PCR primers were then designed and used to detect a MET amplification-associated chromosomal rearrangement in the post therapy samples (tissue and plasma) of patient #3 (Figure S3, lower panel).
Figure 3. Monitoring MET amplification in circulating tumor DNA during anti-EGFR therapy.
A. Size of liver metastases at segment 1 (S1, red), segment 2–3 (S2–S3, green) and segment 5 (S5, violet) and of lymph node target lesions at the hepatoduodenal ligament (cyan) during panitumumab therapy in patient #2 at the indicated time points, showing response to panitumumab (Pmab) followed by progression. B. Carcinoembryonic antigen (CEA) levels (blue line) and number of Genome Equivalent (GE, red line) obtained from 1 mL of plasma from patient #2, as assessed by Real Time PCR. C. DNA electrophoresis of PCR products using primers designed to detect the presence of the MET associated amplified rearrangement on Chromosome 7. The lower band corresponds to an 89 bp tumor-specific PCR product which is positive only when the re-arrangement is present. A control assay detecting the wild-type locus generated amplicon of 124 bp (upper band) is also shown.
MET amplification is associated with primary resistance to cetuximab in CRC patient-derived tumor xenografts (‘exenopatients’)
KRAS mutations drive both primary and secondary (acquired) resistance to cetuximab and panitumumab. We reasoned that MET gene amplification could also be responsible for primary resistance to EGFR targeted antibodies in CRC. To assess this hypothesis we took advantage of a large collection of patient-derived CRC liver metastasis xenografts (‘xenopatients’), which we previously annotated for their molecular profile and sensitivity to cetuximab (11). The mCRC xenopatients responded to cetuximab with rates and extents analogous to those observed in the clinic: approximately 10% of the cases displayed partial response, 30% had disease stabilization, while 60% progressed on anti-EGFR therapy (11). We previously reported that, in agreement to what found in the clinical setting, mutations in KRAS, BRAF, NRAS or PIK3CA and amplification of HER2 are associated with resistance to cetuximab in CRC xenopatients (10–12). Overall, these biomarkers account for most but not all of the samples in which the anti-EGFR therapy was ineffective. We reasoned that MET amplification could be responsible for some of these unexplained samples and would be mutually exclusive with the other genetic lesions.
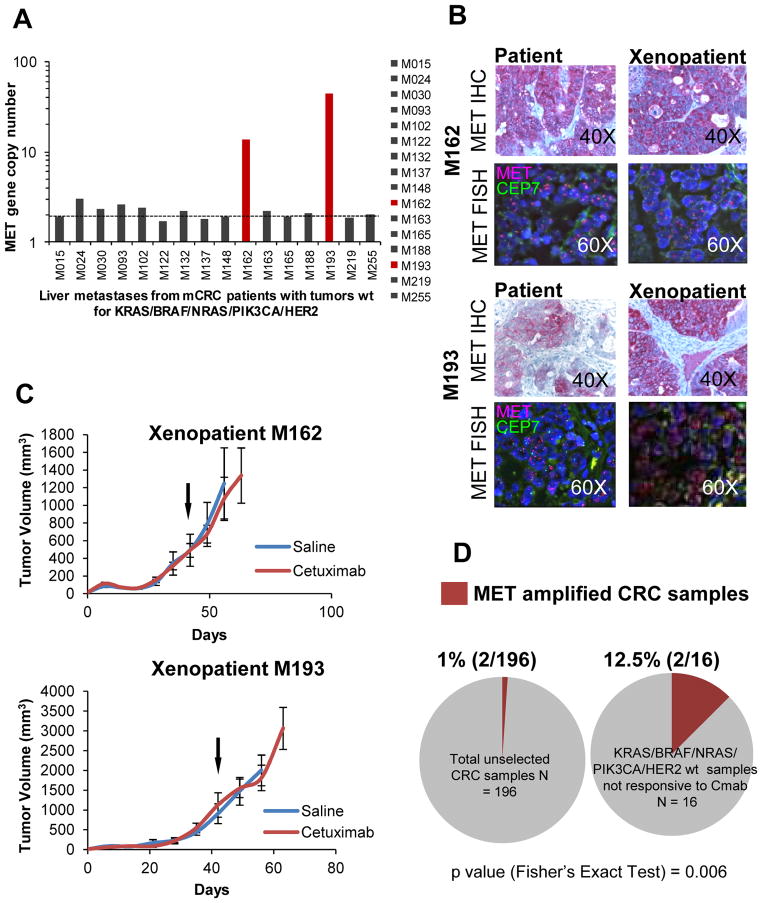
By extending the analyses previously performed on a subset of samples (23), we assessed the MET gene copy status in the entire xenopatient cohort and found that 1% (2/196) of cases carried MET amplification, as determined by real time PCR analysis (Fig 4A). Importantly, amplification of the MET gene and overexpression of the MET protein were confirmed by FISH and IHC, respectively, in both xenografted tumors and their original human counterparts (Fig. 4B). Notably, none of the mCRC patients from whom the ‘xenopatients’ were derived had been exposed to cetuximab or panitumumab, ruling out the possibility that MET amplification could have been positively selected by previous anti-EGFR therapy. Interestingly, treatment with cetuximab was ineffective in mice engrafted with CRC metastatic specimens carrying MET amplification (Fig. 4C). Indeed, the MET amplified cases segregated into the subpopulation of xenopatients resistant to cetuximab and wild type for KRAS, NRAS, BRAF, PIK3CA and HER2 (16 cases, Fig. 4D). Overall, these data indicate that MET amplification characterizes a significant fraction (2/16; 12.5%, Fig. 4D) of cetuximab-resistant cases that are wild-type for KRAS, BRAF, NRAS, PIK3CA and HER2, possibly identifying a new biologically distinct mCRC subpopulation (P= 0.006 by Fisher’s Exact test).
Figure 4. MET amplification is associated with lack of response to cetuximab in a series of CRC patient derived xenografts (‘exenopatients’).
A. Quantitative PCR gene copy number analysis of MET amplification in a series of cetuximab-resistant ‘xenopatients’, which did not carry genetic alterations in genes previously associated with resistance to anti-EGFR therapy (KRAS, BRAF, NRAS, PIK3CA, HER2). Dotted line indicates an estimated copy number of 2. B. FISH and IHC analysis of MET in CRC samples that showed increased MET copy number by qPCR analysis. Patients (left) and corresponding xenopatients (right) are shown. MET (7q31), red; D7Z1 chromosome 7 centromeric probe (7p11.1-q11.1), green. FISH, Original magnification 60x. IHC, original magnification 40x. C. Growth curves in mice cohorts derived from MET-amplified xenopatients, treated with placebo (blue) or cetuximab 20 mg/Kg i.p. twice a week (red). n = 6 for each treatment arm. Arrows indicate treatment start. D. Prevalence of MET amplification in unselected metastatic colorectal cancer samples, according to q-PCR experiments (left), and in cetuximab-resistant (Cmab), genetically selected (without genetic alterations in KRAS, BRAF, NRAS, PIK3CA, HER2) xenopatients (right). P values were calculated by Fisher’s Exact test.
MET activation drives resistance to EGFR inhibitors in preclinical CRC models
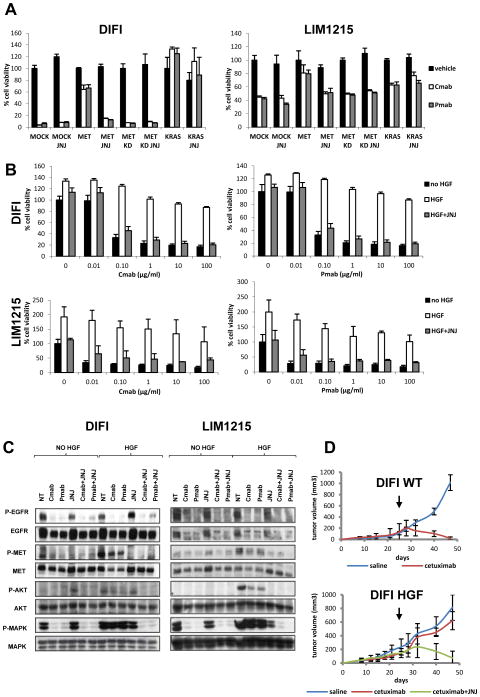
Data obtained in patients samples and in xenografted tumors suggest that amplification of the MET locus, sustaining overexpression of the Met receptor, mediates resistance to EGFR blockade in colorectal cancers. It has been previously established that overexpression constitutively activates the MET receptor and is an oncogenic event in multiple cancer types (24, 25). To formally assess whether MET overexpression alone is causally responsible for cetuximab or panitumumab resistance we performed in vitro and in vivo forward genetic experiments. As model systems we exploited two CRC cell lines, namely DiFi and LIM1215. DiFi cells over-express EGFR as a result of high-level amplification of the EGFR gene locus (26, 27). By contrast, LIM1215 cells express ‘normal’ EGFR levels but are still sensitive to cetuximab or panitumumab (28, 29). Both cell lines are wild type for KRAS, BRAF, NRAS and PIK3CA, paralleling the molecular features of the CRC patients most likely to respond to cetuximab. Ectopic overexpression of the MET receptor was achieved in both cell lines by means of lentiviral-mediated transduction of the corresponding cDNA. As control, transduction with KRAS or a kinase inactive version of MET (MET kinase dead) was employed (Supplementary Fig. S4). As shown in Fig. 5A, MET constitutive activation, due to cDNA transfection and consequent protein overexpression, conferred resistance to cetuximab or panitumumab in proliferation assays to a degree equivalent to that triggered by KRAS. Notably, the ability of wild type MET to drive resistance to anti-EGFR monoclonal antibodies was abolished by the concomitant treatment with the anti-MET inhibitor JNJ-38877605 (30), further confirming that MET-promoted intracellular signaling was driving resistance (Fig. 5A).
Figure 5. MET activation confers resistance to cetuximab or panitumumab in colon cancer cell lines in vitro and in vivo.
A. DIFI and LIM1215 cell lines were transduced with the following lentiviral vectors: MOCK (empty vector), MET, MET KD (kinase dead) or KRAS, then seeded in 96 wells costars and cultured 7 days in the presence of vehicle (NT), cetuximab (Cmab,1 μg /ml) or panitumumab (Pmab, 1 μg /ml), with or without the MET inhibitor JNJ38877605 (250 nM). B. DIFI and LIM1215 cells were treated for 1 week with increasing concentrations of cetuximab or panitumumab, with or without 20 ng/ml of HGF or HGF plus the MET inhibitor JNJ38877605 (250 nM). C. DIFI and LIM1215 cells were pretreated 24 hours with cetuximab (1 μg /ml), panitumumab (1 μg /ml) or JNJ38877605 (250nM), then stimulated for 15 minutes with HGF (80 ng/ml) in the presence of the indicated inhibitors. Whole-cell extracts were then subjected to western blot analysis and probed with the indicated antibodies. D. Wild-type (WT) DIFI cells or DIFI transduced with HGF were subcutaneously injected in NOD-SCID mice. Upon tumor growth, mice were randomized (7 mice per group) and received intra-tumor injection of saline/ cetuximab or intra-tumor injection of cetuximab and oral administration of JNJ38877605. Arrows indicate treatment start.
Another mechanism of MET mediated oncogenic activation is represented by autocrine or paracrine stimulation by its ligand, the Hepatocyte Growth Factor (HGF) (31–34). Additional experiments were therefore performed in DiFi and LIM1215 cells to assess whether HGF-induced MET activation could also endure resistance to cetuximab or panitumumab. The MET inhibitor JNJ-38877605 served as control. Indeed, paracrine activation of MET by HGF was sufficient to confer resistance to anti-EGFR antibodies (Fig. 5B).
These results suggest that HGF/MET-initiated signaling can bypass blockade of the EGFR by the monoclonal antibodies cetuximab and panitumumab. To get a mechanistic insight on how HGF-MET ligand receptor pair could overcome EGFR inhibition we performed biochemical experiments. We found that HGF-mediated MET activation counteracted the cetuximab and panitumumab induced inhibition of MAPK and, to a lesser extent, of AKT (Fig. 5C).
To corroborate the results obtained in vitro we performed in vivo experiments in mouse models. Stable, ectopic transduction of the MET receptor was not conductive in DiFi and LIM1215, as transduced cells lost expression of the MET transgene after few passages. On the other end, we found that long term HGF overexpression was feasible and resulted in constitutive activation of the HGF/MET signaling axis in DiFi cells. Parental and HGF-expressing DiFi cells were therefore subcutaneously injected in immunocompromised mice and allowed to form palpable tumors. At this point, animals received cetuximab for three weeks, which, as expected, profoundly inhibited growth of the parental cells but had minimal or no influence on tumors expressing the MET ligand HGF (Figs. 5D). Co-treatment with cetuximab and the MET inhibitor JNJ-38877605, instead, induced marked tumor regression.
These data suggest that colorectal tumors displaying constitutive activation of MET signaling triggered by either MET amplification or HGF-induced MET activation might be effectively targeted by MET inhibitors.
Patient-derived xenografts with MET amplification respond to MET inhibitors
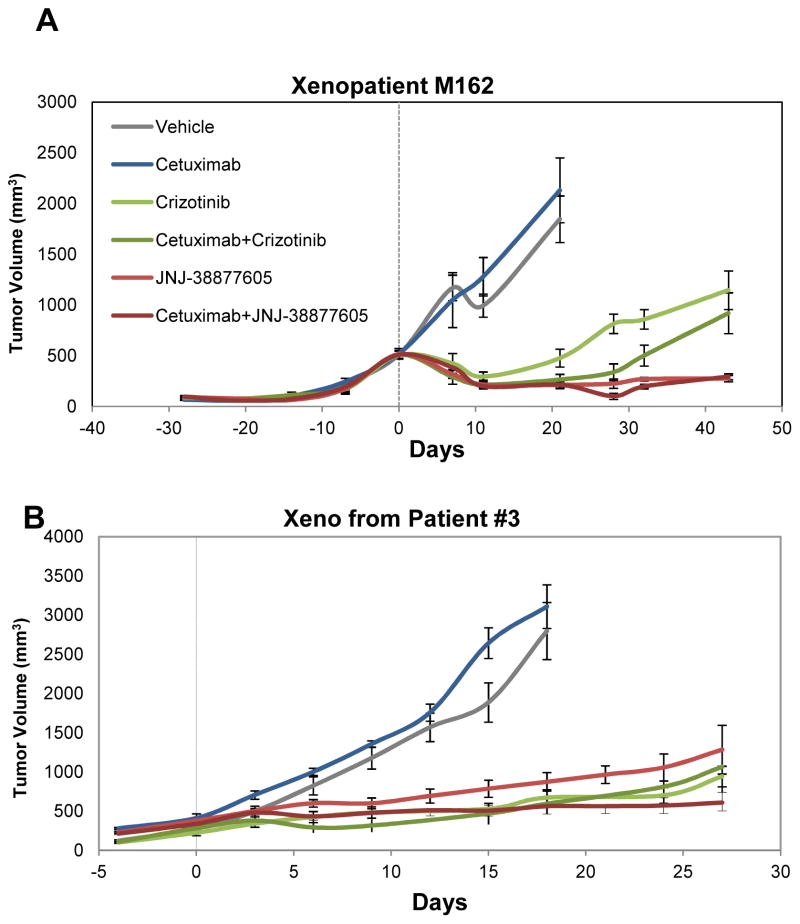
To ascertain the potential clinical relevance of the above described findings,, we decided to adapt the intervention trial executed in CRC cell-line xenografts to patient-derived xenografts, which represent a more reliable proxy of prospective findings in patients. In this pilot study, we employed one case of primary resistance (M162) and one model of secondary resistance (derived from the post treatment tumor tissue of patient #3). As for the choice of therapeutic regimens, we focused on small molecule inhibitors of MET that were administered individually or in combination with cetuximab. We selected JNJ-38877605, the MET-specific tool compound (not in clinical use) already employed in the xenograft experiments with cell lines; and crizotinib, a dual MET/ALK inhibitor that has shown promising anti-tumor activity in MET-amplified esophagogastric adenocarcinomas (35). For each case, the original tumor specimen was serially passaged in vivo. When xenografts reached an average volume of approximately 500 mm3, mice were randomized into six independent treatment cohorts; i) vehicle (placebo); ii) cetuximab alone; iii) JNJ-38877605 alone; iv) crizotinib alone; v) JNJ-38877605 and cetuximab; vi) crizotinib and cetuximab. In case M162 (primary resistance), crizotinib monotherapy produced initial shrinkage followed by slow resumption of tumor growth, which was substantially delayed by co-treatment with cetuximab (Fig. 6A). The efficacy of JNJ-38877605 was more pronounced, with long-lasting abolition of tumor growth even in the absence of cetuximab (Fig. 6A).
Figure 6. Patient-derived xenografts bearing MET amplification respond to MET inhibitors.
Tumor growth curves in MET-amplified xenopatients displaying de novo (A, patient M162 shown in Fig. 4) or secondary (B, patient #3 shown in Fig. 1) resistance to cetuximab. Mice were treated with the indicated modalities. Data are presented as means +/− SEM (error bars) of n=5 (A) or n=6 (B) animals for each treatment arm.
Response of patient #3 xenografts (a model of secondary resistance) to the same regimens was substantially analogous. In concordance with the results obtained in M162, all treatments potently delayed tumor growth. Different from M162, the anti-tumor activity of crizotinib was not enhanced by the addition of cetuximab, and the most effective modality in producing durable disease stabilization proved to be the JNJ-38877605-cetuximab combination (Fig. 6B).
Notwithstanding minor individual differences, these results overall indicate that interception of MET signaling leads to severe impairment of tumor growth in MET-amplified CRCs and provide proof-of-concept for the use of Met inhibitors, alone or in combination with anti-EGFR antibodies, as novel therapeutic opportunities to contrast MET-driven primary and secondary resistance in the clinic.
Discussion
Drugs directed against oncoproteins that sustain the growth of cancer cells have emerged as important therapeutic agents in the treatment of a variety of human malignancies. Among these, inhibitors of receptor tyrosine kinases (antibodies or small molecules) have shown marked clinical activity (1). Unfortunately, the overall value of these agents is substantially limited by the acquisition of drug resistance, which eventually arises in most, if not all, treated patients. Several hypotheses have been put forward to explain why resistance arises. One well-supported possibility, is that the lesions which respond to the treatment are genetically heterogeneous and already contain a large selection of molecular variants from which the drugs simply select those conferring resistance in a classical Darwinian fashion (reviewed in (36)).
In this work, we have studied the mechanisms of acquired resistance to cetuximab and panitumumab, two monoclonal antibodies that inhibit the signaling cascade initiated by the EGFR receptor and in the clinical setting ameliorate the survival of patients with mCRC. Similar to what happens with other targeted agents, mCRC patients whose tumors respond to EGFR targeted antibodies develop acquired resistance within 3–12 months (5, 6). We have recently reported that resistance in this setting can be driven by the selection of oncogenic mutations in the KRAS gene or, less frequently, amplification of the KRAS locus (18). Using a highly sensitive digital PCR approach (BEAMing), we were able to detect the emergence of KRAS mutations in patients’ blood months before relapse was evidenced by CT scans. Analysis of plasma samples from patients receiving panitumumab monotherapy likewise showed the emergence of oncogenic KRAS variants in approximately 40% of the cases (17). Together, these analyses suggest that in a large proportion of mCRC patients who respond and then become refractory to cetuximab or panitumumab resistance is caused by emergence of KRAS oncogenic alleles. At the same time they raise the question of which molecular mechanisms drive resistance in the remaining patients.
To tackle this, we used BEAMing to select patients in which KRAS mutations did not emerge during therapy. To investigate novel mechanisms of resistance, genomic DNA was extracted from three cases and subjected to exome sequencing alongside the corresponding pre-treatment neoplastic tissue. High levels of MET amplification were found at relapse. No other gene copy number variations were detected, and next-generation sequencing analysis confirmed that other candidate drivers of resistance (KRAS, NRAS, BRAF, PIK3CA) remained wild type. Post-relapse tissue was also analyzed by FISH, which confirmed MET amplification, and by IHC that showed high levels of MET protein expression.
The MET gene, encoding the tyrosine kinase receptor for Hepatocyte Growth Factor, has been shown to have an oncogenic role in several human tumors, where it becomes constitutively activated as a consequence of gene amplification, overexpression, activating mutations or autocrine stimulation (37). MET and its ligand HGF have been previously implicated in acquired resistance to targeted therapies. For example, MET amplification is found in approximately 5–20% of the EGFR mutated lung cancers that respond and then progress on erlotinib/gefitinib based therapies (38–40). Notably, MET amplification appears to arise in tumors with pre-existing clones of MET-amplified cells, which undergo positive selection during gefitinib and erlotinib therapy (41). The same appears to be true for patient #3 in our cohort, as FISH analyses identified the presence of rare MET amplified cells in the sample prior to cetuximab exposure.
If mCRC patients became resistant to anti-EGFR antibodies as a result of the emergence of MET amplification in their tumors, we expected that this genetic event could be detected in their circulation during the therapy. To assess this possibility we developed a PCR based assay to detect the presence of the MET amplicon in circulating, cell-free, DNA. The test was applied to longitudinal blood samples from patient #2 and showed that MET amplification was present as early as 3 months after initiation of therapy. As a blood draw prior to anti-EGFR therapy was not available for this patient, we could not assess the status of the MET amplicon in the plasma at baseline. Notably, however, when the same analysis was applied to the pre-treatment tissue, MET amplification was detected although at low level. These findings relative to patient #2, together with the IHC and FISH analyses performed on the pre-treatment tissue of patient #3, support the hypothesis that anti-EGFR therapy selects MET amplified (cetuximab and panitumumab resistant) pre-existing clones. During treatment, MET amplified cells would then become the leading population eventually limiting the efficacy of further anti-EGFR therapies. If confirmed in larger datasets, these results support the use of blood tests to monitor the emergence of MET amplification in patients undergoing anti-EGFR therapies. This approach may drive the early initiation of MET inhibitors in those patients who respond to cetuximab and panitumumab and do not display emergence of KRAS mutations in blood tests during anti-EGFR therapy.
The discovery of MET amplification as a mechanism of secondary resistance to cetuximab prompted us to hypothesize that the same genetic alteration might be responsible for de-novo resistance to EGFR targeted antibodies in CRCs. Indeed, we found that xenografted tumors (‘xenopatients’) carrying MET amplification did not respond to cetuximab and that this molecular alteration was mutually exclusive with mutations in KRAS, BRAF, NRAS, PIK3CA and with HER2 amplification. It should be noted that the prevalence of MET amplification in untreated mCRC was low - around 1% (2/196 samples) in our cohort, consistent with the frequency (1/193 cases) reported by the recent TCGA consortium (42), as well as in previous studies (43, 44). For this reason, as described for other low prevalence driver alterations (10, 45), the clinical validation of MET amplification as a biomarker of resistance to anti-EGFR therapy in mCRC will require very large retrospective (and possibly prospective) studies.
Cells of epithelial–endothelial origin widely express the MET receptor, which is essential for embryonic development and tissue repair. Hepatocyte growth factor, the ligand for the MET receptor, is expressed mainly in cells of mesenchymal origin, although some tumors appear to express both HGF and MET (25). Whether amplification-driven, MET overexpression is sufficient to fully activate its oncogenic properties remains a controversial matter. In general it appears that addition of HGF can further trigger MET-initiated signaling. Our functional analysis indeed indicates that HGF plays an important role in driving MET mediated resistance to anti-EGFR monoclonal antibodies in CRC cells. Furthermore, in agreement with recent studies (29, 46, 47), we found that HGF stimulation is sufficient to confer cetuximab and panitumumab resistance both in vitro and in vivo. These findings support the possibility that HGF overexpression by cancer cells or the surrounding stroma might be an independent mechanism of acquired (or primary) resistance to cetuximab. This could be particularly relevant in patients with hepatic metastasis as the liver is a known reservoir for HGF (48).
The most relevant aspect of our work is that MET (as opposed to KRAS) is an actionable target. Multiple agents, some of which already approved for clinical use, have been developed to target MET or HGF, including crizotinib, which was originally designed to inhibit MET and only later found to be active for patients with mutant ALK or ROS-1 (30, 49). HGF-directed antibodies are also being evaluated in clinical trials with some early encouraging results (30). The preclinical trial in which we treated two CRC xenopatients carrying MET amplification is encouraging. In both instances Met inhibitors (including the clinically approved drug crizotinib) were effective. Our results therefore support the initiation of clinical trials based on MET inhibitors in the subset of CRC patients with MET amplification (de novo or acquired). Importantly, one of the treated xenopatients was obtained from a biopsy (patient #3). This individual is currently alive, further underscoring the translational impact of these results.
Due to limited tissue availability, we have been unable to perform whole-genome analysis and identify putative mechanisms of resistance in one of the seven patients (patient #4) analyzed in this study. We confirmed the absence of MET gene amplification by FISH, and a candidate-biomarker approach ruled out the presence of mutations or amplifications in known EGFR pathway components (KRAS, BRAF, NRAS, HRAS, PIK3CA, EGFR, HER2). It is therefore likely that mechanisms other than KRAS or MET oncogenic alterations could be responsible for the emergence of resistance to EGFR antagonists in a fraction of mCRC cases. Based on our current finding we forecast that this additional mechanism will be based on the deregulation of genes involved in EGFR signaling that have been previously been associated to ‘de novo’ resistance to anti-EGFR therapies.
When considered together, KRAS mutations or MET amplification occur in the large majority of mCRC patients who initially respond and then relapse on anti-EGFR therapies and likely represent major mechanisms of acquired resistance to anti-EGFR therapies. Current evidence suggests that these two molecular alterations occur in a mutually exclusive fashion, thus defining independent patient populations that will likely require different therapeutic approaches once resistance ensues. Nevertheless, our findings do not rule out the possibility that within the same patient distinct metastatic lesions might evolve independent resistance mechanisms.
In conclusion, this work defines MET gene amplification as a novel mechanism of both primary and acquired resistance to cetuximab or panitumumab and identifies a patient population that could immediately benefit from the clinically available MET inhibitors.
Methods
Patients and tumor samples
We retrospectively analyzed 7 patients with histologically confirmed mCRC at Ospedale Niguarda Ca’ Granda (Milan, Italy). Tumor specimens were obtained through protocols approved by the Institutional Review Board of Ospedale Niguarda Ca’ Granda (Milan, Italy, protocols 1014/2009 and 194/2010). All tumor samples were formalin fixed paraffin embedded (FFPE). All patients provided informed consent and samples were procured and the study was conducted under the approval of the Review Boards and Ethical Committees of the Institutions. Patients evaluated in this study were selected based on evidence that treatment outcome could be attributable with the most likelihood to administration of either panitumumab or cetuximab (synergy with irinotecan should be taken into account for those patients treated with cetuximab in combination with irinotecan in the chemorefractory setting). Patients were enrolled in clinical trials or received panitumumab or cetuximab as per label indication. For those patients who were treated with cetuximab in combination with irinotecan, refractoriness to previous irinotecan-based regimens was documented as disease progression during, or within, 6 months of receiving the irinotecan-based regimen (administered for at least 6 weeks). Besides the above-mentioned inclusion criteria, the availability of tumor sample qualitatively and quantitatively suitable for molecular analyses was also a requirement for being considered in the present study. Clinical response was assessed every 6–8 weeks with radiological examination (computerized to modensitometry or magnetic resonance imaging). The Response Evaluation Criteria in Solid Tumors (RECIST) (50) were adopted for evaluation and objective tumor response was classified into partial response (PR), stable disease (SD) and progressive disease (PD). Patients with SD or PD were defined as non-responders (50). Two independent oncologists and radiologists verified in a blinded manner the clinical response for all patients.
Plasma samples collection
At least 4 mL of whole blood were collected by blood draw using EDTA as anticoagulant. Plasma was separated within 5 hours, through two different centrifugation steps (the first at room temperature for 10 min at 1600 g, and the second at 3000 g for the same time and temperature) obtaining 1 mL of plasma. Plasma was stored at −80 °C until DNA extraction.
DNA extraction
Tissue sections of 5 μm thickness were obtained from FFPE tissues and stained with hematoxylin. In order to enrich for malignant cells, neoplastic areas were macro/microdissected from tissue slides by scraping under microscopic guidance. Genomic DNA was extracted with QIAamp DNA FFPE tissue kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol and relative concentration was quantified using the Infinite 200 NanoQuant spectrophotometer (Tecan). Plasma was thawed at room temperature and centrifuged at 16000 g for 5 minutes, in order to remove any cell debris. DNA was extracted using the QIAamp Circulating Nucleic Acid Kit (QIAGEN), using columns with silica-based membrane, tube extenders and a vacuum pump, according to manufacturer’s instructions. The DNA was eluted in two steps with 140 μl RNase free water. DNA concentration was quantified with NanoDrop ND-1000 spectrophotometer (Thermo Scientific). The median concentration was 50 ng/μl, with 260/280 and 260/230 ratios around 2.70 and 0.45 respectively.
Cancer Exome and Genome Sequence Analyses and Digital Karyotyping
Library construction, exome capture, next generation sequencing, and bioinformatic analyses of tumor and normal samples were performed at Personal Genome Diagnostics (Baltimore, MD). In brief, genomic DNA from tumor and normal samples were fragmented and used for Illumina TruSeq library construction (Illumina, San Diego, CA). Exonic regions were captured in solution using the Agilent SureSelect 50 Mb kit (version 4) according to the manufacturer’s instructions (Agilent, Santa Clara, CA). Paired-end sequencing of both exome and genome libraries, resulting in 100 bases from each end of the fragments, was performed using a HiSeq 2000 Genome Analyzer (Illumina, San Diego, CA). The tags were aligned to the human genome reference sequence (hg18) using the Eland algorithm of CASAVA 1.7 software (Illumina, San Diego, CA). The chastity filter of the BaseCall software of Illumina was used to select sequence reads for subsequent analysis. The ELANDv2 algorithm of CASAVA 1.7 software (Illumina, San Diego, CA) was then applied to identify point mutations and small insertions and deletions. Known sequence polymorphisms recorded in dbSNP were removed from the analysis. Potential somatic mutations were filtered and visually inspected as described previously (22). Copy number analyses using next generation Digital Karyotyping and rearrangement analyses were performed as previously described (20–22).
Gene copy number analysis (NGSeq)
Reads’ depths for both samples were calculated filtering the samtools mpileup results for positions actually included in coding exons of RefSeq according to hg18. Averages over overlapping 500,000 bp wide windows were then computed plotted.
Real time PCR for circulating DNA quantification
The extracted circulating DNA was quantified with a Real Time PCR using Human LINE quantification. PCR is performed in a final volume of 10ul, containing 5μl of SYBR MIX (Promega), 0.72μl of 12.5 μM for each forward and reverse LINE primers (F primer : 5′-TCACTCAAAGCCGCTCAACTAC-3′ ; R primer: 5′-TCTGCCTTCATTTCGTTATGTACC-3′), 0.56 μl of water and 3μl of DNA. All reactions are performed in triplicates. Various dilutions of normal human DNA purified from human colonocytes were incorporated in each plate to serve as standards. The analysis of the results obtained by Real Time PCR is used to calculate the number of genome equivalents present in 1 mL of each sample of plasma.
End Point PCR
The amplification was performed in a 10 il PCR reaction, with 2× Phusion Flash PCR Master Mix (NEB, BioLabs) and 0.5 μM of each primer (F: 5′-ggaagagctatgaagcgtga-3′;R: 5′-cacatgctgagagttgaggtct-3′; LIC:5′-gtaaaacgacggccagtaagagctgggaatacaagca -3′). Amplification was carried out using the following cycling conditions: 98°C for 120 s; 3 cycles of 98 °C for 10 s, 69°C for 15 s, 72°C for 15 s; 3 cycles of 98°C for 10 s, 66°C for 15 s, 72°C for 15 s; 3 cycles of 98°C for 10 s, 63°C for 10 s, 72°C for 15 s; 41 cycles of 98°C for 10 s, 63°C for 15 s, 72°C for 15 s.
BEAMing assay
BEAMing was performed as described previously (18). The first amplification was performed in a 50 μl PCR reaction, 1× Phusion high-fidelity buffer, 1.5 U Hotstart Phusion polymerase (NEB, BioLabs), 0.5 μM of each primer with tag sequence, 0.2 mM of each deoxynucleoside triphosphate, and 0.5 mM MgCl2. Amplification was carried out using the following cycling conditions: 98°C for 45 s; 2 cycles of 98 °C for 10 s, 67°C for 10 s, 72°C for 10 s; 2 cycles of 98°C for 10 s, 64°C for 10 s, 72°C for 10 s; 2 cycles of 98°C for 10 s, 61°C for 10 s, 72°C for 10 s; 31 cycles of 98°C for 10 s, 58°C for 10 s, 72°C for 10 s. PCR products were diluted, and quantified using the PicoGreen double-stranded DNA assay (Invitrogen). A clonal bead population is generated performing emPCR. PCR mixture (150 μl) was prepared containing 18 pg template DNA, 40 U platinum Taq DNA polymerase (Invitrogen), 1× platinum buffer, 0.2 mM dNTPs, 5 mM MgCl2, 0.05 μM Tag1 (TCCCGCGAAATTAATACGAC), 8 μM Tag2 (GCTGGAGCTCTGCAGCTA) and 6×107 magnetic streptavidin beads (MyOne, Invitrogen) coated with Tag1 oligonucleotide (dual biotin-TSpacer18-TCCCGCGAAATTAATACGAC). The 150 μl PCR reactions were distributed into the wells of a 96-well PCR plate together with 70 μl of the emusifire oil. The water-in-oil emulsion was obtained by pipetting. The PCR cycling conditions were: 94 °C for 2 min; 50 cycles of 94 °C for 10 s, 58 °C for 15 s, 70 °C for 15 s. All primer sequences are available on request.
MET Immunohistochemistry
MET protein expression was evaluated by immunohistochemistry performed on 3 μm thick tissue sections using a specific MET antibody (Met (D1C2) XPR Rabbit mAB, Cell Signaling Technology, Inc.; dilution 1:1000) and the automated system BenchMark Ultra (Ventana Medical System, Inc., Roche), as previously described (18). Images were captured with the AxiovisionLe software (Zeiss, Gottingen, Germany) using an Axio Zeiss Imager 2 microscope (Zeiss, Gottingen, Germany). A consensus for classification of samples based on expression of MET protein is presently lacking. We therefore used a scoring system similar to the one applied for HER2 protein. More specifically, we classified samples as negative (0, 1+), when no staining or faint staining was present in less than 10% of cells; ambiguous (2+) when moderate staining was present in more than 10% of cells; positive (3+), when a circumferential, basolateral or lateral signal for MET overexpression of protein with strong intensity was present in more than 10% of the cells (Supplementary Figure S5).
MET Fluorescent in situ hybridisation (FISH) analysis
Dual color FISH analysis was performed as previously described (18) using 10 μl of commercial MET amplification probe (Cytocell Ltd, Cambridge, UK) consisting of a 278Kb red probe spanning the MET gene (7q31) and the green centromeric probe for chromosome 7 7D7Z1, 7p11.1-q11.1) provided as control. Samples with a ratio greater than 3 between C-MET gene and chromosome 7 centromere signals, in at least 10% of 100 cells analysed in 10 different fields, were scored as positive for MET gene amplification. Healthy tissue was used as internal negative control.
MET gene copy number by real-time PCR
Quantitative PCR experiments for estimation of MET copy number variations were performed in triplicates using a Human TaqMan Copy Number Assay for MET (assay ID: hs01277655_cn) and the TaqMan Copy Number Reference Assay RNase P (Applied Biosystems, Foster City, CA). Furthermore, in order to exclude an entire chromosome 7 polysomy, the MAD1L1 gene (assay ID: hs00981515_cn), which is located on the p-arm of chromosome 7, was evaluated for CNV. PCR runs were performed using ABI MicroAmp Optical Fast 96-well Reaction Plates on an Applied Biosystems ViiA 7 Real-Time PCR System (Applied Biosystems, Foster City, CA) using 20 ng total gDNA as a template. CNV of the target gene was calculated by the ABI SDS software 1.1 using relative quantification based on the ΔΔCt method and control samples as calibrators.
Cell Culture and Inhibitors
LIM1215 cells were grown in RPMI-1640 medium with 10% FBS and 1 μg/ml insulin; the DiFi cell line was cultured in F12 medium supplemented with 10% FBS. DiFi were a kind gift from Dr J. Baselga (Oncology Department of Vall d’Hebron University Hospital, Barcelona) in November 2004, while LIM1215 were obtained from R. Whitehead (51), with permission from the Ludwig Institute for Cancer Research in November 2009. The genetic identity of the cell lines was confirmed by short tandem repeat (STR) profiling (Cell ID, Promega), which was last repeated in January 2013. HGF was from Peprotech. JNJ38877605 was provided by Janssen, Cetuximab and Panitumumab were obtained from Pharmacy at Niguarda Ca’Granda Hospital, Milan, Italy. MET, MET kinase dead and KRAS constructs were in the p156RRLsin.PPThCMV.MCS.pre lentiviral vector.
Virus preparation and cell transduction
Lentiviral vectors were produced as previously described (52). Cells were subjected to multiple rounds of transduction using 200 ng/ml of p24.
Biological assays
Cell viability was assessed by ATP content using the CellTiter-Glo luminescent assay (Promega), after treatment for 7 days. Measurements were recorded using a Victor-X4 plate reader (PerkinElmer). Data points represent mean + SD of three independent experiments.
Western blot analysis
Cells were pre-incubated over night with 1 μg/ml of cetuximab or panitumumab and /or 250nM of JNJ38877605 in serum-free medium. The day after, they were stimulated with 80 ng/ml of HGF for 15 minutes, in the presence of the above mentioned inhibitors. Cells were lysed in LB buffer (2% SDS, 0.5M Tris-HCl (pH 6.8)). Primary antibodies: anti-Actin (1–9), anti-EGR and anti-RAS (F234) were from Santa Cruz Biotechnology, anti-MET (clone 3D4) was from Invitrogen; antibodies against phosphorylated Met (Tyr1234/1235), phosphorylated EGFR (Tyr1068), phosphorylated ERK (Thr202/Tyr204), phosphorylated AKT (Ser473), total AKT, ERK were from Cell Signaling. Secondary antibodies were from Amersham. Detection was performed with ECL system (Amersham).
Xenograft transplantation experiments
Approximately 5×106 tumor cells were injected subcutaneously in 6-week-old immunodeficient NOD-SCID mice in the presence of 10% matrigel. Tumor treatment (intra-tumor injection of 200 μg of cetuximab twice a week) started when tumor volume reached 200 mm3. The derivation of patient derived tumor grafts (‘xenopatients’) and their profiling for response to treatment with Cetuximab was conducted as previously reported (11, 23, 53). Established tumors (average volume 400 mm3) were treated with the following regimens, either single agent or in combination: cetuximab (Merck-Serono) 20 mg/Kg, twice weekly ip; Crizotinib (Sequoia Research ProductsLtd) 50 mg/kg, daily, os; JNJ-38877605 (Janssen Pharmaceuticals) 50mg/kg, daily, os.Tumor size was evaluated once- or twice-weekly by caliper measurements and approximate volume of the mass was calculated using the formula 4/3π(D/2)(d/2)2, where d is the minor tumor axis and D is the major tumor axis.
All animal procedures were approved by the Ethical Commission of the Institute for Cancer Research and Treatment and bythe Italian Ministry of Health. Tumor volumes are plotted as mean ± SE.
Statistical Analysis
Statistical significance was performed using GraphPad Prism (GraphPad Software). Error bars represent the SE or SD, as indicated in each figure legend. All experiments were repeated at least 3 times.
Supplementary Material
Statement of significance.
Amplification of the MET proto-oncogene is responsible for de novo and acquired resistance to anti-EGFR therapy in a subset of CRCs. As multiple anti-MET therapeutic strategies are available, these findings offer immediate novel opportunities to design clinical studies.
Acknowledgments
Grant Support
The research leading to these results has received funding from the European Community’s Seventh Framework Programme under grant agreement n. 259015 COLTHERES; AIRC 2010 Special Program Molecular Clinical Oncology 5xMille, Project n. 9970; Intramural Grant Fondazione Piemontese per la Ricerca sul Cancro ONLUS (5 per mille 2008 Ministero dell’Istruzione, dell’Università e della Ricerca);. Oncologia Ca’ Granda ONLUS (OCGO) Fondazione (S.S.); Intramural Grant – 5xmille 2008 – Fondazione Piemontese per la Ricerca sul Cancro – ONLUS (A.B, L.T., F.D.N.); AIRC IG grant n. 12812 (A.B.); AIRC IG 10116 (L.T.); AIRC IG 11819 (S.G.); AIRC MFAG 11349 (F.D.N.); MIUR FIRB, Fondo per gli Investimenti della Ricerca di Base – Futuro in Ricerca (A.Be.); ‘Farmacogenomica’ - Fondazione Piemontese per la Ricerca sul Cancro – ONLUS 5 per mille 2009 MIUR (F.D.N.); Swim Across America (L.A.D.), NIH Grant CA121113 (V.E.V); The European Community’s Seventh Framework Programme (SYSCOL) (V.E.V).
The authors thank Barbara Martinoglio and Michela Buscarino at the IRC@C for providing technical support with real-time PCR and Sanger sequencing, respectively, and Paolo Michieli for helpful scientific discussion. We thank Sabrina Arena for providing the KRAS virus constructs and Cristina Basilico for providing the MET KD construct.
Footnotes
Disclosure of Potential Conflicts of Interest
T. Perera is a full-time employee of Janssen Pharmaceutica and a shareholder of Johnson and Johnson, the manufacturer of JNJ38877605. L.A.D. and V.E.V. are co-founders of Inostics and Personal Genome Diagnostics and are members of their Scientific Advisory Boards. L.A.D. and V.E.V. own Inostics and Personal Genome Diagnostics stock, which is subject to certain restrictions under University policy. The terms of these arrangements are managed by the Johns Hopkins University in accordance with its conflict-of-interest policies.
References
- 1.Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. N Engl J Med. 2008;358:1160–74. doi: 10.1056/NEJMra0707704. [DOI] [PubMed] [Google Scholar]
- 2.Rothenberg ML, LaFleur B, Levy DE, Washington MK, Morgan-Meadows SL, Ramanathan RK, et al. Randomized phase II trial of the clinical and biological effects of two dose levels of gefitinib in patients with recurrent colorectal adenocarcinoma. J Clin Oncol. 2005;23:9265–74. doi: 10.1200/JCO.2005.03.0536. [DOI] [PubMed] [Google Scholar]
- 3.Townsley CA, Major P, Siu LL, Dancey J, Chen E, Pond GR, et al. Phase II study of erlotinib (OSI-774) in patients with metastatic colorectal cancer. British Journal of Cancer. 2006;94:1136–43. doi: 10.1038/sj.bjc.6603055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Santoro A, Comandone A, Rimassa L, Granetti C, Lorusso V, Oliva C, et al. A phase II randomized multicenter trial of gefitinib plus FOLFIRI and FOLFIRI alone in patients with metastatic colorectal cancer. Ann Oncol. 2008;19:1888–93. doi: 10.1093/annonc/mdn401. [DOI] [PubMed] [Google Scholar]
- 5.Cunningham D, Humblet Y, Siena S, Khayat D, Bleiberg H, Santoro A, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med. 2004;351:337–45. doi: 10.1056/NEJMoa033025. [DOI] [PubMed] [Google Scholar]
- 6.Van Cutsem E, Peeters M, Siena S, Humblet Y, Hendlisz A, Neyns B, et al. Open-label phase III trial of panitumumab plus best supportive care compared with best supportive care alone in patients with chemotherapy-refractory metastatic colorectal cancer. Journal of Clinical Oncology. 2007;25:1658–64. doi: 10.1200/JCO.2006.08.1620. [DOI] [PubMed] [Google Scholar]
- 7.Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–34. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 8.Karapetis CS, Khambata-Ford S, Jonker DJ, O’Callaghan CJ, Tu D, Tebbutt NC, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–65. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 9.Di Nicolantonio F, Martini M, Molinari F, Sartore-Bianchi A, Arena S, Saletti P, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26:5705–12. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- 10.De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753–62. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 11.Bertotti A, Migliardi G, Galimi F, Sassi F, Torti D, Isella C, et al. A molecularly annotated platform of patient-derived xenografts (“xenopatients”) identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer Discovery. 2011;1:508–23. doi: 10.1158/2159-8290.CD-11-0109. [DOI] [PubMed] [Google Scholar]
- 12.Yonesaka K, Zejnullahu K, Okamoto I, Satoh T, Cappuzzo F, Souglakos J, et al. Activation of ERBB2 signaling causes resistance to the EGFR-directed therapeutic antibody cetuximab. Science translational medicine. 2011;3:99ra86. doi: 10.1126/scitranslmed.3002442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Montagut C, Dalmases A, Bellosillo B, Crespo M, Pairet S, Iglesias M, et al. Identification of a mutation in the extracellular domain of the Epidermal Growth Factor Receptor conferring cetuximab resistance in colorectal cancer. Nature Medicine. 2012;18:221–3. doi: 10.1038/nm.2609. [DOI] [PubMed] [Google Scholar]
- 14.Lu Y, Li X, Liang K, Luwor R, Siddik ZH, Mills GB, et al. Epidermal growth factor receptor (EGFR) ubiquitination as a mechanism of acquired resistance escaping treatment by the anti-EGFR monoclonal antibody cetuximab. Cancer Res. 2007;67:8240–7. doi: 10.1158/0008-5472.CAN-07-0589. [DOI] [PubMed] [Google Scholar]
- 15.Benavente S, Huang S, Armstrong EA, Chi A, Hsu KT, Wheeler DL, et al. Establishment and characterization of a model of acquired resistance to epidermal growth factor receptor targeting agents in human cancer cells. Clinical Cancer Research. 2009;15:1585–92. doi: 10.1158/1078-0432.CCR-08-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li C, Iida M, Dunn EF, Ghia AJ, Wheeler DL. Nuclear EGFR contributes to acquired resistance to cetuximab. Oncogene. 2009 doi: 10.1038/onc.2009.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diaz LA, Jr, Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537–40. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–6. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coquelle A, Pipiras E, Toledo F, Buttin G, Debatisse M. Expression of fragile sites triggers intrachromosomal mammalian gene amplification and sets boundaries to early amplicons. Cell. 1997;89:215–25. doi: 10.1016/s0092-8674(00)80201-9. [DOI] [PubMed] [Google Scholar]
- 20.Leary RJ, Kinde I, Diehl F, Schmidt K, Clouser C, Duncan C, et al. Development of personalized tumor biomarkers using massively parallel sequencing. Science translational medicine. 2010;2:20ra14. doi: 10.1126/scitranslmed.3000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leary RJ, Sausen M, Kinde I, Papadopoulos N, Carpten JD, Craig D, et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Science translational medicine. 2012;4:162ra54. doi: 10.1126/scitranslmed.3004742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sausen M, Leary RJ, Jones S, Wu J, Reynolds CP, Liu X, et al. Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma. Nat Genet. 2013;45:12–7. doi: 10.1038/ng.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galimi F, Torti D, Sassi F, Isella C, Cora D, Gastaldi S, et al. Genetic and expression analysis of MET, MACC1, and HGF in metastatic colorectal cancer: response to met inhibition in patient xenografts and pathologic correlations. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:3146–56. doi: 10.1158/1078-0432.CCR-10-3377. [DOI] [PubMed] [Google Scholar]
- 24.Gherardi E, Birchmeier W, Birchmeier C, Vande Woude G. Targeting MET in cancer: rationale and progress. Nature reviews Cancer. 2012;12:89–103. doi: 10.1038/nrc3205. [DOI] [PubMed] [Google Scholar]
- 25.Trusolino L, Bertotti A, Comoglio PM. MET signalling: principles and functions in development, organ regeneration and cancer. Nature reviews Molecular cell biology. 2010;11:834–48. doi: 10.1038/nrm3012. [DOI] [PubMed] [Google Scholar]
- 26.Wu X, Fan Z, Masui H, Rosen N, Mendelsohn J. Apoptosis induced by an anti-epidermal growth factor receptor monoclonal antibody in a human colorectal carcinoma cell line and its delay by insulin. J Clin Invest. 1995;95:1897–905. doi: 10.1172/JCI117871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moroni M, Veronese S, Benvenuti S, Marrapese G, Sartore-Bianchi A, Di Nicolantonio F, et al. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: a cohort study. The lancet oncology. 2005;6:279–86. doi: 10.1016/S1470-2045(05)70102-9. [DOI] [PubMed] [Google Scholar]
- 28.Jhawer M, Goel S, Wilson AJ, Montagna C, Ling YH, Byun DS, et al. PIK3CA mutation/PTEN expression status predicts response of colon cancer cells to the epidermal growth factor receptor inhibitor cetuximab. Cancer Res. 2008;68:1953–61. doi: 10.1158/0008-5472.CAN-07-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liska D, Chen CT, Bachleitner-Hofmann T, Christensen JG, Weiser MR. HGF rescues colorectal cancer cells from EGFR inhibition via MET activation. Clinical Cancer Research. 2011;17:472–82. doi: 10.1158/1078-0432.CCR-10-0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peters S, Adjei AA. MET: a promising anticancer therapeutic target. Nature reviews Clinical oncology. 2012;9:314–26. doi: 10.1038/nrclinonc.2012.71. [DOI] [PubMed] [Google Scholar]
- 31.Naldini L, Vigna E, Narsimhan RP, Gaudino G, Zarnegar R, Michalopoulos GK, et al. Hepatocyte growth factor (HGF) stimulates the tyrosine kinase activity of the receptor encoded by the proto-oncogene c-MET. Oncogene. 1991;6:501–4. [PubMed] [Google Scholar]
- 32.Otsuka T, Takayama H, Sharp R, Celli G, LaRochelle WJ, Bottaro DP, et al. c-Met autocrine activation induces development of malignant melanoma and acquisition of the metastatic phenotype. Cancer Research. 1998;58:5157–67. [PubMed] [Google Scholar]
- 33.Michieli P, Basilico C, Pennacchietti S, Maffe A, Tamagnone L, Giordano S, et al. Mutant Met-mediated transformation is ligand-dependent and can be inhibited by HGF antagonists. Oncogene. 1999;18:5221–31. doi: 10.1038/sj.onc.1202899. [DOI] [PubMed] [Google Scholar]
- 34.Kentsis A, Reed C, Rice KL, Sanda T, Rodig SJ, Tholouli E, et al. Autocrine activation of the MET receptor tyrosine kinase in acute myeloid leukemia. Nature Medicine. 2012;18:1118–22. doi: 10.1038/nm.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lennerz JK, Kwak EL, Ackerman A, Michael M, Fox SB, Bergethon K, et al. MET amplification identifies a small and aggressive subgroup of esophagogastric adenocarcinoma with evidence of responsiveness to crizotinib. J Clin Oncol. 2011;29:4803–10. doi: 10.1200/JCO.2011.35.4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turner NC, Reis-Filho JS. Genetic heterogeneity and cancer drug resistance. The lancet oncology. 2012;13:e178–85. doi: 10.1016/S1470-2045(11)70335-7. [DOI] [PubMed] [Google Scholar]
- 37.Comoglio PM, Giordano S, Trusolino L. Drug development of MET inhibitors: targeting oncogene addiction and expedience. Nature reviews Drug discovery. 2008;7:504–16. doi: 10.1038/nrd2530. [DOI] [PubMed] [Google Scholar]
- 38.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–43. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 39.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and Histological Evolution of Lung Cancers Acquiring Resistance to EGFR Inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cappuzzo F, Janne PA, Skokan M, Finocchiaro G, Rossi E, Ligorio C, et al. MET increased gene copy number and primary resistance to gefitinib therapy in non-small-cell lung cancer patients. Ann Oncol. 2009;20:298–304. doi: 10.1093/annonc/mdn635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turke AB, Zejnullahu K, Wu YL, Song Y, Dias-Santagata D, Lifshits E, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cappuzzo F, Varella-Garcia M, Finocchiaro G, Skokan M, Gajapathy S, Carnaghi C, et al. Primary resistance to cetuximab therapy in EGFR FISH-positive colorectal cancer patients. Br J Cancer. 2008;99:83–9. doi: 10.1038/sj.bjc.6604439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Inno A, Di Salvatore M, Cenci T, Martini M, Orlandi A, Strippoli A, et al. Is there a role for IGF1R and c-MET pathways in resistance to cetuximab in metastatic colorectal cancer? Clinical colorectal cancer. 2011;10:325–32. doi: 10.1016/j.clcc.2011.03.028. [DOI] [PubMed] [Google Scholar]
- 45.Valtorta E, Misale S, Sartore-Bianchi A, Nagtegaal ID, Paraf F, Lauricella C, et al. KRAS gene amplification in colorectal cancer and impact on response to EGFR-targeted therapy. Int J Cancer. 2013 doi: 10.1002/ijc.28106. [DOI] [PubMed] [Google Scholar]
- 46.Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500–4. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilson TR, Fridlyand J, Yan Y, Penuel E, Burton L, Chan E, et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. 2012;487:505–9. doi: 10.1038/nature11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Michalopoulos GK, DeFrances MC. Liver regeneration. Science. 1997;276:60–6. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- 49.Timofeevski SL, McTigue MA, Ryan K, Cui J, Zou HY, Zhu JX, et al. Enzymatic characterization of c-Met receptor tyrosine kinase oncogenic mutants and kinetic studies with aminopyridine and triazolopyrazine inhibitors. Biochemistry. 2009;48:5339–49. doi: 10.1021/bi900438w. [DOI] [PubMed] [Google Scholar]
- 50.Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92:205–16. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 51.Whitehead RH, Macrae FA, St John DJ, Ma J. A colon cancer cell line (LIM1215) derived from a patient with inherited nonpolyposis colorectal cancer. Journal of the National Cancer Institute. 1985;74:759–65. [PubMed] [Google Scholar]
- 52.Vigna E, Naldini L. Lentiviral vectors: excellent tools for experimental gene transfer and promising candidates for gene therapy. The journal of gene medicine. 2000;2:308–16. doi: 10.1002/1521-2254(200009/10)2:5<308::AID-JGM131>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 53.Migliardi G, Sassi F, Torti D, Galimi F, Zanella ER, Buscarino M, et al. Inhibition of MEK and PI3K/mTOR suppresses tumor growth but does not cause tumor regression in patient-derived xenografts of RAS-mutant colorectal carcinomas. Clin Cancer Res. 2012;18:2515–2525. doi: 10.1158/1078-0432.CCR-11-2683. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.