Abstract
Gene silencing mediated by RNA interference (RNAi) was first discovered in Caenorhabditis elegans, and was subsequently recognized in various other organisms. In mammalian cells, RNAi can be induced by small interfering RNAs (siRNAs). In earlier studies, our group developed a vector-based system for expression of siRNA under control of a polymerase III promoter, the U6 promoter, which can induce RNAi in living cells. We here describe a system for controlling the U6 promoter-driven expression of siRNA using the Cre–loxP recombination system. We constructed a ‘Cre-On’ siRNA expression vector which could be switched on upon excision catalyzed by Cre recombinase, which was delivered to cells directly from the medium as a fusion protein. An examination of the effectiveness of RNAi against a reporter gene revealed that addition of TAT-NLS-Cre (where NLS is a nuclear localization signal and TAT is a peptide of 11 amino acids derived from HIV) to the medium resulted in plasmid recombination, generation of siRNA and suppression of reporter activity. This system should allow us to induce RNAi in a spatially, temporally, cell type-specifically or tissue-specifically controlled manner and potentiate the improved application of RNAi in both an experimental and a therapeutic context.
INTRODUCTION
RNA interference (RNAi) is a post-transcriptional gene silencing phenomenon induced by double-stranded RNA (dsRNA) that was first discovered in Caenorhabditis elegans (1). In RNAi, it appears that dsRNAs are cleaved by a member of the RNase III family (Dicer) into small interfering RNAs (siRNAs) of 21 or 22 nt in length (2–4), which in turn induce the degradation of the target mRNA, with resultant suppression of expression of the target gene. This phenomenon has been found in evolutionarily diverse organisms, such as plants, a nematode, the fruit fly and a protozoan (5–8). In the case of mammalian cells, it was reported initially that dsRNAs cause the non-specific degradation of mRNA, but now it is clear that 21 or 22 nt RNAs with 2 or 3 nt 3′ overhangs, known as small interfering RNAs (siRNAs), induce RNAi in cultured mammalian cells without inducing the dsRNA-dependent non-specific inhibition of protein synthesis (2,9,10). Various groups, including our own, have developed vector-mediated systems for specific RNAi in mammalian cells using polymerase (pol) III promoters such as the U6, H1 and tRNAVal promoters (11–18). Furthermore, both synthetic siRNAs and small hairpin RNAs transcribed in vivo can successfully suppress the expression of transgenes and an endogenous gene in adult mice (19–21).
Today, RNAi shows considerable promise as both an experimental and a therapeutic tool. Suppression of the expression of a gene of interest by RNAi has several positive features. Such a system is easy to design, exhibits strong site specificity and has a strong suppressive effect. Moreover, only low concentrations of siRNA are required. To make RNAi a more efficient tool, for example for the study of genes that are essential for survival, and for cell type-specific, tissue-specific and time–course experiments, it is necessary to control the expression of siRNA both spatially and temporally. siRNA expression systems using a pol II or a pol III promoter that can be regulated by tetracycline have been reported (22–26) but, to our knowledge, there have been no reports of the control of expression of pol III promoter-based siRNA expression vectors using a Cre–loxP system with exogenously introduced Cre recombinase fusion proteins.
The Cre–loxP system is a system that is widely used in reverse genetics. The Cre recombinase is a site-specific recombinase encoded by bacteriophage P1 that recognizes and promotes recombination at loxP sites (27), which consist of two 13 bp repeats, each separated by an 8 bp spacer region. Direct repeats of loxP result in excision, while inverted repeats cause inversion of the sequence placed between them (28). Cre-mediated recombination can be achieved in various kinds of eukaryotic cell, such as yeast (29), plant (30) and mammalian cells (31). Furthermore, Cre recombinase can also be stably expressed in transgenic mice (32,33).
In addition to the expression of Cre recombinase in vivo, Cre recombinase can also be delivered directly into cells as a purified protein. It was reported recently that fusion of the TAT sequence and a nuclear localization signal (NLS) to Cre recombinase promotes the uptake by cells of recombinant Cre recombinase from the medium (34). TAT is an arginine-rich peptide of 11 amino acids derived from HIV that is able to cross cell membranes (35,36). It is taken up in a rapid, concentration-dependent manner by a wide variety of cells (37–39). Furthermore, Cre recombinase fused with TAT-NLS retains the capacity for efficient catalysis of recombination in mammalian cells (34).
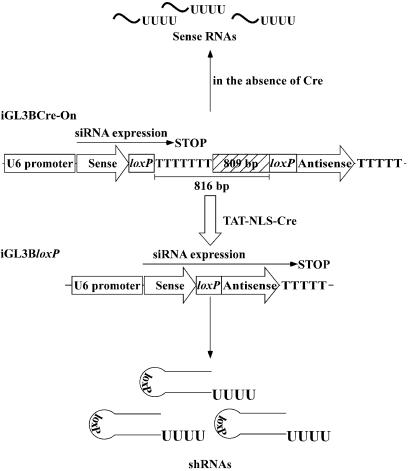
In this study, our goal was to switch on the expression of a pol III promoter-based siRNA expression vector exogenously using recombinant TAT-NLS-Cre recombinase (TAT-NLS-Cre). The Cre-On siRNA expression vector was prepared by inserting, between two loxP sites, a region that prohibits complete transcription of the siRNA coding sequence, namely by inserting a region that consists of a stretch of T residues to stop transcription and an 809 bp linker fragment (Fig. 1). Thus, in the absence of Cre recombinase, the only products of transcription are incomplete small RNA fragments that cannot induce RNAi. Upon recombination catalyzed by Cre recombinase, the region that interferes with transcription is removed and complete and active siRNAs are produced (40). We demonstrate here, using a luciferase reporter system, that the TAT-NLS-Cre-mediated recombination of the construct allowed us to switch on RNAi.
Figure 1.
Strategy for controlling the expression of siRNA with TAT-NLS-Cre. In the absence of TAT-NLS-Cre, transcription stops just after the sense sequence, yielding incomplete siRNAs. Thus, RNAi cannot occur. In the presence of TAT-NLS-Cre, recombination occurs and the region between loxP sites is excised. As a result, complete stem-type siRNAs are formed and induce silencing of the target gene.
MATERIALS AND METHODS
Construction of siRNA expression vectors targeted against firefly luciferase
As a positive control, we used the iGL3B vector, which expresses 21 nt hairpin-type siRNAs with a 9 nt loop, as described previously (41). We also constructed the iGL3BloxP vector, which included a sense–loxP–antisense sequence by inserting a sense–loxP–antisense fragment that had been amplified by PCR into the pU6 vector (11). The sense and antisense sequences used to make iGL3BloxP were the same as those of iGL3B (the target sequence is: 5′-gtg cgc tgc tgg tgc caa ccc-3′), while the sequence of loxP was 5′-ata act tcg tat agc ata cat tat acg aag tta t-3′.
To generate the iGL3BCre-On vector, we amplified a sense–loxP–TTTTTTT–ApaI–NotI–loxP–antisense fragment by PCR (the sense and antisense sequences were the same as those of iGL3B) and inserted it into the pU6 vector. Then we inserted an 809 bp unrelated fragment (coding sequence of the neomycin resistance gene) into the ApaI and NotI sites between two loxP sites.
As a negative control, we used pU6iRed, an siRNA expression vector against red fluorescent protein (DsRed2) constructed in our laboratory for another purpose.
Construction of the vector for expression of TAT–NLS–Cre recombinase (pTriEx™-3 Hygro-TAT-NLS-Cre) and TAT–NLS–EGFP (pTriEx™-3 Hygro-TAT-NLS-EGFP)
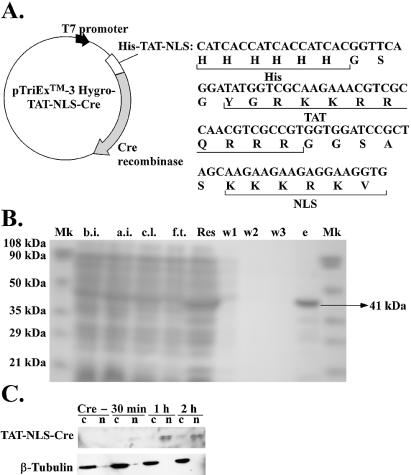
First, we constructed the pEF9-NLS-Cre vector, a vector to express recombinant NLS–Cre recombinase protein (NLS–Cre) inside mammalian cells. We inserted the Kozak–NheI–NLS–XbaI–EcoRI–NotI sequence, generated by PCR-based subcloning, into the BamHI and ApaI sites of pEF9 (42). The sequence encoding Cre recombinase was excised from pMM23 (a kind gift from Dr D.Ow) and inserted at the XbaI and EcoRI sites to generate pEF9-NLS-Cre. For the expression of TAT-NLS-Cre in Escherichia coli, we constructed pTriEx-3 Hygro-TAT-NLS-Cre. To do this, we first constructed pTriEx-3 Hygro-TAT by generating a His–TAT–NheI-containing fragment (His refers to a six histidine amino acid sequence) by PCR-based subcloning and inserting it into the NcoI and NotI sites of pTriEx-3 Hygro (Novagen, Madison, WI). To generate pTriEx-3 Hygro-TAT-NLS-Cre, we excised the NLS-Cre sequence from pEF9-NLS-Cre and inserted it at the NheI and NotI sites of pTriEx-3 Hygro-TAT. The sequences of TAT and NLS are shown in Figure 2A.
Figure 2.
Construction of vectors for the expression of NLS-Cre. (A) Vector for the expression of TAT-NLS-Cre recombinase protein, pTriEx-3 Hygro-TAT-NLS-Cre. Cre recombinase was expressed from a T7 promoter and sequences encoding a histidine tag, TAT and NLS were fused upstream of the sequence that encoded Cre recombinase. (B) PAGE of TAT-NLS-Cre recombinase expressed in E.coli (41 kDa). Mk, markers; b.i., before induction; a.i., after induction by IPTG; c.l., cleared lysate; f.t., flow-through; Res, resin after binding; w1–w3, washes 1–3; e, eluate. The concentration of TAT-NLS-Cre in the eluate was 0.15 µg/µl. (C) Western blotting of cell extracts. c, cytosolic fraction; n, nuclear fraction; Cre –, without TAT-NLS-Cre. Cells were incubated in medium supplemented with TAT-NLS-Cre for the indicated times. Cre recombinase was detected in nuclear extracts, while β-tubulin was detected in cytosolic extracts.
As a negative control, we also constructed pTriEx-3 Hygro-TAT-NLS-EGFP, which could express TAT–NLS–EGFP fusion protein. To construct this vector, we first generated a NLS–EGFP fragment by amplifying the EGFP sequence from pEGFP-c2 vector (Clontech, Palo Alto, CA) with a primer containing NLS sequence and inserted it at the NheI and NotI sites of pTriEx-3 Hygro-TAT.
Expression and purification of TAT–NLS–Cre, TAT–NLS–EGFP and residual E.coli lysate
The plasmid pTriEx-3 Hygro-TAT-NLS-Cre or pTriEx-3 Hygro-TAT-NLS-EGFP was introduced into E.coli strain RosettaBlue(DE3)pLacI (Novagen) and expression of fusion proteins was induced by the addition to the culture medium of isopropyl β-d-thiogalactoside (IPTG). Escherichia coli was cultured in LB medium that contained 1% glucose, 34 µg/ml chloramphenicol, 12.5 µg/ml tetracycline and 100 µg/ml ampicillin at 37°C until absorbance at 600 nm reached 0.7. Expression of the recombinant protein was induced by IPTG (final concentration 1 mM) and incubation was continued for 3–4 h at 37°C. After cells had been harvested by centrifugation, cell pellets were resuspended in 3 ml/g wet wt lysis buffer [100 mM NaH2PO4, 10 mM Tris–HCl (pH 8.0), 300 mM NaCl, 10 mM imidazole] plus 2 mg/ml lysozyme, 5 U/ml Benzonase® Nuclease (Novagen) and Complete, EDTA-free (Roche, Mannheim, Germany) and incubated on ice for 30 min. Cleared lysates were obtained after sonication and centrifugation. To purify the recombinant proteins, the cleared lysates and Ni–NTA–agarose (Qiagen, Hilden, Germany) were mixed and rotated overnight at 4°C. The next day, the mixtures were poured into Poly-Prep® chromatography columns (Bio-Rad, Hercules, CA) and the flow-throughs were discarded. The columns were washed three times with 10 bed vol of washing buffer [100 mM NaH2PO4, 10 mM Tris–HCl (pH 8.0), 300 mM NaCl, 20 mM imidazole]. After elution with elution buffer [100 mM NaH2PO4, 10 mM Tris–HCl (pH 7.4), 300 mM NaCl, 250 mM imidazole], recombinant proteins were dialyzed against a 1:1 (v/v) mixture of Dulbecco’s modified Eagle’s medium (Sigma, St Louis, MO) and phosphate-buffered saline (PBS) (pH 7.4) containing 0.1% Pluronic F-68 solution (Sigma) and 100 U/ml Antibiotic-Antimycotic mixture (Invitrogen, Carlsbad, CA). The final preparations were analyzed by SDS–PAGE on a 15% polyacrylamide gel, and the results are shown in Figure 2B.
For residual E.coli lysate, pTriEx-3 Hygro empty vector was introduced into E.coli strain RosettaBlue(DE3)pLacI (Novagen) and cultured in LB medium that contained 1% glucose, 34 µg/ml chloramphenicol, 12.5 µg/ml tetracycline and 100 µg/ml ampicillin at 37°C until absorbance at 600 nm reached 0.7. After addition of IPTG to a final concentration of 1 mM, E.coli were harvested and treated in the same way as that of pTriEx-3 Hygro-TAT-NLS-Cre or pTriEx-3 Hygro-TAT-NLS-EGFP.
Transfection, transduction and dual luciferase reporter assay
Cells were cultured in 96-well tissue culture plates to 60% confluency. They were then transfected with 94 ng/well siRNA expression vectors targeted against firefly luciferase or of the control vector, plus 12.5 ng firefly luciferase expression vector (pGL3; Promega, Madison, WI) and 3.75 ng Renilla luciferase expression vector (pRL-RSV) (42). Transfections were performed with Effectene (Qiagen) as described by the manufacturer. Four hours after transfection, TAT–NLS–Cre, TAT–NLS–EGFP, residual E.coli lysate or the dialysis medium was added as indicated. Luciferase activities were analyzed 24 h after transfection with the Dual Luciferase System (Promega).
Separation of nuclei from the cytosol and western blotting analysis
Cells were cultured in 24-well tissue culture plates to 80% confluency. After incubation with TAT–NLS–Cre (final concentration 10 µg/ml) or dialysis medium for the indicated times, cells were washed twice with PBS and treated with trypsin–EDTA (Invitrogen) for 15 min at 37°C. Cells were then suspended in PBS and centrifuged (3000 r.p.m., 10 min, 4°C). Supernatants were discarded and the pellets were resuspended in hypotonic buffer [10 mM HEPES (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, plus Complete, EDTA-free (Roche)]. After incubation for 2 min on ice, NP-40 (Wako, Osaka, Japan) was added to a final concentration of 0.4% and suspensions were centrifuged (3000 r.p.m., 10 min, 4°C). Supernatants (containing cytosolic proteins) were collected and the pellets were washed with hypotonic buffer. To obtain nuclear proteins, equal volumes of the hypotonic buffer and a high salt buffer [0.4 mM EDTA (pH 8.0), 1.5 mM MgCl2, 30 mM HEPES (pH 7.9), 800 mM NaCl, 1% NP-40, plus Complete, EDTA-free (Roche)] were added to the pellets and the mixtures were incubated on ice for a total of 30 min with vortex mixing at 10 min intervals. After centrifugation (15 000 r.p.m., 30 min, 4°C), supernatants (containing nuclear proteins) were collected. Total proteins were quantified using a DC Protein Assay Kit (Bio-Rad) with bovine serum albumin as the standard protein.
For western blotting analysis, samples (10 µg total protein) were fractionated by SDS–PAGE (15% polyacrylamide) and bands of proteins were transferred to an Immobilon™-PSQ transfer membrane (Millipore, Billerica, CA). After blocking in TBS-T (Tris-buffered saline containing 0.1% Tween) containing 3% skimmed milk, the membrane was incubated for 1 h with polyclonal antibodies raised in rabbit against Cre recombinase (1:1000 dilution in TBS-T plus 0.3% skimmed milk) (Novagen) and for 1 h with antibodies against rabbit Ig conjugated to horseradish peroxidase (1:1000 dilution in TBS-T plus 0.3% skimmed milk) (Amersham Biosciences, Little Chalfont, UK). As a control for the separation of nuclear and cytosolic fractions, we also performed western blotting by incubating for 1 h with monoclonal antibodies raised in mouse against β-tubulin (anti-β-tubulin, clone AA2, 1:1000 dilution in TBS-T plus 0.3% skimmed milk) (Upstate) and 1 h with antibodies against mouse Ig conjugated to horseradish peroxidase (1:1000 dilution in TBS-T plus 0.3% skimmed milk) (Amersham Biosciences). The results of western blotting are shown in Figure 2C.
Amplification by PCR of transfected plasmids
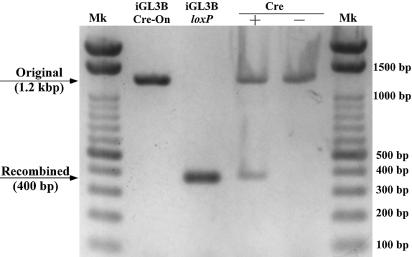
Cells were grown in 24-well tissue culture plates, transfected with 375 ng iGL3B Cre-On plasmid, 50 ng pGL3 and 15 ng pRL-RSV and incubated with TAT–NLS–Cre (30 µg/ml) or dialysis medium as described above. Then, 22 h later, cells were resuspended in Passive Lysis Buffer (Promega) and centrifuged (3000 r.p.m., 10 min, 4°C). After two washes with Tris–EDTA buffer (TE) [10 mM Tris–HCl (pH 8.0) plus 1 mM EDTA], the pellets were resuspended in TE and boiled (65°C for 10 min and 99°C for 5 min). Then 4–6 µl of each sample was used as template for PCR, which was performed with Ex Taq™ (Takara Shuzo, Shiga, Japan). The results are shown in Figure 3.
Figure 3.
Fractionation by gel electrophoresis of products of PCR amplified from iGL3BloxP (positive control), iGL3BCre-On (negative control), Cre + (nuclei from cells transfected with iGL3BCre-On and incubated with TAT-NLS-Cre, final concentration 30 µg/ml) and Cre – (nuclei from cells transfected with iGL3BCre-On and incubated with dialysis medium). Mk, size markers. Products of PCR from cells incubated with TAT-NLS-Cre protein yielded both ‘Original’ and ‘Recombined’ bands, while cells incubated with dialysis medium yielded only the ‘Original’ band. These results indicate that recombination occurred only in the presence of the exogenously added TAT-NLS-Cre.
RESULTS
In this study, our goal was to establish an exogenously Cre controllable U6 promoter-based siRNA expression vector. Since Cre recombinase catalyzes the excision of sequences between loxP sites, our strategy was to place, between two loxP sites, an insert that inhibited expression of complete siRNAs to generate a Cre-On system. Schematic representatives of the switches for the expression of siRNAs are shown in Figure 1. In this scheme, two loxP sites (with a fragment of 816 bp inserted between them) are placed between sense and antisense regions of the siRNA encoding sequence. The 816 bp insert starts with TTTTTTT, which causes termination of transcription, since TTTT is a terminator sequence for the U6 promoter in mammalian cells. As a result, only sense RNAs, which have no suppressive effect, can be transcribed in the absence of Cre recombinase. In the presence of Cre recombinase, recombination occurs and the inserted fragment, including the termination sequence, is excised. The excision of this fragment allows transcription of full, functional siRNAs since, now, only a 34 bp loxP site connects the sense and antisense sequences, yielding short hairpin RNAs (shRNAs) (Fig. 1).
Construction of Cre recombinase controllable siRNA expression vectors
To study the possibility of controlling the expression of siRNA with Cre recombinase, we used three types of siRNA expression vector directed against firefly luciferase, namely iGL3B, iGL3BloxP and iGL3BCre-On, the first two of which were used as positive controls. An siRNA expression vector directed against DsRed2 (pU6iRed) was also used as an unrelated negative control. iGL3B is a vector that produces 21 nt hairpin-type siRNA with a 9 nt loop, constructed in our laboratory as described previously (41). We designed iGL3BCre-On and iGL3BloxP on the basis of the sense and antisense sequences of iGL3B. iGL3BCre-On was designed for the Cre-On system (Fig. 1) and iGL3BloxP was similar to iGL3B, with the exception that the 9 nt loop was replaced by a 34 nt loxP sequence. In other words, iGL3BloxP had exactly the same structure as that of recombined iGL3BCre-On (Fig. 1, after recombination). This vector was constructed to examine the efficiency with which recombined iGL3BCre-On induced RNAi against the targeted gene for firefly luciferase. A comparison of efficiency between iGL3B and iGL3BloxP is shown below in Figure 4A (compare lanes 5 and 6). As discussed below, at least at the concentration of plasmid used, the efficiencies of iGL3B and iGL3BloxP were almost the same. However, at a lower concentration of plasmids, iGL3BloxP with a larger 34 nt loop had a significantly lower activity than iGL3B with a 9 nt loop, indicating that the size of the loop within the shRNAs has some influence on the activity.
Figure 4.
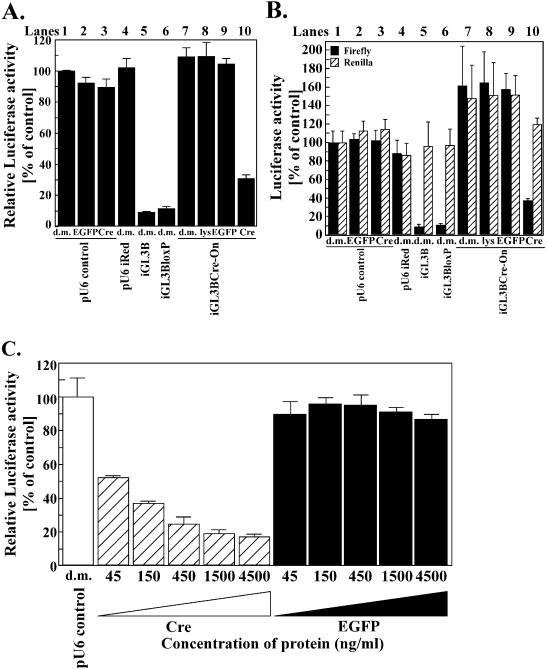
(A) Luciferase activity due to iGL3BCre-On. Lane 1, negative control (d.m., dialysis medium); lane 2, pU6 control and 4.5 µg/ml TAT-NLS-EGFP (EGFP); lane 3, pU6 control and 4.5 µg/ml TAT-NLS-Cre (Cre); lane 4, pU6iRed and dialysis medium; lane 5, iGL3B and dialysis medium; lane 6, iGL3BloxP and dialysis medium; lane 7, iGL3BCre-On and dialysis medium; lane 8, iGL3BCre-On and residual E.coli lysate (lys); lane 9, iGL3BCre-On and 4.5 µg/ml TAT-NLS-EGFP; lane 10, iGL3BCre-On and 4.5 µg/ml TAT-NLS-Cre. Compared with the result for the negative control (pU6 control and incubation with dialysis medium), the luciferase activity in the cells transfected with iGL3BCre-On and incubated with TAT-NLS-Cre was suppressed by ∼70%, while incubation with dialysis medium, with TAT-NLS-EGFP or with residual E.coli lysate did not suppress luciferase activity. (B) Firefly and Renilla luciferase activities of the results shown in (A). The activity of Renilla was not affected by the addition of recombinant protein, dialysis medium or residual E.coli lysate. (C) The extent of suppression of luciferase activity decreased with decreases in the concentration of TAT-NLS-Cre, while incubation with TAT-NLS-EGFP at various concentrations failed to have any effect on luciferase activity, which was the same as the control (incubated with dialysis medium). The results were calculated by normalizing the activity due to firefly luciferase to that due to Renilla luciferase. Then the results were calculated as percentages of the control value. Each bar indicates an average value and vertical bars indicate standard errors of triplicate assays.
We chose the firefly gene for luciferase as the target of siRNA for the following reasons. First, the efficiency of gene silencing by siRNA depends to a large extent on the choice of target site. In this study, our siRNA expression vectors were directed against the target site of iGL3B, which has been shown to be very susceptible to RNAi. Since we knew that we would be unlikely to achieve 100% recombination, we hoped to induce successful RNAi by choosing a very susceptible target, even if not all of the siRNA expression vectors underwent recombination. Second, the choice of firefly luciferase as reporter allowed us to perform quantitative assays using the standard dual luciferase system (11).
Expression and purification of TAT–NLS–Cre
To use NLS–Cre in cells, especially in the nucleus, we decided to deliver the NLS–Cre recombinant protein directly rather than delivering its expression vector. However, in this case it is necessary to enhance the entry of NLS–Cre into cells. It has been reported that NLS can, itself, function as a protein transduction domain (PTD) and promote the cellular uptake of recombinant protein, but the combination of both TAT and NLS is more efficient (34). Therefore, we constructed pTriEx-3 Hygro-TAT-NLS-Cre, which encodes the 41 kDa fusion protein TAT–NLS–Cre recombinase (TAT–NLS–Cre) that includes an N-terminal histidine tag and is expressed in E.coli (Fig. 2A). We purified the fusion protein from a lysate of E.coli cells by Ni chelate affinity chromatography and checked the purity of the protein by SDS–PAGE (Fig. 2B, 41 kDa).
Confirmation of the localization and activity of TAT–NLS–Cre
To determine whether the TAT–NLS–Cre protein could be used to control the expression of siRNAs by the strategy described above, we first confirmed the localization and ability of TAT–NLS–Cre to catalyze recombination. After incubating HeLa S3 cells in medium supplemented with the TAT–NLS–Cre protein for the indicated times, we washed the cells twice with PBS, incubated them with trypsin in order to eliminate TAT–NLS–Cre attached to cell membranes and prepared cytosolic and nuclear fractions. Then we performed western blotting of these fractions with antibodies against Cre recombinase to confirm the localization of TAT–NLS–Cre. The results are shown in Figure 2C. It appeared that a 1 h incubation was sufficient to allow entry of TAT–NLS–Cre into nuclei. Incubation for 1 h increased the concentration of TAT–NLS–Cre in nuclei but no further increase was apparent after incubation for 2 h. TAT–NLS–Cre was detected in the nuclear fraction, whereas β-tubulin, which was used as a control for separation of the nuclear and cytosolic fractions, was detected in the cytosolic fraction, indicating that there was no detectable cytosolic contamination in the nuclear fraction.
To examine whether TAT–NLS–Cre retained the activity of Cre recombinase and could catalyze the recombination of loxP sites, we transfected HeLa S3 cells with iGL3BCre-On and incubated them with TAT–NLS–Cre or with dialysis medium under the conditions indicated above. Plasmids were then extracted from the cells and amplified by PCR. If recombination had occurred, the extracted plasmids should have had exactly the same structure as that of iGL3BloxP, yielding a PCR product of ∼400 bp (indicated by ‘Recombined’ in Fig. 3). If recombination had not occurred, the extracted plasmids should have retained the initial structure, yielding a PCR product of ∼1.2 kb (indicated by ‘Original’ in Fig. 3). As shown in Figure 3, PCR analysis of cells incubated with dialysis medium yielded a fragment of 1.2 kb, while PCR analysis of cells incubated with TAT–NLS–Cre yielded both 1.2 kb and 400 bp fragments. From these results we conclude that although we had not achieved a recombination efficiency of 100%, TAT–NLS–Cre did have the ability to enter cells, to find its way to the nucleus and to catalyze recombination.
The suppressive effect of iGL3BCre-On can be switched on by NLS–Cre in a dose-dependent manner
Finally, we examined whether we could control the expression of iGL3BCre-On using TAT–NLS–Cre recombinant protein. Four hours after co-transfecting HeLa S3 cells with siRNA expression vectors or the mock vector (pU6 control) together with expression vectors for firefly luciferase and Renilla luciferase, we added TAT–NLS–Cre or, as a control, TAT–NLS–EGFP, residual E.coli lysate, obtained by the same procedure as TAT–NLS–Cre, or dialysis medium (for the dialysis of TAT–NLS–Cre) to the medium. We then analyzed luciferase activities on the following day and calculated the luciferase activities relative to those obtained with the pU6 control (Fig. 4A, lane 1). As shown in Figure 4A, incubation of cells transfected with iGL3BCre-On with TAT–NLS–Cre (lane 10) resulted in ∼70% suppression of firefly luciferase activity.The corresponding suppression obtained with positive controls was 80–90% (Fig. 4A, lanes 5 and 6).
In contrast, the corresponding set of incubation experiments indicated that the addition of TAT–NLS–Cre to the pU6 empty vector (negative control) did not cause any suppression (lanes 1–3). Similarly, the addition of TAT–NLS–EGFP (lane 9) or residual E.coli lysate (lane 8) to the effector plasmid, iGL3BCre-On, showed no effect. Moreover, cells transfected with pU6iRed (an unrelated siRNA expression vector against DsRed2, lane 4) also showed no suppressive effect, indicating that the suppression of expression of the target gene was not due to a non-specific effect of dsRNA or TAT–NLS–Cre itself.
Finally, we examined whether the dose of TAT–NLS–Cre affects the switching of iGL3BCre-On by incubating iGL3BCre-On-transfected HeLa S3 cells with TAT–NLS–Cre at various concentrations. As shown in Figure 4C, increasing the concentration of TAT–NLS–Cre increased the efficiency of switching of iGL3BCre-On. We also incubated iGL3BCre-On-transfected HeLa S3 cells with TAT–NLS–EGFP at various concentrations as negative controls. TAT–NLS–EGFP had no suppressive effects on luciferase activity at any of the tested concentrations.
Taken together with the fact that TAT–NLS–Cre could successfully enter the nucleus and cause recombination between loxP sites, the present results clearly demonstrate that iGL3BCre-On had been switched on by the exogenously supplied TAT–NLS–Cre recombinant protein.
DISCUSSION
We have here described one of the first examples of the control of the expression of siRNA in a pol III system by Cre recombinase (43) and the first example using TAT–NLS–Cre recombinase fusion protein. We have demonstrated that our Cre-On siRNA expression vector was switched on only in the presence of Cre recombinase and that the gene suppressive activity depended on the concentration of Cre recombinase.
The use of Cre recombinase to control the expression of siRNA has several advantages. First, Cre recombinase can be expressed stably, and both tissue type- and cell type-specifically and its expression is heritable in mammals, such as mice (32,33,44,45). Thus, it should be possible to control the expression of siRNA in a tissue-specific manner using a pol III promoter such as the U6 promoter, whose expression is constitutive and ubiquitous. It has been reported that siRNA can also be expressed using a pol II system (22,46), which might allow tissue-specific expression. However, a pol III system might have some advantages as a consequence of its high level of activity (∼4 × 105 copies of the pol III transcript per cell) (47). Second, the expression of Cre can be regulated by tetracycline (45), a hormone (48), a steroid (49) and an interferon (50), if the Cre coding region is fused to a ligand-binding domain specific for the corresponding compound. Such regulation of expression of Cre would allow control of the level of expression of Cre and, as a consequence, of siRNA.
Transgenic Cre recombinase can be expressed in cells and organisms, but problems do occur. In this study, we first tried to express NLS–Cre encoded by pEF9-NLS-Cre inside HeLa S3 cells and to control the expression of the Cre-On siRNA expression vector, but we failed to observe the expected results. Possible reasons for this failure are as follows. Although the results presented in this work are highly reproducible, the results of co-transfection experiments using reporter plasmids and a NLS–Cre-expressing plasmid gave much larger error bars (not shown). Thus, trans effects between the pol II promoters of NLS–Cre (the EF-1α promoter) and reporters (pGL3 and pRL-RSV) on co-transfected plasmids had probably occurred (51).
Furthermore, since time is required for the expression of NLS–Cre and recombination of siRNA plasmids, it is possible that the reporter proteins (firefly and Renilla luciferases) might have been expressed before recombination could take place, masking any effects due to switching on the expression of siRNA. The reporter proteins might remain stable for some time and luciferase activities might not have been affected directly, even though no new corresponding mRNAs were produced. It is possible that selection of a suitable promoter for each experimental system and establishment of a cell line or transgenic animal that stably expresses NLS–Cre so that recombination can occur directly after transfection of siRNA expression vectors might overcome these problems. An alternative is to use NLS–Cre fused to the TAT peptide, as we demonstrated in the present study. TAT-fused recombinant proteins move rapidly into a wide range of cells, solving the problem of promoters and the time required for expression of NLS–Cre. Moreover, it is then no longer necessary to establish a suitable stable NLS–Cre-expressing cell line for each experimental system.
In conclusion, we have demonstrated that expression of siRNA under the control of a pol III promoter can be regulated by Cre recombinase. We were able to demonstrate delivery of Cre recombinase into nuclei after incubating cells in medium supplemented with TAT–NLS–Cre. Moreover, delivery should also be possible after establishment of stable Cre recombinase-expressing cell lines or transgenic organisms. The choice of delivery system will depend on the needs of each experiment. Although siRNA technology will not replace the conventional recombination-mediated knockout, it complements the conventional technology because analysis of housekeeping genes might become possible by spatio-temporal control of gene expression, as described in this paper. Collectively, our work has opened up new possibilities for using and controlling the use of siRNAs in research and therapeutics.
Acknowledgments
ACKNOWLEDGEMENTS
This study was partly supported by grants from the National Institute of Advanced Industrial Science and Technology (AIST), Japan. The authors wish to thank Dr D. Ow, Plant Gene Expression Center, California, for kindly providing plasmid pMM23.
REFERENCES
- 1.Fire A., Xu,S., Montgomery,M.K., Kostas,S.A., Driver,S.E. and Mello,C.C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- 2.Kawasaki H., Suyama,E., Iyo,M. and Taira,K. (2003) siRNAs generated by recombinant human Dicer induce specific and significant but target site-independent gene silencing in human cells. Nucleic Acids Res., 31, 981–987. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3.Provost P., Dishart,D., Doucet,J., Frendewey,D., Samuelsson,B. and Radmark,O. (2002) Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J., 21, 5864–5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang H., Kolb,F.A., Brondani,V., Billy,E. and Filipowicz,W. (2002) Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J., 21, 5875–5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fire A. (1999) RNA-triggered gene silencing. Trends Genet., 15, 358–363. [DOI] [PubMed] [Google Scholar]
- 6.Sharp P.A. (2001) RNA interference 2001. Genes Dev., 15, 485–490. [DOI] [PubMed] [Google Scholar]
- 7.Hammond S.M., Caudy,A.A. and Hannon,G.J. (2001) Post-transcriptional gene silencing by double-stranded RNA. Nat. Rev. Genet., 2, 110–119. [DOI] [PubMed] [Google Scholar]
- 8.Zamore P.D. (2001) RNA interference: listening to the sound of silence. Nature Struct. Biol., 8, 746–750. [DOI] [PubMed] [Google Scholar]
- 9.Caplen N.J., Parrish,S., Imani,F., Fire,A. and Morgan,R.A. (2001) Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc. Natl Acad. Sci. USA, 98, 9742–9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elbashir S.M., Harborth,J., Lendeckel,W., Yalcin,A., Weber,K. and Tuschl,T. (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature, 411, 494–498. [DOI] [PubMed] [Google Scholar]
- 11.Miyagishi M. and Taira,K. (2002) U6 promoter-driven siRNAs with four uridine 3′ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat. Biotechnol., 20, 497–500. [DOI] [PubMed] [Google Scholar]
- 12.Brummelkamp T.R., Bernards,R. and Agami,R. (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science, 296, 550–553. [DOI] [PubMed] [Google Scholar]
- 13.Kawasaki H. and Taira,K. (2003) Short hairpin type of dsRNAs that are controlled by tRNAVal promoter significantly induce RNAi-mediated gene silencing in the cytoplasm of human cells. Nucleic Acids Res., 31, 700–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paddison P.J., Caudy,A.A., Bernstein,E., Hannon,G.J. and Conklin,D.S. (2002) Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev., 16, 948–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee N.S., Dohjima,T., Bauer,G., Li,H., Li,M.J., Ehsani,A., Salvaterra,P. and Rossi,J. (2002) Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol., 20, 500–505. [DOI] [PubMed] [Google Scholar]
- 16.Sui G., Soohoo,C., Affar,el B., Gay,F., Shi,Y., Forrester,W.C. and Shi,Y. (2002) A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl Acad. Sci. USA, 99, 5515–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paul C.P., Good,P.D., Winer,I. and Engelke,D.R. (2002) Effective expression of small interfering RNA in human cells. Nat. Biotechnol., 20, 505–508. [DOI] [PubMed] [Google Scholar]
- 18.Yu J.Y., DeRuiter,S.L. and Turner,D.L. (2002) RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc. Natl Acad. Sci. USA, 99, 6047–6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carmell M.A., Zhang,L., Conklin,D.S., Hannon,J.G. and Rosenquist,T.A. (2003) Germline transmission of RNAi in mice. Nature Struct. Biol., 10, 91–92. [DOI] [PubMed] [Google Scholar]
- 20.Hasuwa H., Kaseda,K., Einarsdottir,T. and Okabe,M. (2002) Small interfering RNA and gene silencing in transgenic mice and rats. FEBS Lett., 532, 227–230. [DOI] [PubMed] [Google Scholar]
- 21.Rubinson D.A., Dillon,C.P., Kwiatkowski,A.V., Sievers,C., Yang,L., Kopinja,J., Zhang,M., McManus,M.T., Gertler,F.B., Scott,M.L. and Parijs,L.V. (2003) A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nature Genet., 33, 401–406. [DOI] [PubMed] [Google Scholar]
- 22.Xia H., Mao,Q., Paulson,H.L. and Davidson,B.L. (2002) siRNA-mediated gene silencing in vitro and in vivo. Nat. Biotechnol., 20, 1006–1010. [DOI] [PubMed] [Google Scholar]
- 23.Miyagishi M., Sumitomo,H., Miyoshi,H., Kawakami,Y. and Taira,K. (2004) Optimization of an siRNA-expression system with an improved hairpin and its significant suppressive effects in mammalian cells. J. Gene Med., in press. [DOI] [PubMed] [Google Scholar]
- 24.van de Wetering M., Oving,I., Muncan,V., Pon Fong,M.T., Brantjes,H., van Leenen,D., Holstege,F.C., Brummelkamp,T.R., Agami,R. and Clevers,H. (2003) Specific inhibition of gene expression using a stably integrated, inducible small-interfering-RNA vector. EMBO Rep., 4, 609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsukura S., Jones,P.A. and Takai,D. (2003) Establishment of conditional vectors for hairpin siRNA knockdowns. Nucleic Acids Res., 31, e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Czauderna F., Santel,A., Hinz,M., Fechtner,M., Durieux,B., Fisch,G., Leenders,F., Arnold,W., Giese,K., Klippel,A. and Kaufmann,J. (2003) Inducible shRNA expression for application in a prostate cancer mouse model. Nucleic Acids Res., 31, e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sternberg N. and Hamilton,D. (1981) Bacteriophage P1 site-specific recombination. I. Recombination between loxP sites. J. Mol. Biol., 150, 467–486. [DOI] [PubMed] [Google Scholar]
- 28.Abremski K., Hoess,R. and Sternberg, N (1983) Studies on the properties of P1 site-specific recombination: evidence for topologically unlinked products following recombination. Cell, 32, 1301–1311. [DOI] [PubMed] [Google Scholar]
- 29.Sauer B. (1987) Functional expression of the cre-lox site-specific recombination system in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol., 7, 2087–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Odell J., Caimi,P., Sauer,B. and Russell,S. (1990) Site-directed recombination in the genome of transgenic tobacco. Mol. Gen. Genet., 223, 369–378. [DOI] [PubMed] [Google Scholar]
- 31.Sauer B. and Henderson,N. (1988) Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc. Natl Acad. Sci. USA, 85, 5166–5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lakso M., Sauer,B., Mosinger,B.,Jr, Lee,E.J., Manning,R.W., Yu,S.H., Mulder,K.L. and Westphal,H. (1992) Targeted oncogene activation by site-specific recombination in transgenic mice. Proc. Natl Acad. Sci. USA, 89, 6232–6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orban P.C., Chui,D. and Marth,J.D (1992) Tissue- and site-specific DNA recombination in transgenic mice. Proc. Natl Acad. Sci. USA, 89, 6861–6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peitz M., Pfannkuche,K., Rajewsky,K. and Edenhofer,F. (2002) Ability of the hydrophobic FGF and basic TAT peptides to promote cellular uptake of recombinant Cre recombinase: a tool for efficient genetic engineering of mammalian genomes. Proc. Natl Acad. Sci. USA, 99, 4489–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Green M. and Loewenstein,P.M. (1988) Autonomous functional domains of chemically synthesized human immunodeficiency virus tat trans-activator protein. Cell, 55, 1179–1188. [DOI] [PubMed] [Google Scholar]
- 36.Frankel A.D. and Pabo,C.O. (1988) Cellular uptake of the tat protein from human immunodeficiency virus. Cell, 55, 1189–1193. [DOI] [PubMed] [Google Scholar]
- 37.Ford K.G., Darling,D., Souberbielle,B. and Farzaneh,F. (2000) Protein transduction: a new tool for the study of cellular ageing and senescence. Mech. Ageing Dev., 121, 113–121. [DOI] [PubMed] [Google Scholar]
- 38.Nagahara H., Vocero-Akbani,A.M., Snyder,E.L., Ho,A., Latham,D.G., Lissy,N.A., Becker-Hapak,M., Ezhevsky,S.A. and Dowdy,S.F. (1998) Transduction of full-length TAT fusion proteins into mammalian cells: TAT-p27Kip1 induces cell migration. Nature Med., 4, 1449–1452. [DOI] [PubMed] [Google Scholar]
- 39.Schwarze S.R., Hruska,K.A. and Dowdy,S.F. (2000) Protein transduction: unrestricted delivery into all cells? Trends Cell Biol., 10, 290–295. [DOI] [PubMed] [Google Scholar]
- 40.Taira K. and Miyagishi,M. (2001) siRNA expression system and method for producing functional gene knock-down cell using the system. Japanese patent application 2001-363385.
- 41.Miyagishi M. and Taira,K. (2003) Strategies for generation of an siRNA expression library directed against the human genome. Oligonucleotides, 13, 325–333. [DOI] [PubMed] [Google Scholar]
- 42.Miyagishi M., Fujii,R., Hatta,M., Yoshida,E., Araya,N., Nagafuchi,A., Ishihara,S., Nakajima,T. and Fukamizu,A. (2000) Regulation of Lef-mediated transcription and p53-dependent pathway by associating β-catenin with CBP/p300. J. Biol. Chem., 275, 35170–35175. [DOI] [PubMed] [Google Scholar]
- 43.Fritsch L., Martinez,L.A., Sekhri,R., Naguibneva,I., Gerard,M., Vandromme,M., Schaeffer,L. and Harel-Bellan,A. (2004) Conditional gene knock-down by CRE-dependent short interfering RNAs. EMBO Rep., 5, 178–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gu H., Marth,J.D., Orban,P.C., Mossmann,H. and Rajewsky,K. (1994) Deletion of a DNA polymerase b gene segment in T cells using cell type-specific gene targeting. Science, 265, 103–106. [DOI] [PubMed] [Google Scholar]
- 45.Utomo A.R.H., Nikitin,A.Y. and Lee,W.H. (1999) Temporal, spatial and cell type-specific control of Cre-mediated DNA recombination in transgenic mice. Nat. Biotechnol., 17, 1091–1096. [DOI] [PubMed] [Google Scholar]
- 46.Kato Y. and Taira,K. (2003) Expression of siRNA from a single transcript that includes multiple ribozymes in mammalian cells. Oligonucleotides, 13, 335–344. [DOI] [PubMed] [Google Scholar]
- 47.Weinberg R.A. and Penman,S. (1968) Small molecular weight monodisperse nuclear RNA. J. Mol. Biol., 38, 289–304. [DOI] [PubMed] [Google Scholar]
- 48.Metzger D., Clifford,J., Chiba,H. and Chambon,P. (1995) Conditional site-specific recombination in mammalian cells using a ligand-dependent chimeric Cre recombinase. Proc. Natl Acad. Sci. USA, 92, 6991–6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kellendonk C., Tronche,F., Monaghan,A.P., Angrand,P.O., Stewart,F. and Schütz,G. (1996) Regulation of Cre recombinase activity by the synthetic steroid RU 486. Nucleic Acids Res., 24, 1404–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuhn R., Schwenk,F., Aguet,M. and Rajewsky,K. (1995) Inducible gene targeting in mice. Science, 269, 1427–1429. [DOI] [PubMed] [Google Scholar]
- 51.Farr A. and Roman,A. (1992) A pitfall of using a second plasmid to determine transfection efficiency. Nucleic Acids Res., 20, 920. [DOI] [PMC free article] [PubMed] [Google Scholar]