Description
A 17-year-old man, the third child of physically normal parents, was diagnosed with a case of cleft hand (lobster hand) in our outpatient department during a routine visit. He was born with deformed hands and feet in the form of syndactyly of thumb with the index finger in the upper limbs. He had a median cleft of the hands (figures 1–3). There was no facial dysmorphism in the patient. He had a history of congenital heart disease (ventricular septal defect with aortic regurgitation) and seronegative hypothyroididsm. The two siblings of the index case were physically normal. Our patient most probably had autosomal mode of inheritance. The genetic analysis of the patient showed mutation in TP63 gene (R280C mutation) causing split hand/foot malformation syndrome (SHFM) type 4.
Figure 1.
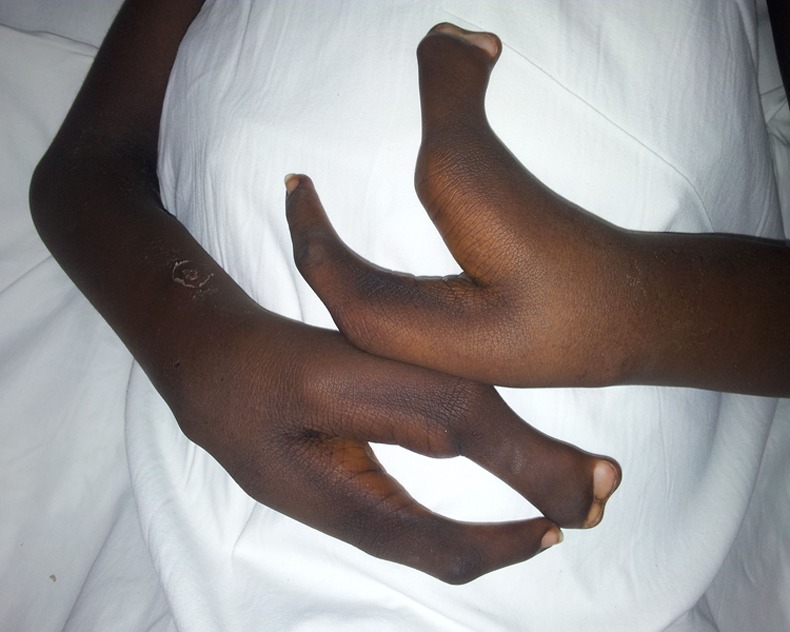
Image showing both claw hands.
Figure 2.
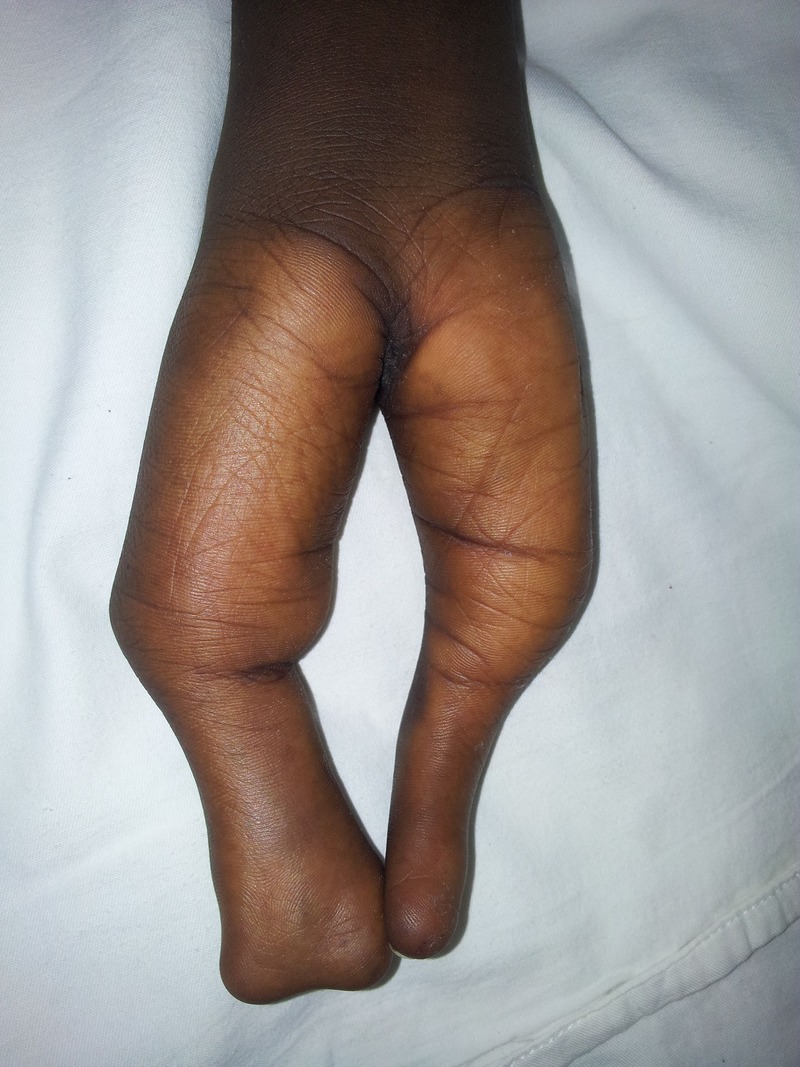
Image of the supine claw hand.
Figure 3 .
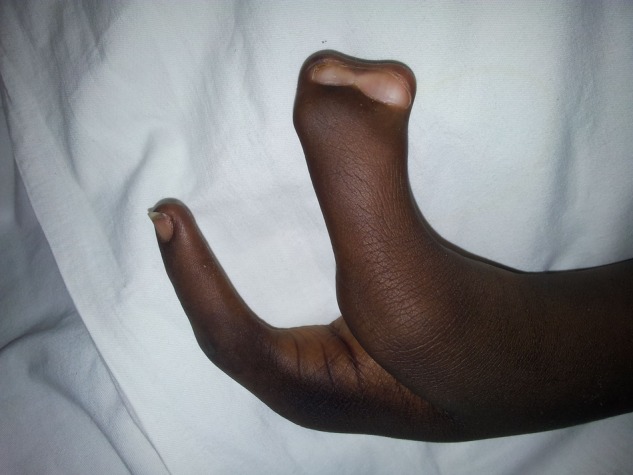
Image showing prone claw hand. Note the fusion of thumb and index finger and two distinct nail beds of the fused fingers.
Discussion
Ectrodactyly–ectodermal dysplasia–cleft syndrome is a rare orthopaedic malformation with incidence of around 1 in 90 000 live births.1 It has various names including split hand–split foot–ectodermal dysplasia–cleft syndrome or split hand, cleft hand or lobster claw hand/foot. It is called lobster claw hand/foot as there is a median cleft in upper and lower limbs because of the absence of central digital rays, giving the affected limbs the appearance of lobster claws.2 It is characterised by the deficiency or absence of one or more central digits of the hand or foot. It is thought to arise because of a wedge-shaped defect of the apical ectoderm of the limb buds.3 The associated anomalies include tibial aplasia, learning disabilities, ectodermal and craniofacial findings, orofacial clefting, renal abnormalities such as vesicoureteral reflux, recurrent urinary tract infection (UTI), missed or abnormal teeth, enamel hypoplasia and conductive hearing loss.4 5 There are several classifications for cleft hand but that described by Manske and Halikis is most commonly used.6 There are five types of SHFM syndrome based on the chromosomal associations and genes thought to be responsible for SHFM. Table 1 shows different types of SHFM with their chromosomal locations and the candidate genes responsible for them. The only proven mutations known to underlie SHFM in humans have been found in the TP63 gene, which encodes a homologue of the tumour-suppressor p53 gene.7 The p63 gene plays a very important role during embryonic development and does not behave like a tumour suppression gene. The TP63 gene has at least six different isoforms. Of these different isoforms a few are transcriptional activators similar to p53, whereas some other isoforms which have repressive activity towards p53-driven and p63-driven gene expressions have also been isolated.8 The first report of mutations in the TP63 gene was seen in patients with ectrodactyly-ectodermal dysplasia-cleft (EEC) syndrome9 followed by various case reports of TP63 mutations in patients with isolated ectrodactyly.10 11 Various types of TP63 mutations have been identified in non-syndromic SHFM families and isolated patients.11
Table 1.
Different types of Split hand/foot malformation (SHFM) with their chromosomal locations and candidate genes
| Disorder | Chromosomal location | Candidate gene(s) |
|---|---|---|
| SHFM1 | 7q21 | DLX5, DLX6, DSS1 |
| SHFM2 | Xq26 | FGF13, TONDU |
| SHFM3 | 10q24 | Dactylin, SUFU, BTRC |
| SHFM4 | 3q27 | TP63 |
| SHFM5 | 2q31 | DLX1, DLX2 |
These include:
K193E
K194E
Q634X
E639X
R280C
R280H
InsP154
The majority of these mutations lead to amino acid substitutions in the DNA-binding domain that are predicted to abrogate interaction with the DNA and, hence, lead to reduction in transactivation activity. The R280C and R280H mutations are usually associated with ectrodactyly, ectodermal dysplasia and cleft lip/palate (EEC) syndrome but have also been reported with SHFM syndrome.11
Learning points.
Patients with claw hand syndrome must be evaluated with ECHO, hearing screening and thyroid screening for early diagnosis and treatment.
Parents are to be counselled for malformations in the next pregnancy and regular antenatal scans are to be advised for the mother.
These patients should undergo genetic analysis to identify the mutations involved in the disease.
Footnotes
Competing interests: None.
Patient consent: Obtained.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1. Vadoma. http://en.wikipedia.org/wiki/Vadoma#column-one#column-one.
- 2. %183600 split-hand/foot malformation 1; SHFM1. http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?cmd=entryandid=183600.
- 3.Zlotogora J. On the inheritance of the split hand/split foot malformation. Am J Med Genet 1994;53:29–32 [DOI] [PubMed] [Google Scholar]
- 4. Kay SP, McCombe D. Central hand deficiences. In: Green DP, Pederson MD, Hotchkiss RN, Wolf SW, eds. Green's Operative Hand Surgery. 5th ed. Philadelphia: Elsevier Churchill Livingstone, 2005:1404–15.
- 5.Brunner HG, Hamel BC, Van Bokhoven H. The p63 gene in EEC and other syndromes. J Med Genet 2002;39:377–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manske PR, Halikis MN, Surgical classification of central deficiency according to the thumb web. J Hand Surg 1995;20:687–97 [DOI] [PubMed] [Google Scholar]
- 7.Duijf PH, van Bokhoven H, Brunner HG. Pathogenesis of split-hand/split-foot malformation. Hum Mol Genet 2003;12:51–60 [DOI] [PubMed] [Google Scholar]
- 8.Yang A, Kaghad M, Wang Y, et al. p63, a p53 homolog at 3q27–29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell 1998;2:305–16 [DOI] [PubMed] [Google Scholar]
- 9.Celli J, Duijf P, Hamel BC, et al. Heterozygous germline mutations in the p53 homolog p63 are the cause of EEC syndrome. Cell 1999;99:143–53 [DOI] [PubMed] [Google Scholar]
- 10.Ianakiev P, Kilpatrick MW, Toudjarska I, et al. Split-hand/split-foot malformation is caused by mutations in the p63 gene on 3q27. Am J Hum Genet 2000;67:59–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Bokhoven H, Hamel BC, Bamshad M, et al. p63 Gene mutations in eec syndrome, limb-mammary syndrome, and isolated split hand-split foot malformation suggest a genotype-phenotype correlation. Am J Hum Genet 2001;69:481–92 [DOI] [PMC free article] [PubMed] [Google Scholar]


