Abstract
Background & objectives:
CYP4F2 and γ-glutamyl carboxylase (GGCX) have small but significant roles in the maintenance dose of coumarinic oral anticoagulants (COAs). CYP4F2 1347 G>A and GGCX 12970 C>G polymorphisms have been used in the pharmacogenetic dosing algorithms of warfarin for Caucasians and Chinese populations. India has a large population with multiple ethnic groups but there are no reports about the frequencies of these polymorphisms in north Indians. In the present study, we aimed to find out the allelic frequencies of CYP4F2 1347 G>A and GGCX 12970 C>G polymorphisms in a north Indian population and relate these to daily maintenance drug dose requirements of COA.
Methods:
CYP4F2 1347 G>A and GGCX 12970 C>G polymorphisms were genotyped by polymerase chain reaction - restriction fragment length polymorphism (PCR-RFLP) protocols and Taqman SNP discrimination assays in healthy volunteers (n=102) and patients (n=225) receiving acenocoumarol, an oral anticoagulant, after cardiac valve replacement surgery.
Results:
In healthy volunteers, the allele frequencies for CYP4F2 1347 G>A and GGCX 12970 C>G were 43.14 and 1.43 per cent, respectively. No significant differences in mean weight normalized doses of acenocoumarol were found for these CYP4F2 and GGCX genotypes. Binary logistic regression analysis revealed no significant association of any of the genotypes or alleles with the dosing phenotypes for both the SNPs.
Interpretation & conclusions:
We report distinct frequencies of CYP4F2 1347 G>A and GGCX 12970 C>G polymorphisms in north Indians but these polymorphisms did not have significant bearing on maintenance dose of acenocoumarol oral anticoagulant in cardiac valve replacement patients.
Keywords: Allele frequency, anticoagulant, CYP2C9, GGCX, north Indians, pharmacogenetics, polymorphism
Warfarin, phenprocoumon and acenocoumarol are the most commonly prescribed oral anticoagulants (COA). In the last few years, there have been many reports linking the variability in the requirement of inter-individual warfarin dosage with polymorphisms and rare coding variants in vitamin K epoxide reductase complex subunit 1 (VKORC1) and cytochrome P450-2C9 (CYP2C9)1,2,3. The VKORC1 and CYP2C9 genes code for the enzymes involved in the pharmacodynamics and pharmacokinetics of warfarin, respectively. A small but significant association of CYP4F2 1347 G>A (rs2108622) single nucleotide polymorphism (SNP) in maintenance dose of oral anticoagulants has also been reported4,5,6.
CYP4F2 is a vitamin K cycle related enzyme that metabolizes vitamin K1 to hydroxyvitamin K1. Caldwell et al showed that an exonic polymorphism in CYP4F2 polymorphism explained ~2 per cent variation in the maintenance dose requirement of Warfarin in a Genome wide association study (GWAS) carried out on Caucasian patients7. Gamma glutamyl carboxylase (GGCX) is also a key component in vitamin K cycle and GGCX gene has been extensively studied for genetic variants that could affect warfarin dosing. One GGCX variant (rs11676382) had a small but significant effect on warfarin maintenance dose and it explained 2 per cent variance in warfarin dose in Caucasians8.
In the Indian context, not much has been reported about the frequency distribution of these polymorphisms and their association with doses of oral anticoagulants. HapMap data show wide ethnicity specific differences in the allelic distribution of these two polymorphisms9. Therefore, an attempt was made in the present study to establish the allele and genotype frequencies of CYP4F2 1347 G>A and GGCX 12970 C>G in a north Indian population. Another objective was to compare north Indian CYP4F2 1347 G>A and GGCX 12970 C>G minor allele frequencies with those of other populations in HapMap project data. Finally, the effect of variant genotypes on maintenance doses of acenocoumarol requirements was evaluated in cardiac valve replacement patients.
Material & Methods
The protocol of the study and procedures employed were reviewed and approved by the Institutional ethics committee of Sanjay Gandhi Post Graduate Institute of Medical Sciences (SGPGIMS), Lucknow, India. The study was carried out at the departments of Genetics and CVTS, SGPGIMS, and department of CTVS, King George's Medical University, Lucknow. Written informed consent was obtained from all the participants.
Subjects and DNA extraction: Healthy subjects were studied for the information of frequency distribution of the two SNPs and patients for analysing the association of genetic background with the acenocoumarol dosing phenotypes. The sample size was calculated by Quanto software (http://hydra.usc.edu/gxe). For this study, 102 unrelated healthy volunteers (males = 57, females = 45; mean age = 38.5 ± 9.9 yr) and 225 cardiac valve replacement patients receiving maintenance doses of acenocoumarol (males = 151, females = 74; mean age = 37.7 ± 1.3 yr) were selected. Patients had undergone surgery for aortic/mitral or double valve replacement. In patients, there were more number of cases of mitral valve regurgitation (n=146) followed by aortic (n=44) and double valve regurgitation (n=35). The patients were recruited consecutively over a period of about one and half year from March 2010 to August 2011. Only patients maintaining INR (International Normalized Ratio) values in the range of 2.0 to 3.5 for the three visits during three months were enrolled for this study. The three visits were consecutive and the minimum difference between the two PT-INR tests was two wk. All the participants were inquired about their place of residence for the last three generations, food habits and mother tongue (Hindi or related languages). Based on this information their north Indian ethnicity was confirmed. All recruited subjects belonged to northern Indian states of Uttar Pradesh, Bihar, Madhya Pradesh, Rajasthan, Punjab and Haryana.
From each participant, 3 ml blood sample was collected in ethylenediaminetetraacetic acid (EDTA) and standard salting out method was used to extract the genomic DNA from peripheral blood leukocytes10. DNA was quantified by NanoDrop Analyser (ND-1000) spectrophotometer (NanoDrop Technologies, Wilmington, USA). The ratio of absorbance at 260 and 280 nm of DNA was between 1.7 and 1.9. The isolated DNA was stored in Tris-EDTA buffer (pH 8.0) at -70°C.
CYP4F2 1347 G>A genotyping: Genotyping for CYP4F2 1347 G>A was performed by polymerase chain reaction (PCR) and restriction fragment length polymorphism (RFLP) according to Deng et al11. Genomic DNA was amplified by PCR using the forward (5′ATCAACCCGTTCCCACCT3′) and reverse (5′ATCAACCCGTTCCCACCT3′) primers. A 491 bp amplicon was generated in 25 μl PCR mix consisting of 10 mM Tris-HCl, pH 8.3, 1.25 mM MgCl2, 50mM KCl, 200 mM dNTPs, 0.2 mM of each of the primers, 2.5 U Taq polymerase (Bangalore Genei, Bangalore, India), and 1 µl (100 ng) of genomic DNA. PCR was performed by initial denaturation for 5 min at 94°C, 30 cycles of denaturation for 30 sec at 94°C, annealing for 30 sec at 59.8 °C, extension for 1 min at 72 °C and 10 min of extension at 72 °C. The PCR amplicon (10 µl PCR product) was digested with 2 units of restriction enzyme PvuII (Fermentas, MA, USA) overnight at 37°C. Ethidium bromide staining was used to visualize digested product on 2 per cent agarose gels. Presence of G nucleotide at position 1347 produced two restriction fragments of 313 and 178 bp. The mutant allele (A at 1347 nucleotide position) produced a single band of 491 bp in homozygous condition and three bands of 491, 313 and 178 bp in heterozygous condition.
GGCX 12970 C>G genotyping: Genotyping for GGCX 12970 C>G polymorphism was also carried out by PCR-RFLP protocol using the primers: forward, 5’GCTTCTTGTTGCGAAAGCTCTAT3’ and reverse, 5’CAAACACTTGGGAACAGTTAGCT3’8. The PCR conditions were similar to those used for CYP4F2 1347 G>A genotyping. PCR product obtained was of 1288 bp. The PCR amplicon (10 µl PCR product) was digested with 2 units of restriction enzyme HindIII (Fermentas, MA, USA) overnight at 37°C. Genotyping was performed by visualizing the ethidium bromide stained 2 per cent agarose gels. Wild type GGCX 12970 CC genotype showed two bands of 1206 and 82 bp. Mutant allele showed three bands of 1288, 1206, 82 bp in heterozygous and single band of 1288 bp in homozygous state. To confirm the PCR-RFLP genotyping, GGCX 12970 C>G polymorphism was also genotyped using Taqman SNP discrimination assays custom designed by Applied Biosystems (Applied Biosystems, Foster City, CA). Accumulation of specific PCR product by hybridization and cleavage of a double-labelled fluorogenic probe during amplification was detected by Taqman probe based 5’ nuclease PCR assay in ABI 7500 Real-time PCR. For quality control, 5 per cent dummy duplicates and blank controls were also taken up along with the samples in each experiment; 100 per cent concordance in results was found for the PCR-RFLP and Taqman probe based genotyping.
Statistical analysis: Genotype and allele frequencies in the study population were checked for Hardy-Weinberg equilibrium. Means of maintenance acenocoumarol doses for different genotypes were calculated along with standard deviations. For associating genotypes and alleles with drug dose requirements, all the patients were divided into three quartiles according to the weekly therapeutic doses: drug sensitive (0 - 0.28 mg/kg/wk), intermediate (0.29 - 0.40 mg/kg/wk) and resistant (0.41 mg/kg/wk and above). Binary logistic regression analysis was used to associate the genotypes and alleles with these classes. All statistical analysis was done by SPSS version 17.0 software (SPSS Japan, Tokyo, Japan).
Results
Allele and genotype frequencies of CYP4F2 1347 G>A polymorphism: The minor allele frequency for the CYP4F2 1347 G>A polymorphism was found to be 43.14 per cent in the healthy volunteers (n = 102). Of the 102 subjects, 18 were homozygous for CYP4F2 (1347 AA) and 52 subjects were heterozygous having both the alleles (1347 GA) (Table I). The genotype frequencies of CYP4F2 1347 G>A polymorphism were found to be in Hardy-Weinberg equilibrium.
Table I.
Genotype and allele frequencies of CYP4F2 1347 G>A and GGCX 12970 C>G in healthy north Indian population
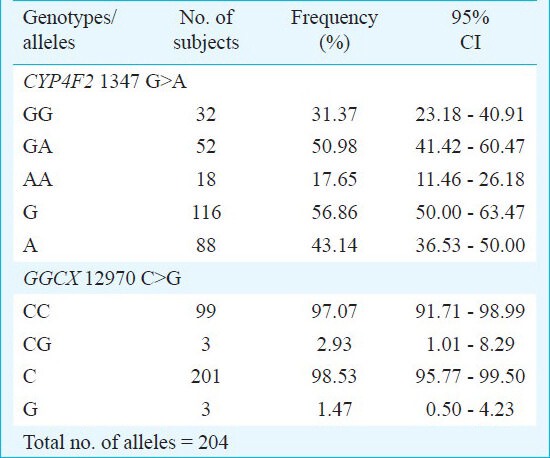
Allele and genotype frequencies of GGCX 12970 C>G polymorphism: None of the healthy volunteers were homozygous for the GGCX 12970 GG variant genotype. Of the 102 subjects, 99 subjects were homozygous (CC) and three were heterozygous (CG) for GGCX 12970 C>G polymorphism (Table I). The GGCX 12970 G mutant allele frequency in north Indians was found to be 1.47 per cent. The genotypes of GGCX 12970 G>A polymorphism also followed Hardy-Weinberg equilibrium.
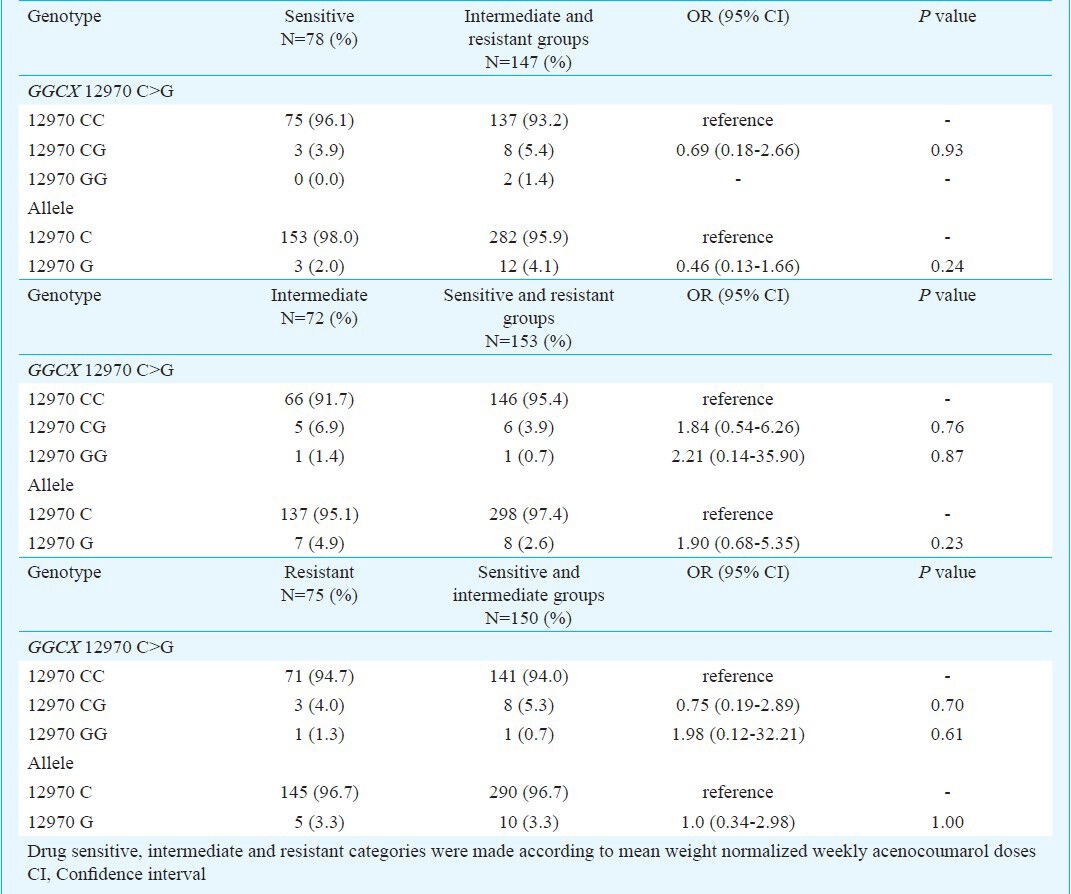
Association of genotypes and alleles with acenocoumarol maintenance dose requirements: In patients, CYP4F2 1347 G>A and GGCX 12970 C>G polymorphisms were related to acenocoumarol maintenance dose requirements at two levels. Firstly, the genotypes were compared with respect to mean weight normalized drug dose requirements. Of the 225 patients, 71, 112 and 42 were having CYP4F2 1347 GG, GA and AA genotypes, respectively. No significant differences in mean weight normalized acenocoumarol doses were found for these CYP4F2 genotypes (Table II). The numbers of patients with GGCX 12970 CC and CG genotypes were 211 and 14 and none of them carried variant GGCX 12970 G allele in homozygous condition. In this case also, the mean weight normalized acenocoumarol doses were comparable among wild type and mutant alleles (Table II). Secondly, binary logistic regression analysis revealed that none of the genotypes and alleles of CYP4F2 1347 G>A and GGCX 12970 C>G polymorphisms were significantly associated with drug sensitive (weekly dose: 0 - 0.28 mg/kg), intermediate (weekly dose: 0.29 - 0.40 mg/kg) and resistant patients (weekly dose: 0.41 mg/kg and above) (Tables III and IV).
Table II.
Relationship between CYP4F2 1347 G>A and GGCX 12970 C>G SNPs with maintenance dose of acenocoumarol in patients receiving maintenance drug doses
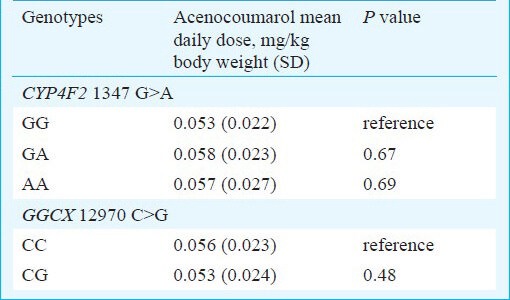
Table III.
Association between CYP4F2 1347 G>A polymorphism and acenocoumarol sensitive/resistant/intermediate dose groups in patients receiving maintenance drug doses
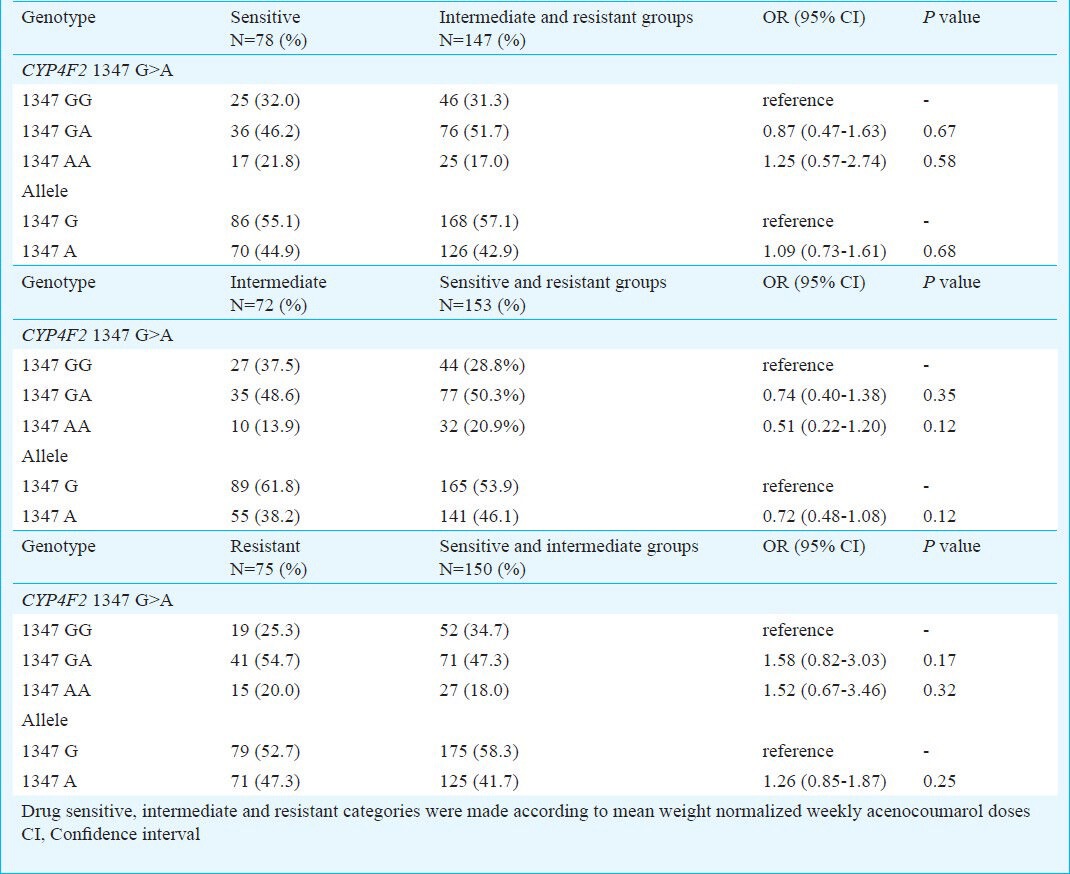
Table IV.
Association between GGCX 12970 C>G polymorphism and acenocoumarol sensitive/resistant/intermediate dose groups in patients receiving maintenance drug doses.
Discussion
The CYP4F2 1347 G>A minor allele frequency in the north Indian population (43.14%) was significantly different from Caucacians, Africans and East Asians. The frequency of CYP4F2 1347 A allele was closest to that in Gujarati Indians of Texas, USA. East Asians (i.e. Han Chinese and Japanese) and Caucasians have nearly similar frequencies of about 23 to 21 per cent for the CYP4F2 1347 A allele. Africans have the least CYP4F2 1347 A allele frequency of 5.1 per cent and largely differed from north Indians (Table V)9. In a recent study, Zeng et al12 reported the CYP4F2 1347 A allele frequency to be 25 per cent for Bai and Tibetan populations. The CYP4F2 1347 G>A minor allele frequency was 4.4, 8.7 and 24 per cent in American Indians, Mozambicans and Brazilians, respectively13.
Table V.
Minor allele frequencies of CYP4F2 1347 G>A and GGCX 12970 C>G polymorphism in north Indian and the HapMap selected populations9
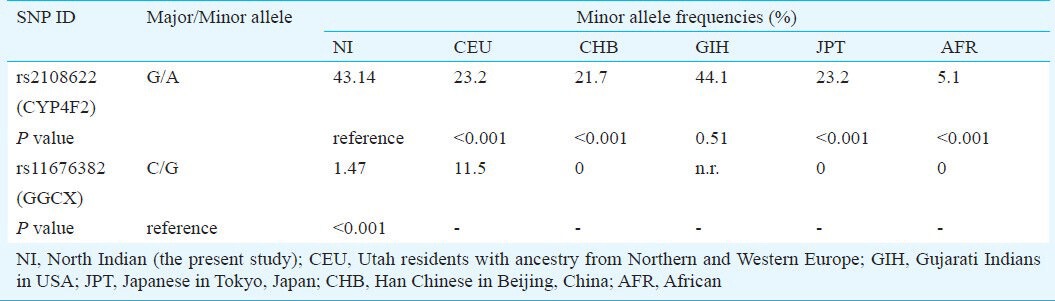
The frequency of GGCX 12970 C>G minor allele was also compared with five major populations from the HapMap project9. The GGCX 12970 C>G minor allele frequency was 0 per cent in East Asians and Africans. Caucasians have a higher frequency of 11.5 per cent. There are no reports about this polymorphism for Gujarati Indians in HapMap data. The GGCX 12970 C>G minor allele frequency in north Indians (1.47%) was found to be different than that in Caucasian population.
Effect of CYP4F2 1347 G>A and GGCX 12970 C>G on maintenance dose of coumarinic oral anticoagulant: Previous studies have shown that VKORC1-1639 G>A, CYP2C9*2, *3 genotypes, clinical (e.g., age, weight and body surface area) and environmental (e.g. concomitant medications and diet) factors explain approximately 50 per cent variation in maintenance dose requirements of oral anticoagulants2,3,14. However, individual contribution of all these genetic and non-genetic factors varies among different ethnic populations. Therefore, these variables have been modeled into ethnicity specific algorithms for better prediction of therapeutic dose of oral anticoagulants3,15,16,17. Earlier reports show that in Caucasian patients, CYP4F2 1347 G>A and GGCX 12970 C>G polymorphisms have very small effects on maintenance dose requirements of oral anticoagulant warfarin8,18. However, interethnic differences in allelic distribution suggest that these markers might have a role in the populations where these are more frequent and thereby affect the performance of COA dosing algorithms. For example, results of this study show a high prevalence of variant allele in case of CYP4F2 1347 C>G polymorphism in north India. Recently, we have generated a pharmacogenetic dosing algorithm for acenocoumarol and have shown that this allele explains about 3.5 per cent of variability in maintenance dose requirements. GGCX 12970 C>G polymorphism has lesser contribution in our north India specific dosing algorithm (~1.7%)19. The small contributions were expected due to very low frequency in case of GGCX. CYP4F2 is not directly involved in the vitamin K cycle; therefore, instead of high frequency its contribution is lower in the dosing algorithm. We observed a small contribution of CYP4F2 1347 G>A and GGCX 12970 C>G polymorphisms in our pharmacogenetic dosing algorithm19. COA dosing algorithms containing these two markers have been proposed for other populations also. For example, clinical and genetic factors (including CYP4F2 1347 G>A polymorphism) explain about 42 and 43.4 per cent of variance in warfarin maintenance dose in Han-Chinese and Japanese patients, respectively6,20. A multivariate regression model based on VKORC1, CYP2C9, CYP4F2 and clinical factors explained 58 per cent of dosage variance in Caucasians21. About 2 per cent of total variance in warfarin dose is explained by GGCX 12970 C>G polymorphism in the Caucasians8.
In conclusion, the genotypes and alleles of CYP4F2 1347 G>A and GGCX 12970 C>G polymorphisms did not show significantly association with the maintenance dose requirements of acenocoumarol in the binary logistic regression analysis in patients with cardiac valve replacement. However, multi-ethnic nature of Indian population suggests for more such studies in different parts of the country. This will help scientists and clinicians to develop better treatment protocols for individual patients based on their genetic makeup.
Acknowledgment
Authors acknowledge the research grant support from Department of Biotechnology, Government of India, New Delhi.
References
- 1.Wadelius M, Pirmohamed M. Pharmacogenetics of warfarin: current status and future challenges. Pharmacogenomics J. 2007;7:99–111. doi: 10.1038/sj.tpj.6500417. [DOI] [PubMed] [Google Scholar]
- 2.Limdi NA, Veenstra DL. Warfarin pharmacogenetics. Pharmacotherapy. 2008;28:1084–97. doi: 10.1592/phco.28.9.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schelleman H, Limdi NA, Kimmel SE. Ethnic differences in warfarin maintenance dose requirement and its relationship with genetics. Pharmacogenomics. 2008;9:1331–46. doi: 10.2217/14622416.9.9.1331. [DOI] [PubMed] [Google Scholar]
- 4.Takeuchi F, McGinnis R, Bourgeois S, Barnes C, Eriksson N, Soranzo N, et al. A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet. 2009;5:e1000433. doi: 10.1371/journal.pgen.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teichert M, van Schaik RH, Hofman A, Uitterlinden AG, de Smet PA, Stricker BH, et al. Genotypes associated with reduced activity of VKORC1 and CYP2C9 and their modification of acenocoumarol anticoagulation during the initial treatment period. Clin Pharmacol Ther. 2009;85:379–86. doi: 10.1038/clpt.2008.294. [DOI] [PubMed] [Google Scholar]
- 6.Cha PC, Mushiroda T, Takahashi A, Kubo M, Minami S, Kamatani N, et al. Genome-wide association study identifies genetic determinants of warfarin responsiveness for Japanese. Hum Mol Genet. 2010;19:4735–44. doi: 10.1093/hmg/ddq389. [DOI] [PubMed] [Google Scholar]
- 7.Caldwell MD, Awad T, Johnson JA, Gage BF, Falkowski M, Gardina P, et al. CYP4F2 genetic variant alters required warfarin dose. Blood. 2008;111:4106–12. doi: 10.1182/blood-2007-11-122010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rieder MJ, Reiner AP, Rettie AE. Gamma-glutamyl carboxylase (GGCX) tagSNPs have limited utility for predicting warfarin maintenance dose. J Thromb Haemost. 2007;5:2227–34. doi: 10.1111/j.1538-7836.2007.02744.x. [DOI] [PubMed] [Google Scholar]
- 9.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 10.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng S, Zhu G, Liu F, Zhang H, Qin X, Li L, et al. CYP4F2 gene V433M polymorphism is associated with ischemic stroke in the male Northern Chinese Han population. Prog Neuropsychopharmacol Biol Psychiatry. 2010;34:664–8. doi: 10.1016/j.pnpbp.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 12.Zeng WT, Zheng QS, Huang M, Cen HJ, Lai Y, Chen WY, et al. Genetic polymorphisms of VKORC1, CYP2C9, CYP4F2 in Bai, Tibetan Chinese. Pharmazie. 2012;67:69–73. [PubMed] [Google Scholar]
- 13.Vargens DD, Damasceno A, Petzl-Erler ML, Suarez-Kurtz G. Combined CYP2C9, VKORC1 and CYP4F2 frequencies among Amerindians, Mozambicans and Brazilians. Pharmacogenomics. 2011;12:769–72. doi: 10.2217/pgs.11.35. [DOI] [PubMed] [Google Scholar]
- 14.Hirsh J, Dalen J, Anderson DR, Poller L, Bussey H, Ansell J, et al. Oral anticoagulants: mechanism of action, clinical effectiveness, and optimal therapeutic range. Chest. 2001;119(Suppl 1):8S–21S. doi: 10.1378/chest.119.1_suppl.8s. [DOI] [PubMed] [Google Scholar]
- 15.Gage BF, Eby C, Johnson JA, Deych E, Rieder MJ, Ridker PM, et al. Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clin Pharmacol Ther. 2008;84:326–31. doi: 10.1038/clpt.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu AH, Wang P, Smith A, Haller C, Drake K, Linder M, et al. Dosing algorithm for warfarin using CYP2C9 and VKORC1 genotyping from a multi-ethnic population: comparison with other equations. Pharmacogenomics. 2008;9:169–78. doi: 10.2217/14622416.9.2.169. [DOI] [PubMed] [Google Scholar]
- 17.Klein TE, Altman RB, Eriksson N, Gage BF, Kimmel SE, Lee MT, et al. International Warfarin Pharmacogenetics Consartium. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009;360:753–64. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cooper GM, Johnson JA, Langaee TY, Feng H, Stanaway IB, Schwarz UI, et al. A genome-wide scan for common genetic variant with a large influence on warfarin maintenance dose. Blood. 2008;112:1022–7. doi: 10.1182/blood-2008-01-134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rathore SS, Agarwal SK, Pande S, Singh SK, Mittal T, Mittal B. Therapeutic dosing of acenocoumarol: proposal of a population specific pharmacogenetic dosing algorithm and its validation in north indians. PLoS One. 2012;7:e37844. doi: 10.1371/journal.pone.0037844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang R, Li L, Li C, Gao Y, Liu W, Hu D, et al. Impact of CYP2C9*3, VKORC1-1639, CYP4F2rs2108622 genetic polymorphism and clinical factors on warfarin maintenance dose in Han-Chinese patients. J Thromb Thrombolysis. 2012;34:120–5. doi: 10.1007/s11239-012-0725-7. [DOI] [PubMed] [Google Scholar]
- 21.Wells PS, Majeed H, Kassem S, Langlois N, Gin B, Clermont J, et al. A regression model to predict warfarin dose from clinical variables and polymorphisms in CYP2C9, CYP4F2, and VKORC1: Derivation in a sample with predominantly a history of venous thromboembolism. Thromb Res. 2010;125:e259–64. doi: 10.1016/j.thromres.2009.11.020. [DOI] [PubMed] [Google Scholar]


