Abstract
Background & objectives:
Carbapenemase-producing Enterobacteriaceae isolates have been increasingly identified worldwide. Though molecular data regarding New Delhi metallo-beta-lactamase-1 (NDM-1) producers are available, data regarding their rate of infection in a hospital setting and percentage among different clinical isolates are scarce. Hence, this study was undertaken to determine the occurrence of blaNDM-1 gene among clinical isolates of multidrug resistant Gram-negative bacilli (MDRGNB) in a tertiary care centre in Bangalore, Karnataka, India.
Methods:
A total of 74 MDRGNB isolates were studied. These were screened for MBL production by phenotypic assays such as double disk synergy test (DDST) and Modified Hodge's test (MHT). PCR was performed for the molecular detection of the gene and antibiograms were confirmed by automated bacteriology system.
Results:
Of the 74 MDRGNB isolates, 34 were positive for blaNDM-1 gene. All isolates were resistant to aztreonam and two isolates were resistant to tigecycline. Complete resistance to the tested carbapenems was seen in 28 (82.35%) of the positive isolates whereas variable carbapenem resistance was seen in six (17.64%) of the positive clinical isolates. Of the total 34 PCR positive isolates, 33 (97.05%) NDM-1 producers were identified by DDST and 26 (76.47%) by MHT as producers of MBL.
Interpretation & conclusions:
A high percentage of plasmid encoded NDM was noted in MDRGNB. Phenotypic and molecular screening should be employed along with routine antimicrobial susceptibility testing to reflect the true number of metallo-beta-lactamase producers.
Keywords: Double disk synergy test, Modified Hodge's test, multidrug resistant Gram-negative bacteria, New Delhi metallo β lactamase-1
In 1965, the first report of a plasmid-encoded beta-lactamase in a Gram-negative bacterium appeared from Greece1. Metallo-beta-lactamases are enzymes that break down beta lactam drugs2. New Delhi metallo β lactamase-1 is a type of carbapenemase produced by certain strains of bacteria, and is able to inactivate all β-lactams except aztreonam3. Most NDM-1-positive strains also express the CMY-4 and CTX-M-15 β-lactamases, which confer resistance to all β-lactams. Thus it provides resistance against all compounds that contain a beta-lactam ring such as penicillins, cephalosporins, and the carbapenems. NDM-1 enzyme has been found clinically in Enterobacteriaceae and Acinetobacter baumannii4.
The gene that encodes for NDM-1 is called blaNDM-1 and has been identified on bacterial chromosomes and plasmids. Plasmids carrying the blaNDM-1 gene also carry a number of other genes conferring resistance to all aminoglycosides, macrolides, and sulphamethoxazole, thus making these isolates multidrug resistant or, because of other non-plasmid-mediated resistances, resistant in some cases to all antibiotics5. Plasmids carrying the gene for this carbapenemase, blaNDM-1, can have up to 14 other antibiotic resistance determinants and can transfer this resistance to other bacteria, resulting in multidrug-resistant or extremely drug-resistant phenotypes. blaNDM-1 gene has been reported to be carried on plasmids ranging from 140 to 400 kb in most isolates6.
In view of the increasing reports of NDM-1 producing strains from India and around the world7,8,9,10, the present work was conducted to examine the occurrence of NDM-1 gene among nosocomial isolates of MDRGNB from a tertiary care centre in southern India.
Material & Methods
Bacterial isolates: The total number of patients in this study was 61. In some cases more than one type of clinical sample (blood, urine, tracheal aspirate) was drawn from the same patient, thereby, giving a total of 70 clinical samples. The repeat samples from the same patient giving the same culture were counted for only once and the repeat samples of the same type from the same patient giving different cultures were counted individually as this indicated newly acquired infections. The 70 clinical samples on culture yielded 74 MDRGNB, due to the growth of two different isolates each from four of the clinical samples. The 74 MDRGNB were isolated based on conventional laboratory diagnostic techniques during September 2011 to February 2012 in the Neuromicrobiology department, National Institute of Mental Health and Neuro Sciences (NIMHANS), Bangalore, India. The Gram-negative isolates resistant to 1st and 2nd line antibiotics by standard disk diffusion test11 were considered multidrug resistant, stocked in semisolid agar tubes and used for further characterization. The following antibiotics (concentration/disk in µg) were considered; ampicillin (10), amikacin (30), gentamycin (10), ciprofloxacin (5), ofloxacin (5), cefotaxime (30), ceftriaxone (30), ceftazidime (30), imipenem (10), cefoperazone+sulbactam (75/10), piperacillin+tazobactam (100/19), aztreonam (30), cefipime (30), tobramycin (19) (Hi-Media, Mumbai, India). The isolates were further identified at the species level and antibiograms generated by Vitek 2 Compact 60 (bioMerieux, Germany) automated bacteriology system.
Phenotypic assay: Modified Hodge's test (MHT) was performed according to the standard Clinical and Laboratory Standards Institute (CLSI) guidelines for the detection of carbapenemase in Enterobacteriaceae11; 0.5 McFarland of negative control E.coli ATCC 25922 was uniformly swabbed onto Muller Hinton Agar (MHA) and test isolate was streaked as a straight line from the edge of the imipenem (Ipm) disk (10 µg), to the edge of the plate. An indentation in the growth of the negative control towards the imipenem disk on either side of the test isolate was considered as positive for the production of beta lactamase (BL+ve) by the test isolate. Klebsiella pneumoniae BAA 2156 was used as positive control.
Double disk synergy test (DDST) was performed using two Ipm disks (10 µg), one containing 10 μl of 0.1 M (292 μg) anhydrous EDTA (Sigma Chemicals, St. Louis, MO). Disks were placed 25 mm apart and an increase in zone diameter of > 4 mm around the IPM-EDTA disk compared to that of the IPM disk alone was considered positive for MBL production12. These isolates were considered to be of the MBL+ve phenotype.
Detection of NDM-1 gene: Plasmid DNA was extracted from all 74 isolates by alkaline lysis method and 1µl of the isolated DNA was subjected to PCR with specific primers; NDM - Forward (5’-GGGCAGTCGCTTCCAACGGT) and NDM-Reverse (5’-GTAGTGCTCAGTGTCGGCAT) that amplified 475bp internal fragment of the gene13. K. pneumoniae ATCC BAA2156 was used as a positive control. DNA fragments were visualized by electrophoresis on 2 per cent agarose gel at 100 V for 1 h in 1X Tris acetate EDTA (TAE) containing 0.05 mg/l ethidium bromide. The samples were run alongside a 100 bp ladder that served as a molecular weight marker and an amplified product corresponding to 475 bp was considered positive (Figure A & B). The PCR amplicons including that of the positive control strain were sent for sequencing to Eurofins Genomics Pvt. Ltd., Bangalore, India.
Fig. A.
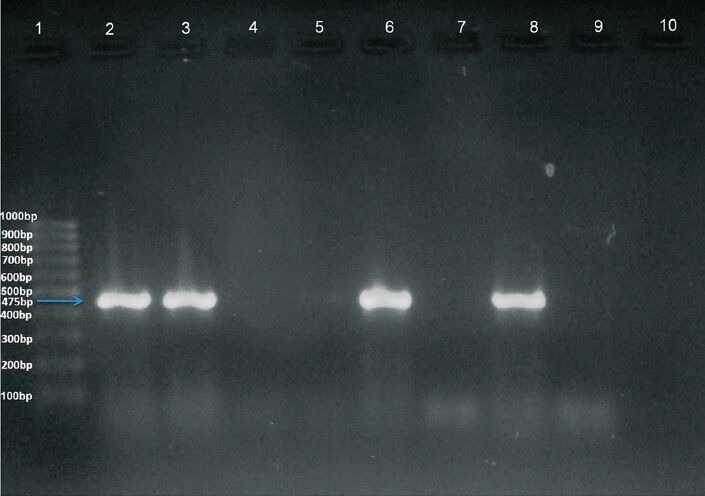
Amplified DNA after PCR with specific primers. Lane 1: 100 bp ladder. Lane 2: Positive control (Klebsiella pneumoniae BAA2156). Lanes 3, 6 and 8: Clinical isolates showing positive result. Lanes 4, 5, 7 and 9: Clinical isolates showing negative result. Lane 10: negative control (Escherichia coili 25922).
Fig. B.
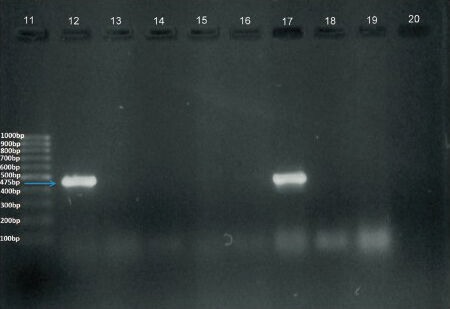
Amplified DNA after PCR with specific primers. Lane 11: 100 bp ladder. Lane 12: Positive control (Klebsiella pneumoniae BAA2156). Lanes 13 to 16, 18, 19: Clinical isolates showing negative result. Lanes 17: Clinical isolates showing positive band. Lane 20: (Escherichia coili 25922).
The study protocol was approved by the ethics committee of NIMHANS, Bangalore.
Statistical analysis: All data were analyzed using SPSS 16.0 statistical software (SPSS Inc., Chicago, Illinois, USA). Chi-square test was done and P< 0.05 was considered significant.
Results
During the study period, 4976 samples were screened and of these 74 (1.48%) were MDRGNB. Of the 74 MDRGNB isolates, 34 were positive for blaNDM-1 gene by PCR. Sequencing of all 34 PCR amplicons showed that it was part of the blaNDM-1 gene. Phenotypically MBL production was seen in 69 of 74 (93.24%) of the MDR isolates. Of the 34 PCR positive isolates, 33 (97.05%) of NDM-1 producers were identified by DDST, 26 (76.47%) by MHT and all (100%) were flagged by Vitek 2 Compact 60 as producers of MBL (Table I).
Table I.
Comparison of conventional and automated phenotypic assay

All 34 PCR positive isolates were resistant to aztreonam among which two isolates were also resistant to tigecycline. The number of positive isolates completely resistant to the tested carbapenems was 28 of 34 (82.35%) whereas six (17.64%) isolates showing variable carbapenem resistance. Respiratory tract infection (RTI) and urinary tract infection (UTI) were the two main types of infections observed in the study. NDM positive MDRGNB caused 33.33 per cent (13/39) of the total RTIs and 60 per cent (9/15) of the total UTIs.
Of the 70 clinical samples, 32 grew Gram-negative bacteria positive for the gene. The different clinical samples were tracheal aspirate (n=39), urine (n=15), blood (n=2), central line tips (n=3), pus (n=2), wound swabs (n=4), CSF (n=3) and subgaleal collection (n=2). The different clinical isolates cultured from these samples were Acinetobacter baumannii (n=20), Proteus vulgaris (n=2), Pseudomonas aeruginosa (n=4), Alcaligenes fecalis (n=1), Citrobacter freundii (n=1), Enterobacter cloacae (n=4), Roultella ornitholytica (n=1), Pseudomonas putida (n=1), Providencia rettgeri (n=19), Escherichia coli (n=10), Klebsiella pneumoniae (n=10) and Burkholderia cepacia (n=1) (Table II).
Table II.
Type and percentage occurrence of different multidrug resistant strains isolated with the blaNDM-1 gene
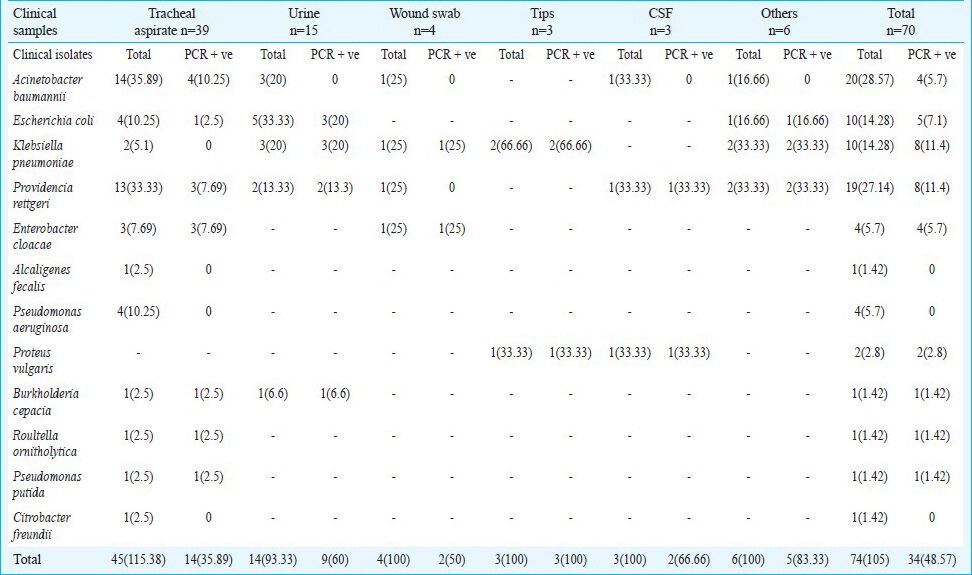
The blood samples (n=2) yielded two NDM positive P. rettgeri. The subgaleal collection (n=2) yielded A. baumannii negative for NDM-1 and K. pneumoniae positive for the gene. The pus samples (n=2) yielded NDM positive K. pneumoniae and E. coli.
The mean age of the 61 patients was 32.40 yr (3-75yr) with 39 (63.93%) males and 22 (36%) females. The rate of infection was 16/22 (72.7%) in females, which was significantly (P< 0.05) higher than males 15/39 (38.46%).
The infection due to MDRGNB resolved in 58 of the 61 patients. The remaining three patients succumbed to death.
Discussion
Non lactose fermenting Gram-negative bacteria such as Providencia rettgeri and Acinetobacter baumannii, widely associated with long term care facilities, were the most common organisms isolated in this study. Respiratory tract infections were the most common infections observed being caused by multidrug resistant P. rettgeri and A. baumannii. Urinary tract infections formed 21.42 per cent of the infections observed in the study. Multidrug resistant E. coli was the major causative organism of UTIs followed by A. baumannii 3/15 (20%) and K. pneumonia. An isolate of K. pneumoniae co-producing NDM-1 with Klebsiella pneumonia carbapenemase-2 (KPC-2), showing wide-spectrum resistance to β-lactams, aminoglycosides, fluoroquinolones, co-trimoxazole, nitrofurantoin and tigecycline has been reported in a hospital in Chennai14. All K. pneumoniae isolates and 60 per cent of the E. coli isolates causing UTI harboured the gene. However, the gene was absent in the A. baumannii isolates causing UTIs. Patients with long term indwelling catheters such as in the ICUs often have been reported to have UTIs caused by E. coli, P. aeruginosa, Proteus mirabilis, Providencia stuartii, Morganella morganii and A. baumannii15.
Of the three CSF samples, two that showed the growth of MDRGNB were positive for the gene. A MDR R. ornitholytica isolate from tracheal aspirate and a B. cepacia isolate from urine sample were positive for the gene. Pseudomonas putida harbouring the gene has been obtained from sewage sample5. However, in our study a MDR P. putida isolate with NDM-1 gene was isolated from tracheal aspirate.
In the present study, 48.57 per cent isolates were positive for the gene by PCR, of which 11.4 per cent were P. rettgeri and K. pneumoniae each. In a study4 conducted in India, the percentage of Enterobacteriaceae harbouring the gene was 30 per cent in Chennai, 13 per cent in Haryana and 44 per cent in the UK with E. coli and K. pneumoniae being in majority3. In our study, the 14.7 per cent of E. coli isolates were NDM-1 positive. While the MHT has been regarded non specific for the detection of metallo-beta-lactamases16, in the present study MHT detected 26 of the 34 NDM-1 positive isolates. The DDST detected 33 of the 34 isolates that were positive for the gene. However, the isolate that was missed by DDST was detected as a producer of beta-lactamase by MHT and Vitek 2 C 60. Therefore, both the phenotypic assays bear significance in the detection of MBLs and help in the preliminary screening of the NDM-1 positive genotypes.
Variable carbapenem resistance was seen in 17.64 per cent of the NDM-1 positive isolates, being susceptible to one of the carbapenems tested or of the intermediate susceptibility type. However, 82.35 per cent were completely resistant to the tested carbapenems. In a study conducted from four local general hospitals in Singapore, the isolates were completely resistant to the second- and third-generation cephalosporins tested as well as carbapenems17. Most of the NDM-1 producers have been reported to remain susceptible only to colistin and tigecycline. However, a high tigecycline resistance of 43.18 per cent has also been reported4. In our study, two isolates were resistant to tigecycline and all 34 were resistant to aztreonam indicating the presence of other resistance mechanisms.
The 61 subjects were on combination drugs and in 58 the infection resolved eventually. The repeated culture of their clinical samples after treatment yielded no growth. This study had certain limitations. The presence of only plasmid encoded MBL was studied. Other metallo-beta-lactamases such as VIM1/VIM2 and carbapenemases such as KPCs were not targeted in the study. Further sequencing of complete plasmids and pulse field gel electrophoresis (PFGE) profiling may be required to better understand the genetic relatedness and molecular epidemiology of these isolates. The accumulation of a number of critically ill patients within a relatively small area and use of sophisticated invasive machinery such as ventilators and catheters increase the proportion of patients who are unusually susceptible to infection and who become reservoirs for their spread. Rapid spread of carbapenem-resistant enterobacterial species in a hospital in Mumbai has been reported18.
The NDM-1 gene spreading in Enterobacteriaceae is an alarming risk because these novel multidrug-resistant bacteria could disseminate worldwide very quickly thus putting an end to our current pharmacopoeia. Early identification of cases of NDM-related infections and prevention of their spread by implementing screening, hygiene measures and isolation of the carriers are needed. Therefore, their detection along with routine antimicrobial susceptibility test (AST) should be performed. Phenotypic tests though specific, do not differentiate between chromosomal and plasmid encoded genes and hence genotypic characterization should be considered.
Acknowledgment
The authors thank Dr K.K. Kumaraswamy for kindly providing the primer sequence and Dr K. Thennarasu, Professor, Department of Biostatistics, NIMHANS, Bangalore for statistical analysis and Eurofins Genomics Pvt Ltd, Bangalore, for sequencing.
Footnotes
Conflict of interest: None.
References
- 1.Datta N, Kontomichalou P. Penicillinase synthesis controlled by infectious R factors in Enterobacteriaceae. Nature. 1965;208:239–41. doi: 10.1038/208239a0. [DOI] [PubMed] [Google Scholar]
- 2.Carfi A, Pares S, Duée E, Galleni M, Duez C, Frère JM, et al. The 3-D structure of a zinc metallo-beta-lactamase from Bacillus cereus reveals a new type of protein fold. EMBO J. 1995;14:4914–21. doi: 10.1002/j.1460-2075.1995.tb00174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walsh TR, Toleman MA, Poirel L, Nordmann P. Metallo-β-lactamases: the quiet before the storm? Clin Microbiol Rev. 2005;18:306–25. doi: 10.1128/CMR.18.2.306-325.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karthikeyan K, Thirunarayan MA, Krishnan P. Coexistence of blaOXA-23 with blaNDM-1 and armA in clinical isolates of Acinetobacter baumannii from India. J Antimicrob Chemother. 2010;65:2253–4. doi: 10.1093/jac/dkq273. [DOI] [PubMed] [Google Scholar]
- 5.Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis. 2010;10:597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walsh TR, Weeks J, Livermore DM, Toleman MA. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis. 2011;11:355–62. doi: 10.1016/S1473-3099(11)70059-7. [DOI] [PubMed] [Google Scholar]
- 7.Poirel L, Yilmaz M, Istanbullu A, Arslan F, Mert A, Bernabeu S, et al. Spread of NDM-1-producing Enterobacteriaceae in a neonatal intensive care unit, Istanbul, Turkey. Antimicrob Agents Chemother. 2014;58:2929–33. doi: 10.1128/AAC.02047-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pasteran F, Albornoz E, Faccone D, Gomez S, Valenzuela C, Morales M, et al. Emergence of NDM-1-producing Klebsiella pneumoniae in Guatemala. J Antimicrob Chemother. 2012;67:1795–7. doi: 10.1093/jac/dks101. [DOI] [PubMed] [Google Scholar]
- 9.Nemec A, Krizova L. Carbapenem-resistant Acinetobacter baumannii carrying the NDM-1 gene, Czech Republic, 2011. Euro Surveill. 2012;17 pii=2012. [PubMed] [Google Scholar]
- 10.Chen Y, Zhou Z, Jiang Y, Yu Y. Emergence of NDM-1-producing Acinetobacter baumannii in China. J Antimicrob Chemother. 2011;66:1255–9. doi: 10.1093/jac/dkr082. [DOI] [PubMed] [Google Scholar]
- 11.Wayne, PA: CLSI; 2011. Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing; 21st informational supplement. [Google Scholar]
- 12.Franklin C, Liolios L, Peleg Y. Phenotypic detection of carbapenem-susceptible metallo-β-lactamase-producing Gram-negative bacilli in the clinical laboratory. J Clin Microbiol. 2006;44:3139–44. doi: 10.1128/JCM.00879-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manchanda V, Rai S, Gupta S, Rautela RS, Chopra R, Rawat DS, et al. Development of TaqMan real-time polymerase chain reaction for the detection of the newly emerging form of carbapenem resistance gene in clinical isolates of Escherichia coli, Klebsiella pneumoniae, and Acinetobacter baumannii. Indian J Med Microbiol. 2011;29:249–53. doi: 10.4103/0255-0857.83907. [DOI] [PubMed] [Google Scholar]
- 14.Kumarasamy K, Kalyanasundaram A. Emergence of Klebsiella pneumoniae isolate co-producing NDM-1 with KPC-2 from India. J Antimicrob Chemother. 2012;67:243–4. doi: 10.1093/jac/dkr431. [DOI] [PubMed] [Google Scholar]
- 15.Hooton TM. The epidemiology of urinary tract infection and the concept of significant bacteriuria. Infection. 1990;18(Suppl):S40–3. doi: 10.1007/BF01643424. [DOI] [PubMed] [Google Scholar]
- 16.Thomson KS. Extended-spectrum β-lactamase, AmpC, and carbapenemase issues. J Clin Microbiol. 2010;48:1019–25. doi: 10.1128/JCM.00219-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teo J, Ngan G, Balm M, Jureen R, Krishnan P, Lin R. Molecular characterization of NDM-1 producing Enterobacteriaceae isolates in Singapore hospitals. Western Pac Surveill Response J. 2012;3:19–24. doi: 10.5365/WPSAR.2011.2.4.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muir A, Weinbren MJ. New Delhi metallo-β-lactamase: a cautionary tale. J Hosp Infect. 2010;75:239–40. doi: 10.1016/j.jhin.2010.02.005. [DOI] [PubMed] [Google Scholar]


