Abstract
Background & objectives:
Urinary tract infections (UTI) are a serious health problem affecting millions of people each year. Although appreciable work on various aspects of UTI including aetiology per se has been done, information on the emerging pathogens like necrotoxigenic Escherichia coli (NTEC) is largely lacking in India. In the present study E. coli isolates from patients with urinary tract infection from northeastern India were investigated for detection and characterization of NTEC.
Methods:
E. coli isolated and identified from urine samples of patients with UTI were serotyped. Antibiogram was determined by disc diffusion test. Plasmid profile was also determined. Virulence genes of NTEC (cnf1, cnf2, pap, aer, sfa, hly, afa) were detected by PCR assay. E.coli isolates carrying cnf gene (s) were identified as NTEC.
Results:
A total of 550 E. coli were isolated and tested for the presence of cnf genes. Of these, 84 (15.27%) belonged to NTEC. The cnf1 gene was present in 52 (61.9%) isolates, cnf2 in 23 (27.4%) and 9 (10.7%) carried both cnf1 and cnf2 genes. All the NTEC strains were found to harbour the pap and aer genes. Serogroup O4 was found to be the most common among the 12 serogroups identified amongst the NTEC isolates. Majority of the isolates (96.4%) were sensitive to furazolidone and were highly resistant to ampicillin. NTEC were found to harbour different numbers of plasmids (1 to 7). No association was observed between the number of plasmids and the antibiotic resistance of the isolates.
Interpretation & conclusions:
The results of the present study showed that about 15 per cent of E. coli isolates associated with UTI belonged to NTEC. More studies need to be done from other parts of the country.
Keywords: Antibiogram, Escherichia coli, NTEC, plasmid profile, serotyping, UTI, virulence genes
Escherichia coli is a component of the normal gut flora in warm blooded animals, human beings and birds. However, some strains are pathogenic and cause gastrointestinal illness and extra-intestinal infections like urinary tract infection. The pathogenecity depends on the expression of an array of virulence factors produced by E. coli. Toxigenic strains of E. coli are primarily of three types - Enterotoxigenic E. coli (ETEC), Shiga toxigenic E. coli (STEC) and necrotoxigenic E. coli (NTEC). Two different types of NTEC have been reported: NTEC1 and NTEC2 depending on the toxin they produce1,2. Cytotoxic necrotizing factor 1 (CNF1) is produced by NTEC1 and cytotoxic necrotizing factor 2 (CNF2) is produced by NTEC2. CNF1 is chromosomally encoded, whereas CNF2 is coded by genes located on the Vir plasmid1,3.
NTEC strains were reported for the first time in neonatal enteritis4. Along with producing CNF1 and CNF2 these strains may produce other toxins like cytolethal distending toxin (CDT), haemolysin (hly), P fimbriae (pap), afimbrial adhesins (afa), S fimbriae (sfa) and others5. These CNF toxins cause enlargement and multinucleation of cultured Vero and HeLa cells and necrosis in rabbit skin. CNF2 also induces necrosis in mouse footpad and moderate fluid accumulation in rabbit illeal loops6. Their role in severe dysenteric syndromes, both in man and animals, is substantiated by several clinical reports7,8. The combined production of several powerful toxins (haemolysin, CNF, CDT) by NTEC strains makes them potentially aggressive pathogens. Moreover, NTEC1 markers from man and animals appear to be highly related according to available molecular markers, which indicate that domestic animals could constitute important reservoirs of NTEC strains which are pathogenic for humans2.
UTI is the most common infection in patients with a chronic indwelling bladder catheter; bacteriuria is essentially unavoidable in this patient group9. E. coli are the most common cause of community-acquired urinary tract infection (UTI) and are responsible for 70-90 per cent of the estimated 150 million UTIs diagnosed annually10. The prevalence of CNF producing gene (cnf) in E. coli associated with UTI has been reported widely11.
Although several studies have been conducted on E. coli strains isolated from UTI patients in India12,13,14, but no comprehensive study has been done on the association of CNF-producing E. coli strains with human UTI diseases in India. Therefore, such an association was investigated in the study by searching for cnf genes and other associated genes among 550 E. coli isolates from patients with urinary tract infections by PCR, with further characterization by serogrouping, plasmid profiling and multiple drug resistance patterns.
Material & Methods
Collection of samples and isolation of E. coli: This study was conducted in the department of Biotechnology, Gauhati University, Guwahati, Assam, India. Urine samples were randomly collected aseptically from patients identified with urinary tract infection from various hospitals in Guwahati, Assam (Down Town Hospital, Gauhati Medical College Hospital and Dispur Hospital) from female patients coming from different parts of northeast India. The patients were in the age group of 18-50 yr of age. The samples were collected from January, 2009 to December, 2010. The study was approved by the Institutional Ethics Committee. A total of 573 urine samples were collected from patients with UTI and 100 from apparently healthy individuals showing no symptoms of UTI were collected as controls. The control samples were collected from healthy female attendants in the age group of 18-50 yr who were contacted when they accompanied the patients to the hospitals. Their number was restricted to 100 because most healthy individuals were reluctant to provide samples. E. coli was isolated from the samples as per the standard technique15. Colony count of E. coli was done as per the standard technique15. The isolates were identified on the basis of standard morphological and biochemical tests16.
Preparation of E. coli DNA for PCR assay: For rapid detection of virulence genes, isolated bacterial cultures were inoculated into 2 ml Luria Bartani (L-B) broth and incubated at 37°C under constant shaking for 24 h. After incubation, 1 ml broth culture was taken in a 1.5 ml microcentrifuge tube and centrifuged at 11,200 g for 10 min. The pellet was washed twice in sterile normal saline solution (NSS) (0.85% NaCl) and resuspended in 400 μl of nuclease-free sterile distilled water and boiled for 10 min followed by immediate chilling. Cell debris was removed by centrifugation at 2800 g for 5 min. The supernatant was used as template DNA for PCR.
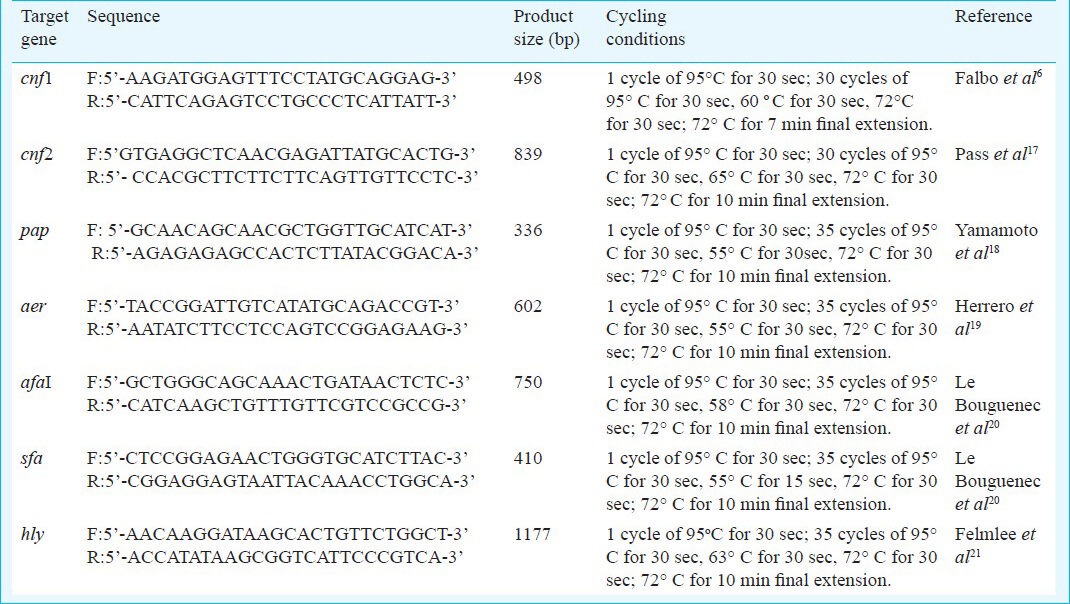
Detection of NTEC: A multiplex PCR was carried out using two sets of oligonucleotide primers for cnf1 and cnf26,17. The PCR mixture of 25.0 μl contained 1X PCR buffer, 1.5 mM of MgCl2, each primer within the 2 primer sets at a concentration of 40 nM, 200 μM each of dNTPs, 1.0 U of Taq DNA polymerase and 2.0 μl of template DNA. Primers and amplification conditions for the pathogenic gene coding regions used are shown in Table I. Amplified products were separated by agarose gel (2% agarose in 1X Tris-borate-EDTA buffer) electrophoresis at 5v/cm for 2 h and stained with ethidium bromide (0.5 μg/ml). Standard molecular size marker (100 bp DNA ladder) was included in each gel. DNA fragments were observed by ultraviolet transilluminator and photographed in a gel documentation system (Alpha Imager, Germany). The PCR was performed three times to ensure the repeatability of the technique and to make sure that isolates were correctly assigned to respective patterns. The E. coli isolates that harboured either cnf1 and / or cnf2 gene were identified as NTEC and further studied by serotyping, antibiogram, presence of other virulence factors related to NTEC and plasmid profiling.
Table I.
Primers and cycling conditions used in the amplification of specific gene responsible for toxin production
Serotyping of NTEC: E. coli isolates were serotyped based on their somatic (O) antigens at National Salmonella and Escherichia Centre, Central Research Institute, Kasauli, India.
Antibiotic resistance patterns: NTEC isolates were tested for their susceptibility to antimicrobial drugs, by disc diffusion test16 by using commercially available biodiscs (HiMedia, Mumbai, India) which included ampillicin (10 μg), gentamicin (10 μg), streptomycin (10 μg), tetracycline (30 μg), ciprofloxacin (5 μg), furazolidone (50 μg), neomycin (30 μg), kanamycin (30 μg) and nitrofurantoin (300 μg).
Detection of virulence genes by PCR and RAPD analysis: PCR was carried out using five sets of oligonucleotide primers for pap, aer, afa, sfa and hly genes for the NTEC isolates18,19,20,21. The PCR mixture of 25.0 μl contained 1X PCR buffer, 1.5 mM of MgCl2, 40 nM of the specific primer pairs, 200 μM each of dNTPs, 1.0 U of Taq DNA polymerase and 2.0 μl of template DNA. Primers and amplification conditions for the virulence genes coding regions used are shown in Table I. Amplified products were separated by agarose gel (2% agarose in 1X Tris-borate-EDTA buffer) and photographed.
Plasmid profiling of NTEC isolates: Plasmid DNA was extracted and profiled by alkaline lysis method as described by Sambrook et al22.
Statistical analysis: Statistical analysis was performed using SPSS software for Windows, ver.15 (SPSS, IBM, USA). Chi-square was used to evaluate the variables correlation.
Results & Discussion
Of the 573 urine samples collected from patients suffering from UTI, 550 (95.98%) were positive for E. coli and 23 (4.02%) were found negative for E. coli. Colony count of E.coli equaling to > 105 per ml of urine sample was taken to be positive. No significant variation was found on the basis of age of the patients or the different hospitals from which the samples were obtained for the recovery of E. coli. No E. coli could be isolated from the urine samples of the apparently healthy individuals. Of the 550 E. coli isolates, 84 (16.8%) carried at least one or other cnf genes. This percentage was less than what was reported by Landraud et al11, who found 34 per cent of the isolates from UTI producing CNF. Of the 84 NTEC isolates, 52 (61.9%) harboured cnf1 gene (Fig. 1), 23 (27.4%) harboured cnf2 (Fig. 2) and nine (10.7%) carried both cnf1 and cnf2 genes. Our results showed the occurrence of E. coli isolates in human (95.98%) UTI cases which differed from an earlier study where E. coli was found in 50 to 90 per cent of UTI cases23. Such differences in occurrence rate may be attributed to various factors viz. hygienic conditions, geographical and environmental conditions.
Fig. 1.
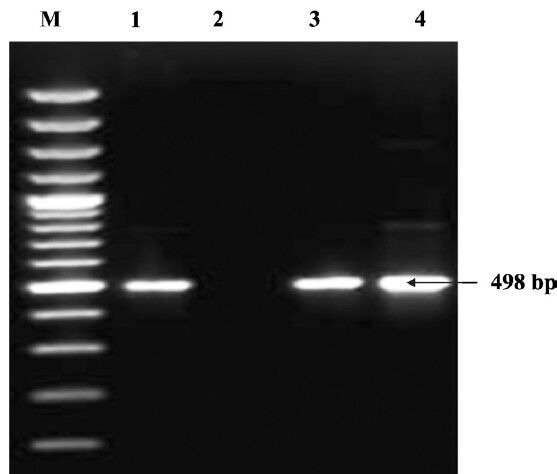
Detection of cnf1 gene by PCR. Lane M: DNA ladder (100 bp); Lane 1: positive control (498 bp); Lane 2: Negative control; Lanes 3 & 4: Positive isolates.
Fig. 2.
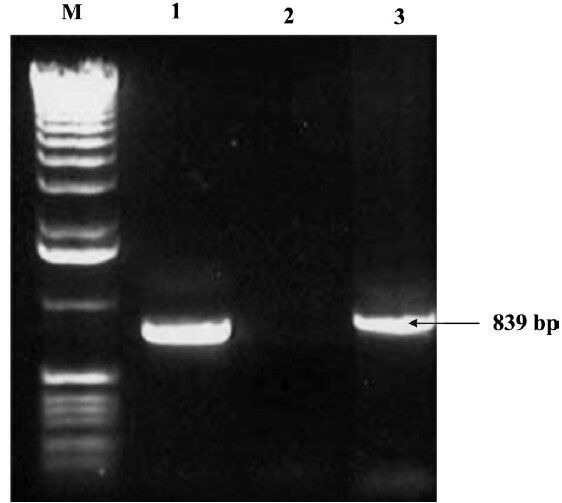
Detection of cnf2 gene by PCR. Lane M: DNA ladder (1 kb); Lane 1: positive control (839 bp); Lane 2: Negative control; Lane 3: Positive isolates.
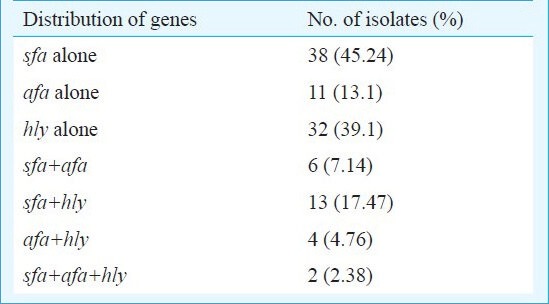
All the 84 NTEC isolates were found to harbour the pap and aer genes. The sfa gene was found in 38 (45.2%) of the NTEC isolates while 11 (13.1%) carried afa and 32 (39.1%) carried the hly gene, respectively. Thirty one (59.61%) of the 52 NTEC1 isolates were found to harbour the hly gene whereas this gene was present in only one (3.8%) of the NTEC2 isolates. However, the presence of different combination of genes varied among different isolates of E. coli (Table II). The presence of these associated genes in NTEC agrees with other reports3,5. The combination of CNF1, α-hly and P-fimbriae genes has been demonstrated in the human UTI strain, J96, possibly as a pathogenicity island24.
Table II.
Distribution of virulence genes among necrotoxigenic E. coli (NTEC) isolates (n=84) from patients with UTI
The NTEC isolates in our study belonged to 12 serogroups, namely O2 (8), O4 (16), O6 (7), O8 (1), O12 (14), O18 (13), O29 (11), O35 (1), O78 (8), O83 (1), O88 (3) and O123 (1). The serogroup O4 was found to be the most prominent serogroup. All the isolated serogroups, in the present study have been reported elsewhere as being among E. coli strains that produce CNF1,3. There was no association between the serotype of the isolates and the presence of virulence genes in the isolates.
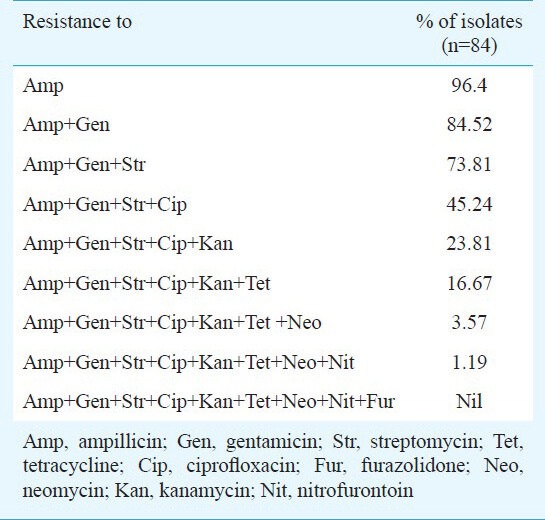
The NTEC isolates were found to have different degrees of resistance towards various antimicrobial agents (Table III). All the isolates were resistant to one or more antimicrobial drugs. None of the 84 isolates were found to be resistant to all the 9 agents tested. The isolates were highly resistant to ampicillin (96.4%) followed by gentamicin (86.9%), streptomycin (78.6%), ciprofloxacin (67.9%). The highest degree of sensitivity (97.6%) was shown by the isolates towards furazolidone. The isolates were less resistant to nitrofurantion (17.8 %), neomycin (19.0%), kanamycin (26.1%) and tetracycline (38.1%). Antibiotic resistance pattern of E. coli isolated from different sources has been reported by several workers25,26. There was no association between the serotype of the isolates and the multidrug resistance of the isolates.
Table III.
Antibiotic resistance patterns among necrotoxigenic E. coli (NTEC) isolated from patients with UTI
All the NTEC isolates were found to harbour different numbers of plasmids. The number of plasmids varied from 1 to 7. The highest number of isolates, 43 (51.2%) harboured two plasmids each. However, no correlation was observed between the number of plasmids and the antibiotic resistance of the isolates.
In conclusion, the results of the present study revealed that a good proportion of E. coli associated with UTI belonged to NTEC. It warrants a systematic study on this important public health problem.
Acknowledgment
The authors thank the Director, National Salmonella & Escherichia Centre, Central Research Institute, Kasauli (HP), India, for serotyping the isolates.
References
- 1.Blanco J, Blanco M, Alonso MP, Blanco JE, Garabal JI, Gonzalez EA. Serogroups of Escherichia coli strains producing cytotoxic necrotizing factors CNF1 and CNF2. FEMS Microbiol Lett. 1992;75:155–9. doi: 10.1016/0378-1097(92)90396-6. [DOI] [PubMed] [Google Scholar]
- 2.De Rycke J, Milon A, Oswald E. Necrotoxic Escherichia coli (NTEC): two emerging categories of human and animal pathogens. Vet Res. 1999;30:221–33. [PubMed] [Google Scholar]
- 3.Mainil JG, Jacquemin E, Pohl P, Fairbrother JM, Ansuini A, Le Bouguenec C, et al. Comparison of necrotoxigenic Escherichia coli isolates from farm animals and from humans. Vet Microbiol. 1999;70:123–35. doi: 10.1016/s0378-1135(99)00134-0. [DOI] [PubMed] [Google Scholar]
- 4.Caprioli A, Falbo V, Roda LG, Ruggeri FM, Zona C. Partial purification and characterization of an Escherichia coli toxic factor that induces morphological cell alterations. Infect Immun. 1983;39:1300–6. doi: 10.1128/iai.39.3.1300-1306.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boquet P. The cytotoxic necrotizing factor 1 (CNF1) from Escherichia coli. Toxicon. 2001;39:1673–80. doi: 10.1016/s0041-0101(01)00154-4. [DOI] [PubMed] [Google Scholar]
- 6.Falbo V, Famiglietti M, Caprioli A. Gene block encoding production of cytotoxic necrotizing factor 1 and hemolysin in Escherichia coli isolates from extraintestinal infections. Infect Immun. 1992;60:2182–7. doi: 10.1128/iai.60.6.2182-2187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paciorek J. Virulence properties of Escherichia coli faecal strains isolated in Poland from healthy children and strains belonging to serogroups O18, O26, O44, O86, O126 and O127 isolated from children with diarrhoea. J Med Microbiol. 2002;51:548–56. doi: 10.1099/0022-1317-51-7-548. [DOI] [PubMed] [Google Scholar]
- 8.Tavechio AT, Marques LR, Abe CM, Gomes TA. Detection of cytotoxic necrotizing factor types 1 and 2 among fecal Escherichia coli isolates from Brazillian children with and without diarrhea. Mem Inst Oswaldo Cruz. 2004;99:81–3. doi: 10.1590/s0074-02762004000100014. [DOI] [PubMed] [Google Scholar]
- 9.Foxman B. Epidemiology of urinary tract infections: incidence, morbidity and economic costs. Am J Med. 2002;113(Suppl 1A):5S–13S. doi: 10.1016/s0002-9343(02)01054-9. [DOI] [PubMed] [Google Scholar]
- 10.Stamm WE, Norrby SR. Urinary tract infections: disease panorama and challenges. J Infect Dis. 2001;183(Suppl 1):S1–4. doi: 10.1086/318850. [DOI] [PubMed] [Google Scholar]
- 11.Landraud L, Gauthier M, Fosse T, Boquet P. Frequency of Escherichia coli strains producing the cytotoxic necrotizing factor (CNF1) in nosocomial urinary tract infections. Lett Appl Microbiol. 2000;30:213–6. doi: 10.1046/j.1472-765x.2000.00698.x. [DOI] [PubMed] [Google Scholar]
- 12.Banerjee R, Kapoor KN, Ghatak S. Isolation of verotoxin producing Escherichia coli from diarrhoeal and urinary tract infection patients. J Commun Dis. 1999;31:161–4. [PubMed] [Google Scholar]
- 13.Mahesh E, Ramesh D, Indumathi VA, Punith K, Kirthi R, Anupama HA. Complicated urinary tract infection in a tertiary care centre in south India. Al Ameen J Med Sci. 2010;3:120–7. [Google Scholar]
- 14.Eshwarappa M, Dosegowda R, Aprameya IV, Khan MW, Kumar PS, Kempegowda P. Clinico-microbiological profile of urinary tract infection in south India. Indian J Nephrol. 2011;21:30–6. doi: 10.4103/0971-4065.75226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edwards PR, Ewing WH. 3rd ed. Minneapolis: Burgess Publishing Co; 1972. Identification of enterobacteriaceae. [Google Scholar]
- 16.Cruickshank R, Duguid JP, Marmion BP, Swain RHA. 12th ed. II. London: Churchill Livingstone; 1975. Medical microbiology. [Google Scholar]
- 17.Pass MA, Odedra R, Batt RM. Multiplex PCRs for identification of Escherichia coli virulence genes. J Clin Microbiol. 2000;38:2001–4. doi: 10.1128/jcm.38.5.2001-2004.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamamoto S, Terai A, Yuri K, Kurazono H, Takeda Y, Yoshida O. Detection of urovirulence factors in Escherichia coli by multiplex polymerase chain reaction. FEMS Immunol Med Microbiol. 1995;12:85–90. doi: 10.1111/j.1574-695X.1995.tb00179.x. [DOI] [PubMed] [Google Scholar]
- 19.Herrero M, de Lorenzo V, Neilands JB. Nucleotide sequence of the iucD gene of the pColV-K30 aerobactin operon and topology of its product studied with phoA and lacZ gene fusions. J Bacteriol. 1988;170:56–64. doi: 10.1128/jb.170.1.56-64.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le Bouguenec C, Archambaud M, Labigne A. Rapid and specific detection of the pap, afa, sfa adhesin-encoding operons in uropathogenic Escherichia coli strains by polymerase chain reaction. J Clin Microbiol. 1992;30:1189–93. doi: 10.1128/jcm.30.5.1189-1193.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Felmlee T, Pellett S, Welch RA. Nucleotide sequence of an Escherichia coli chromosomal hemolysin. J Bacteriol. 1985;163:94–105. doi: 10.1128/jb.163.1.94-105.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook J, Frish EF, Maniatis T. 2nd ed. New York: Cold Spring Harbor; 1982. A laboratory mannual. [Google Scholar]
- 23.Steadman R, Topley N. The virulence of Escherichia coli in urinary tract. In: Brumfitt W, Jeremy MT, Miller H, editors. Urinary tract infections. London: Chapman and Hall Publication; 1998. pp. 37–41. [Google Scholar]
- 24.Blum G, Falbo V, Caprioli A, Hacker J. Gene clusters encoding the cytotoxic necrotizing factor type 1, Prs-fimbriae and α-hemolysin from the pathogenicity island II of the uropathogenic Escherichia coli strain J96. FEMS Microbiol Lett. 1995;126:189–95. doi: 10.1111/j.1574-6968.1995.tb07415.x. [DOI] [PubMed] [Google Scholar]
- 25.Tabatabei RR, Nasirian A. Isolation, identification and antimicrobial resistance patterns of E. coli isolated from chicken flocks. Iranian J Pharmacol Ther. 2003;2:39–42. [Google Scholar]
- 26.Aslam M, Service C. Antimicrobial resistance and genetic profiling of Escherichia coli from a commercial beef packing plant. J Food Prot. 2006;69:1508–13. doi: 10.4315/0362-028x-69.7.1508. [DOI] [PubMed] [Google Scholar]


