Abstract
The mouse autosomal dominant mutation Mody develops hyperglycemia with notable pancreatic β-cell dysfunction. This study demonstrates that one of the alleles of the gene for insulin 2 in Mody mice encodes a protein product that substitutes tyrosine for cysteine at the seventh amino acid of the A chain in its mature form. This mutation disrupts a disulfide bond between the A and B chains and can induce a drastic conformational change of this molecule. Although there was no gross defect in the transcription from the wild-type insulin 2 allele or two alleles of insulin 1, levels of proinsulin and insulin were profoundly diminished in the β cells of Mody mice, suggesting that the number of wild-type (pro)insulin molecules was also decreased. Electron microscopy revealed a dramatic reduction of secretory granules and a remarkably enlarged lumen of the endoplasmic reticulum. Little proinsulin was processed to insulin, but high molecular weight forms of proinsulin existed with concomitant overexpression of BiP, a molecular chaperone in the endoplasmic reticulum. Furthermore, mutant proinsulin expressed in Chinese hamster ovary cells was inefficiently secreted, and its intracellular fraction formed complexes with BiP and was eventually degraded. These findings indicate that mutant proinsulin was trapped and accumulated in the endoplasmic reticulum, which could induce β-cell dysfunction and account for the dominant phenotype of this mutation.
Introduction
Diabetes is a major public issue due to its high prevalence and long-term complications (1). The molecular pathogenesis of diabetes, however, remains largely unknown. The common forms of diabetes are syndromes with heterogeneous etiologies, each of which is influenced by polygenic and multiple environmental factors. Therefore, genetic and pathophysiologic analysis of diabetes remains a major challenge. On the other hand, recent progress in the identification of genetic alterations in monogenic disorders has provided clues for understanding the molecular pathogenesis of the common forms with similar phenotypes. There are several rare monogenic forms of diabetic syndromes, both in humans and in rodent models. In humans, there is a syndrome called maturity-onset diabetes of the young (MODY), which is inherited in an autosomal dominant mode (2). The primary lesions in these diseases are in the pancreatic β cells, resulting in decreased insulin secretion. The causal genes of some types of MODY were recently identified (3).
In contrast, most of the monogenic diabetic syndromes in rodent models such as ob, db, agouti, tubby, and fat mice accompany obesity (4). The responsible genes are involved in the regulation of body weight, and their alterations result in increased insulin resistance in peripheral tissues, except in fat mice. Very recently, Yoshioka and colleagues established a monogenic diabetic model, called the Akita mouse (5). This model does not accompany either obesity or insulitis, but is accompanied by a notable pancreatic β-cell dysfunction, which distinguishes this mouse from the other well-characterized animal models. Diabetes in this mouse resembles that of human MODY in terms of early onset, an autosomal dominant mode of inheritance, and primary dysfunction of the β cells. The gene locus is named murine Mody and has been determined to be located on a distal end of Chromosome 7 by linkage analysis (5) and quantitative trait locus analysis (6).
In this study, we demonstrate that the Mody mouse has a missense mutation of the insulin 2 gene (Ins2), which lies on a corresponding area of the Mody locus identified by the genetic analysis. This mutation completely cosegregates with the qualitative phenotype of diabetes in the Mody congenic lines, and it is therefore concluded to be responsible for diabetes in this mouse. The Mody mutation codes insulin 2, whose cysteine residue at the seventh amino acid of the A chain is replaced with tyrosine. This cysteine is involved in the formation of one of the two disulfide bonds between the A and B chains. The disruption of the intramolecular disulfide bond is expected to induce a drastic conformational change of insulin 2. We present evidence that the transport of proinsulin from the endoplasmic reticulum (ER) to the Golgi apparatus is largely blocked. Furthermore, we demonstrate that the mutant proinsulin is accumulated in the ER as complexes with a ER chaperone, BiP (the immunoglobulin heavy chain binding protein) and eventually degraded intracellularly. The Mody mouse highlights the importance of the ER's role in proinsulin metabolism and presents a novel pathological mechanism in diabetes due to mutations of the insulin gene.
Methods
Mice, phenotyping, and pancreatic islet preparation.
Experimental procedures for the treatment of the mice were approved by the Animal Care and Use Review Committee at the Institute for Molecular and Cellular Regulation, Gunma University. C57BL/6J mice were purchased from CLEA Japan, Inc. (Tokyo, Japan). The heterozygous Mody mice, C57BL/6J background, were bred, fed, and phenotyped for diabetes as described previously (5). Because all the findings in this study are based on the heterozygous mice originally described (5), they are referred to as Mody or Akita mice, although homozygous mice have also been characterized elsewhere (6). Blood samples were obtained from the tail vein. Blood glucose levels were determined using a Tidex monitor (Bayer Corp., Elkhart, Indiana, USA). Mody congenic lines have been produced in Akita University since 1995. The diabetic F1 mice were obtained by crossing C57BL/6 Mody+/– male mice with C3H/He female mice. The diabetic offspring were further backcrossed to C3H/He female mice to generate congenic strains. Three congenic lines have been developed by more than 15 successive backcrosses at this point.
All the pancreatic islets used for biochemical analyses and microscopic examinations were derived from age-matched (8–12-week-old) male C57BL/6J and Mody mice. After mice were anesthetized with an intraperitoneal injection of sodium pentobarbital, pancreatic islets were isolated by pancreatic duct injection of 500 U/ml of collagenase solution (type XI; Sigma Chemical Co., St. Louis, Missouri, USA) followed by digestion at 37°C for 40 min with mild shaking (7). Islets were washed several times with HBSS, separated from acinar cells on a discontinuous Ficoll 400 gradient, and then hand-selected under a dissecting microscope.
Direct sequencing and restriction fragment length polymorphism (RFLP) analysis of insulin genes.
Tails were dissected and stored at –80°C or directly lysed in lysis buffer containing 150 μg/ml of proteinase K and 1 mg/ml of pronase E at 50°C overnight. Genomic DNA was isolated by phenol extraction followed by ribonuclease A (RNase A; 50 μg/ml) treatment and ethanol precipitation. The DNA sequencing of the Ins2 gene and the insulin 1 gene (Ins1) was determined by direct sequencing of PCR products. Primers were designed according to the sequences of murine Ins1 and Ins2 (8). For the Ins2 gene, the 5′-flanking sequence plus exon 1, exon 2, and exon 3 were amplified by the following sets of Ins2-specific primers, respectively: 5′-GCTCTGAAGCAAGTATTACA-3′ and 5′-GAGGATAGCAAAAGTTTCAC-3′, 5′-GACTTGAGGTAGGATATAGC-3′ and 5′-TAGTTAGCACTGGGGACAGA-3′, and 5′-TGCTGATGCCCTGGCCTGCT-3′ and 5′-TGGTCCCACATATGCACATG-3′. The amplified 5′-flanking sequence contains a corresponding region sufficient for conferring pancreatic β cell–specific expression in rats (9). For the Ins1 gene, exon 2 covering a full-length coding region was amplified by Ins1 specific primers: 5′-GACTTTAGGGAGAATGTGGG-3′ and 5′-TTCATTCATTATAGAACTCT-3′. PCR amplification was carried out in 50 μl of 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 200 μM each deoxyribonucleotide (dNTP), 2.5 units of Taq DNA polymerase (GIBCO/BRL, Rockville, Maryland, USA), 10 pmol of each oligonucleotide set, and 25–50 ng of genomic DNA. The PCR conditions were, after denaturation at 94°C for 5 min, 94°C for 30 s, 60°–68°C for 30 s, and 72°C for 1 min for 35 cycles with a final extension for 10 min at 72°C. DNA sequencing was performed using the primers described above with an ABI PRISM Cycle Sequencing Kit (Perkin Elmer, Norwalk, Connecticut, USA). For RFLP analysis of Ins2, genomic DNA that contains exon 3 of Ins2 was amplified by PCR as described above. A missense mutation found in one allele of Ins2 in the Mody mouse disrupts a Fnu 4HI site. The PCR products (280 bp) were then digested by Fnu 4HI and electrophoretically separated on a 2.5% agarose gel. The expected size of the digest was 139 + 141 bp (normal) and 280 bp (mutant).
Analysis of the expression levels of Ins1 and Ins2 in islets by reverse transcription (RT)-PCR.
Total RNA from pancreatic islets was prepared using the acid guanidinium thiocyanate-phenol-chloroform method (10) using TRIZOL reagent (GIBCO/BRL). Total RNA (1 μg) from islets derived from 3–5 C57BL/6J and Mody mice was reverse transcribed at 42°C for 1 h in 20 μl of reaction buffer containing 20 μM of each dNTP, 200 U of Superscript II reverse transcriptase (GIBCO/BRL), 8 U of RNasin (Promega Corp., Madison, Wisconsin, USA), and 50 pmol of oligo-(dT)17 primer. The reaction was stopped by heating at 94°C for 5 min. The 5′ and 3′ primers used for insulin and β-actin were 5′-GCTCTCTACCTGGTGTGTGG-3′, 5′-GTTTTATTCATTGCAGAGGG-3′, 5′-CGTAAAGACCTCTATGCCAA-3′, and 5′-AGCCATGCCAATGTTGTCTC-3′, respectively. The insulin primers were selected from common sequences between Ins1 and Ins2 for both transcripts to be amplified with equal efficiencies. The amplified cDNA contains the position of mutation found in the Mody mice. Labeled at 37°C for 30 min with 10 U of T4 polynucleotide kinase (TOYOBO, Osaka, Japan) in the presence of 50 μCi of [γ-32P]ATP (1000–3000 Ci/mmol; Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom) was 100 pmol of 5′ primer. The PCR was performed in 40 μl of reaction buffer containing 200 μM of each dNTP, 2 U of Taq DNA polymerase, 10 pmol of 32P-labeled 5′ and unlabeled 3′ primers, and the RT mixture described above. The expected size of each PCR product was 257 bp for Ins1, 263 bp for Ins2, and 349 bp for β-actin. The radiolabeled products were recovered by ethanol precipitation and resolved on 7% nondenatured polyacrylamide gels in Tris-borate buffer. The gels were dried onto Whatman 3MM chromatography paper (Whatman International Ltd., Maidstone, United Kingdom) and then subjected to autoradiography. The radioactivity of each band was measured using a BAS2000 bio-imaging analyzer (Fuji Photo Film Co., Tokyo, Japan). Through this analysis it was discovered that the original boundaries of the intron 1 of Ins1 and Ins2 (8) are incorrect: It actually begins from 1005–1118 (GTACTC—-TTCCAG).
Antibodies, immunoassay of insulin, immunoblotting, and immunostaining.
Guinea pig antibodies against rat C-peptide, rabbit antibodies to BiP (GRP78), and protein disulfide isomerase (PDI) were purchased from Linco Research Inc. (St. Charles, Missouri, USA), Affinity BioReagents Inc. (Golden, Colorado, USA), and StressGen Biotechnologies Corp. (Victoria, British Columbia, Canada), respectively. Guinea pig anti–porcine insulin antibodies were kind gifts from K. Wakabayashi and H. Kobayashi (Institute for Molecular and Cellular Regulation, Gunma University). Measurements of immunoreactive insulin (IRI) were performed as described previously (11). For immunoblotting, islet proteins or Chinese hamster ovary (CHO) cells were solubilized in RIPA buffer containing 50 mM Tris-HCl (pH 7.4), 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 150 mM NaCl, 5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, and 10 μg/ml of aprotinin, pepstatin A, and leupeptin. After tricine–SDS-PAGE (12), proteins were transferred on an Immobilon-PSQ membrane (Millipore Corp., Bedford, Massachusetts, USA). Immunoblotting was performed using an enhanced chemiluminescence Western blotting detection kit (Amersham Pharmacia Biotech). For immunofluorescence study, pancreatic tissues were fixed by immersion in Bouin's fixative for 2 days. After dehydration and embedding in Paraplast, 4-μm-thick sections were cut and mounted on gelatin-coated slides. The deparaffinized sections were rinsed in distilled water and PBS (10 mM sodium phosphate buffer and 140 mM NaCl, pH 7.5), they were incubated sequentially with 20% normal goat serum, the first antibody, and lissamine rhodamine sulfonyl chloride–labeled affinity-purified donkey anti–rabbit or anti–guinea pig IgG (Jackson Immuno Research Laboratories Inc., West Grove, Pennsylvania, USA) as described previously (13). The sections were examined with an Olympus BX50 microscope equipped with an epifluorescence attachment (Olympus Optical Co., Tokyo, Japan).
Conventional and immunogold electron microscopy.
Mice were anesthetized with sodium pentobarbital and perfused transcardially with PBS, followed by 2.5% glutaraldehyde in 100-mM cacodylate buffer, pH 7.4. The small tissues containing the islets were removed, immersed in the same fixative at 4°C for 2 h, and then postfixed for 1 h at 4°C in 1% osmium tetroxide in the same buffer. They were then dehydrated through a graded ethanol series and embedded in an Epon/Araldite mixture. Ultrathin sections were cut with a Reichert Ultracut-E microtome (Reichert-Jung, Vienna, Austria), and stained with uranyl acetate and lead citrate. They were observed with a JEM 1010 electron microscope at accelerating potential of 80 kV.
For immunogold electron microscopy, tissues were fixed as described above with a mixture of 0.2% glutaraldehyde, 4% paraformaldehyde, and 0.2% picric acid in 100 mM cacodylate buffer, pH 7.4, for 2 h at 4°C. After dehydration in ethanol, they were embedded in LR White (London Resin Co., Basingstoke, United Kingdom) and sectioned. Single- or double-immunogold labeling was performed as described previously (14). For double immunolabeling, two faces of the grids were incubated with anti-insulin and anti–C-peptide antibodies, and then with anti–guinea pig IgG conjugated with different sizes of gold particles (10 nm and 5 nm; British BioCell International, Cardiff, United Kingdom). The immunolabeled sections were fixed with 1% osmium tetroxide, then stained with a mixture of uranyl acetate and methyl cellulose, according to a protocol as described (15).
Construction of expression vectors for insulin 2 and CHO-cell transfection.
Total RNA (1 μg) derived from islets of Mody mice was transcribed as described above. Subsequently, PCR was performed using 5′-GCCAAGCTTAAGTGATCCGCTACAATCAA-3′ and 5′-GCGGGATCCATCGATGCTCATTCAAAGGTTTTATTC-3′. The underlined sequence contains a HindIII site in the former primer and BamHI and ClaI sites in the latter primer. The reaction products were electrophoretically separated on a 3% NuSieve GTG agarose gel (FMC Corp., Rockland, Maine, USA), and fragments of the expected size (460 bp) were purified, digested with HindIII and BamHI, and subcloned into pBluescript SK(+) (Stratagene, La Jolla, California, USA). Clones were sequenced to discriminate wild-type and mutant forms. Each cDNA was subsequently subcloned into pcDNA3 (Invitrogen Corp., Carlsbad, California, USA).
CHO cells were cultured in F12 medium supplemented with 10% fetal calf serum under 5% CO2 atmosphere at 37°C. The cells (80% confluent culture at a density of 4 × 106 cells/10-cm plate) were transfected with 2.5 μg of pcDNA3 vector that harbors wild-type or mutant Ins2 cDNA using DOTAP liposomal transfection reagent (Boehringer Mannheim GmbH, Mannheim, Germany) according to the manufacturer's instruction. Cells were cultured in the presence of G418 at 500 μg/ml. After 10 days, G418-resistant colonies were isolated and grown. Total RNA was isolated from each clone, and expression of insulin was determined by Northern blot analysis.
Pulse chasing and immunoprecipitation.
CHO cells were grown in plastic culture dishes (6-cm diameter) with 5 ml of serum-containing medium. After incubation of methionine-free DMEM for 30 min at 37°C, monolayers were pulsed in 1.2 ml of the same medium containing 100 μCi [35S]methionine (>1000 Ci/mmol; Amersham Pharmacia Biotech) for 30 min at 37°C. For chasing, the labeling medium was replaced with 1.5 ml of complete DMEM supplemented with 1 mM methionine and 1% fetal calf serum. At the indicated time periods the chase medium was removed and incubated with a mixture consisting of an equal amount of guinea pig antisera to C-peptide and insulin. Cells were solubilized on ice for 10 min after addition of 0.5 ml of lysis buffer containing 50 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, pH 7.4, 1% Triton X-100, 150 mM NaCl, 5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, and 10 μg/ml of aprotinin, pepstatin A, and leupeptin. A supernatant of the whole-cell extract was prepared by scraping the cells from the dishes and sedimenting the insoluble material by centrifugation at 15000 g for 15 min. The extracts were then incubated with antibodies described above at 4°C for 2 h. Then the antibodies were bound to Protein A-Sepharose 4FF (Amersham Pharmacia Biotech) at 4°C for 1 h with gentle rotation. The immunoprecipitates were washed twice with the lysis buffer, twice with the lysis buffer containing 1 M NaCl, once with lysis buffer containing 0.1% Triton X-100, and then eluted with tricine-SDS sample buffer containing 100 mM dithiothreitol (DTT) for analysis by tricine–SDS-PAGE (12). A BAS 2000 bio-imaging analyzer was used for analysis of the radioactive image.
For the coimmunoprecipitation study, the immunoprecipitates produced using antibodies to C-peptide and insulin were washed as described above, except for omitting washing by the lysis buffer containing 1M NaCl. They were then eluted with standard SDS sample buffer containing 100 mM DTT and separated by glycine–SDS-PAGE (16). After transfer to an Immobilon-P membrane (Millipore Corp.), immunoblotting was performed using anti-BiP antibodies.
Results
A missense mutation in the Ins2Mody allele.
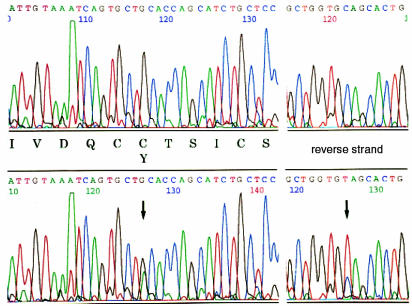
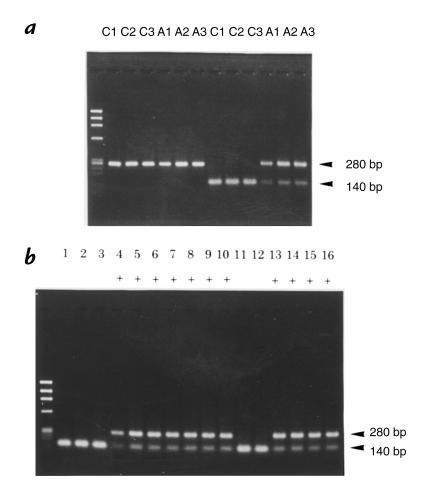
Diabetes of the Akita mouse is caused by a single locus, Mody, and is inherited in an autosomal dominant manner (5). The Mody locus maps to the telomeric region of Chromosome 7, and this area includes an important diabetogenic gene Ins2. Therefore, a possible genomic alteration of Ins2 was initially examined. The three exons and the 5′-flanking region of Ins2 were amplified from genomic DNA from control C57BL/6J and Mody mice. Mody mice had a G→A transition at nucleotide 1907 in exon 3 on one of the two Ins2 alleles (Fig. 1). This mutation changed amino acid Cys96 (TGC) to Tyr (TAC). Cys96 corresponds to the seventh amino acid in the A chain (A7) of mature insulin and forms one of the three intramolecular disulfide bonds with Cys31 located at B7. Disruption of a disulfide bond between the A and B chains is likely to induce a major conformational change in insulin 2 molecules. To determine whether this mutation is responsible for the diabetic phenotype of Mody mice, 10 diabetic and 10 control mice from each line of three Mody mutant C3H/He background congenic lines were examined. Taking advantage of the fact that the mutation disrupts a Fnu 4HI site of the Ins2 gene, RFLP analysis was performed. The PCR products digested with Fnu 4HI were 140 bp (wild-type) and 280 bp (mutant), as expected (Fig. 2a). The genotype of Ins2 and the phenotype for diabetes completely matched in 60 mice from three congenic lines (examples shown in Fig. 2b). Thus, we concluded that this Ins2 mutation induces diabetes in Mody mice.
Figure 1.
Mutation of the Ins2 gene in the Mody mouse. Ins2 exon 3 was amplified using PCR from genomic DNA. The PCR products derived from either control C57BL/6J (upper) or Mody (lower) mice were directly sequenced from both directions. A single G→A transition at nucleotide 1907 of mouse Ins2 gene (8) on one of the two alleles distinguished the Mody allele.
Figure 2.
Genotyping of the Ins2 gene by RFLP analysis. Ins2 exon 3 was amplified using PCR from genomic DNA. The left lane shows ϕX174/Hae III-digested DNA markers. (a) The size of PCR products derived from C57BL/6J (C1, C2, and C3) or Mody mice (A1, A2, and A3) was 280 bp. The mutation found in Mody mice, described in Fig. 1, disrupts an Fnu 4HI site in the exon 3 of Ins2. Digestion with Fnu 4HI did not change the size of the PCR products from the mutated allele (280 bp) but decreased that of the wild-type allele to 140 bp. (b) Representative genotyping of 16 offspring derived from three Mody congenic lines with C3H/He background is shown. Mice with diabetes are shown as “+” under the lane number. The genotype of Ins2 was completely matched with the phenotype in each individual.
Insulin transcription in the islets of Mody mice.
Mice have another functional insulin gene, Ins1. Sequence analysis of the coding region of Ins1 revealed that Mody mice did not have any mutations in Ins1 (data not shown). Therefore, the heterozygous mice should have normal insulin molecules derived from three alleles (one Ins2 and two Ins1). To semiquantify the expression levels of the whole insulin, Ins1 and Ins2 cDNAs were amplified from islet RNA using primers derived from the common sequences of both cDNAs. The total insulin levels in the islets of Mody mice were a little lower than those of the control mice when the amounts of RNA from both mice were adjusted by the levels of β-actin (Fig. 3a). Because Ins1 and Ins2 transcripts should be amplified with equal efficiency by the same primers, the transcript from each allele can be quantified if there are unique restriction enzyme sites (17). Bst EII digestion of the amplified insulin cDNAs separated Ins2 transcripts (111 bp) from Ins1 transcripts (257 bp; Fig. 3b, lanes 3 and 4) and revealed that Ins1 transcripts represent approximately 25% of the total insulin transcripts in both C57BL/6J and Mody mice. Ins2 transcripts from wild-type and mutant alleles in Mody mice were similarly quantified by Fnu 4HI digestion. The transcription levels of wild-type (174 bp) and mutant alleles (263 bp) were similar (Fig. 3b, lanes 5 and 6). These results indicate that there is no gross defect in the transcription from Ins1 and Ins2 alleles in the islets of Mody mice.
Figure 3.
Insulin transcription in the islets. (a) Total RNA from islets was reverse transcribed to cDNA using oligo-(dT)17 primer. β-actin and insulin cDNAs were then amplified by PCR. The cycle numbers used for the PCR were 18 (lanes 1 and 2), 21 (lanes 3 and 4), 24 (lanes 5 and 6), 27 (lanes 7 and 8), and 30 (lanes 9 and 10), respectively. The amounts of cDNA from control C57BL/6J (odd lanes) and Mody mice (even lanes) were adjusted by the levels of amplified β-actin (upper panels). Note that both Ins1 and Ins2 transcripts should be amplified with equal efficiencies because primers were derived from the common sequences between them. The PCR products of Ins2 (263 bp) and Ins1 (257 bp) were not resolved in this gel system. The total insulin levels in the islets of Mody mice were approximately 85%–90% of those of the control mice (lower panels). (b) The insulin transcripts amplified from islet RNA of either C57BL/6J (lanes 1, 3, and 5) or Mody mice (lanes 2, 4, and 6) were run without digestion (lanes 1 and 2). They were then digested with Bst EII for discrimination between Ins1 (257 bp) and Ins2 (111 bp) transcripts (lanes 3 and 4). Similarly, they were digested with Fnu 4HI to separate Ins1 (167 bp), wild-typeIns2 (174 bp), and mutant Ins2 (263 bp) transcripts (lanes 5 and 6). The left lane shows radiolabeled φX174/Hinf I digested DNA markers. Because the PCR products were labeled with an end-labeled 5′ primer, the radioactivity of each band corresponds to the expression level, irrespective of its size. The measurement of the radioactivity of each band revealed that 27% and 73% of the total insulin transcripts in C57BL/6J mice are derived from Ins1 and Ins2, respectively. Similar values, 24% for Ins1 and 76% for Ins2, were obtained from Mody mice. Furthermore, 39% of the total insulin transcripts, which is approximately half the value of total Ins2, were derived from the mutant Ins2 allele in Mody mice, suggesting that both normal and mutant Ins2 alleles are transcribed similarly.
Morphologic studies of the pancreatic islets of Mody mice.
Expression of insulin protein levels was then characterized using immunofluorescence staining (Fig. 4, a and b). The overall size of the islets of Mody mice tended to be smaller, and the intensity of the insulin immunoreactivity was remarkably weaker than that of C57BL/6J mice. The decrease was so prominent that it cannot simply be ascribed to the loss of immunoreactivity of insulin derived from the mutant allele. This indicates that the content of wild-type insulin was also dramatically decreased. A small population of β cells expressed insulin levels comparable to that of normal mice. Immunofluorescence analysis using anti–C-peptide antibodies revealed similar results: a decrease and heterogeneous staining pattern in the islets of Mody mice (Fig. 4, c and d). The amount appeared relatively higher than that of insulin. As anticipated from the nature of the antigen oligopeptide, this antibody recognized both wild-type and mutant proinsulin (see Figs. 7, 8c, 9, and 10), whereas anti-insulin antibody whose antigen is the whole insulin protein did not recognize the mutant (see Fig. 8b). Nevertheless, much weaker C-peptide immunoreactivity in β cells of Mody mice strongly suggests a decrease in the amount of proinsulin, including the wild type.
Figure 4.
Immunofluorescent detection of insulin, C-peptide, and BiP in the islets. Pancreatic islets of C57BL/6J (a, c, and e) and Mody (b, d, and f) mice were incubated with anti-insulin (a and b), anti-C-peptide (c and d), and anti-BiP (e and f) antibodies. Note that the positive staining for insulin, C-peptide, and BiP was found exclusively in the cytoplasm, as revealed by the lissamine rhodamine sulfonyl chloride–labeling method (red). Bar, 50 μm.
Figure 7.
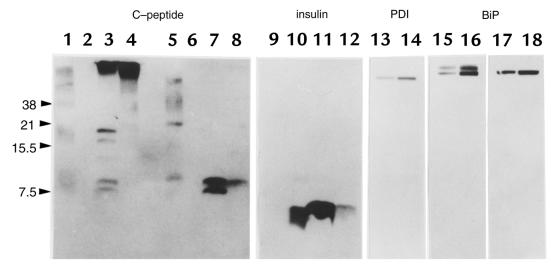
Immunoblotting analysis of the islet proteins. An equal amount of the islet protein (50 μg) from either C57BL/6J (lanes 3 and 7) or Mody mice (lanes 4 and 8) was loaded in each lane onto 16.5% polyacrylamide gel with (lanes 5–8) or without (lanes 1–4) 100 mM DTT. Human proinsulin (lanes 1 and 5) and human insulin (lanes 2 and 6) were loaded as standards. The human C-peptide ran off this tricine–SDS-PAGE system (data not shown). Immunoblotting was performed using anti–C-peptide antibodies (lanes 1–8). On the same membranes, similar analyses were performed using anti-insulin (lanes 9–12, corresponding to lanes 1–4 in the C-peptide immunoblot), anti-PDI (lanes 13 and 14, corresponding to lanes 3 and 4), and anti-BiP antibodies (lanes 15–18, corresponding to lanes 3, 4, 7, and 8). The islet protein lysed by either sample buffer containing 3% SDS (16) or acid-ethanol (26) revealed similar proteins immunoreactive to anti–C-peptide antibodies on nonreducing gels (data not shown). DTT, dithiothreitol; PDI, protein disulfide isomerase.
Figure 8.
CHO cell lines expressing either wild-type or mutant insulin 2. (a) Northern blot analysis of insulin. Total RNA (20 μg) from CHO (lane 1), CHO-Ins2wt (clone w9; lane 2), and CHO-Ins2Mody cells (clone a7; lane 3) were electrophoresed, transferred to a nylon membrane, and hybridized with a mouse Ins2 cDNA probe. (b) Insulin secretion of CHO-Ins2wt (clone w9) and CHO-Ins2Mody (clone a7). Both cells were seeded at a density of 2 × 105 cells/6-cm dish. After 24 h, the cells were incubated with serum-free media for the indicated times. Insulin stored in the cells (black bars) and that released into the media (open bars) were measured using anti-insulin antibodies. Although cell density and time course were different, similar data were obtained from another independent experiment. (c) Proinsulin content and secretion. CHO-Ins2wt (lanes 3 and 8 for clone w7; lanes 4 and 9 for clone w9), and CHO-Ins2Mody cells (lanes 5 and 10 for clone a1; lanes 6 and 11 for clone a3; lanes 7 and 12 for clone a7) were incubated with serum-free media for 24 h. The cells (lanes 3–7) were then solubilized, and the media (lanes 8–12) were concentrated by 10% trichloroacetic acid. These samples were resolved with tricine–SDS-PAGE (16.5% polyacrylamide gel) in a reducing condition (100 mM DTT). Immunoblotting analysis was performed using anti–C-peptide antibodies. Lanes 1 and 2 contain human proinsulin standard and the islet protein from normal C57BL/6J mice, respectively. CHO, Chinese hamster ovary.
Figure 9.
Pulse-chase labeling of CHO cells that express wild-type and mutant insulin. CHO-Ins2wt (clone w9, upper) and CHO-Ins2Mody (clone a7, lower) were labeled for 30 min with [35S]methionine. After chasing for indicated times, immunoprecipitation was performed from either cell extracts (C) or media (M), using a mixture of anti-insulin and anti–C-peptide antibodies. Immune complexes were analyzed with tricine–SDS-PAGE (16.5% polyacrylamide gel).
Figure 10.
Coprecipitation of proinsulin and BiP. Proinsulin was immunoprecipitated from CHO (lane 1), CHO-Ins2wt (clone w9; lane 2), and CHO-Ins2Mody cells (clone a7, lane 3). Immunoprecipitates were loaded on glycine–SDS-PAGE gel (8% polyacrylamide gel), transferred onto an Immobilon-P membrane, and immunoblotted with anti-BiP antibodies. The faint band of BiP immunoprecipitated from CHO cells (lane 1) might be due to its weak affinity to immunoglobulin.
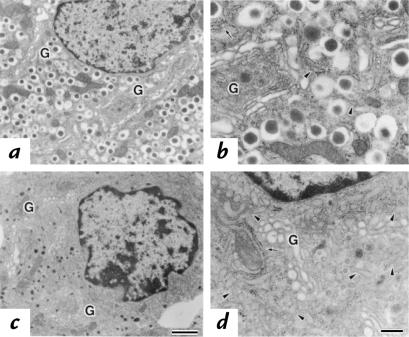
Electron microscopy of control β cells revealed that a large number of dense-core secretory granules filled the entire cytoplasm (Fig. 5, a and b). On the other hand, the size and number of secretory granules were remarkably smaller in Mody mice (Fig. 5, c and d). This accounts for the significant reduction of the β-cell size in mutant mice. Instead, the cytoplasm was filled with the ER. ER without ribosomes was especially prominent, which represents transitional ER located between the Golgi apparatus and the rough ER (Fig. 5d, arrowhead). The lumina of the transitional ER were markedly enlarged and had a more electron-dense appearance. These data suggest a block in the transport of proinsulin from the ER to the Golgi apparatus and an accumulation in the ER.
Figure 5.
Ultrastructural morphology of the islets. Electron micrographs of the β cells were taken from either C57BL/6J (a and b) or Mody (c and d) mice. Parts b and d represent higher magnification of a and c, respectively. Arrows indicate the rough ER, arrowheads represent the transitional ER, and G indicates the Golgi apparatus. Bar, 1 μm (a and c); Bar, 0.25 μm (b and d). ER, endoplasmic reticulum.
This interpretation was further supported by the findings of immunoelectron microscopy using the two-face, double-labeling method. When the ultrathin sections were immunolabeled with gold particles, most of the insulin and C-peptide were found in the secretory granules in the control islets (Fig. 6a). In contrast, significant labeling of C-peptide was observed in the ER in Mody mice, although residual granules exhibited weak immunolabeling of insulin and C-peptide (Fig. 6b). Because the bulk of proinsulin conversion occurs in the immature, clathrin-coated granule (18), the C-peptide immunoreactivity in the ER should represent that of proinsulin.
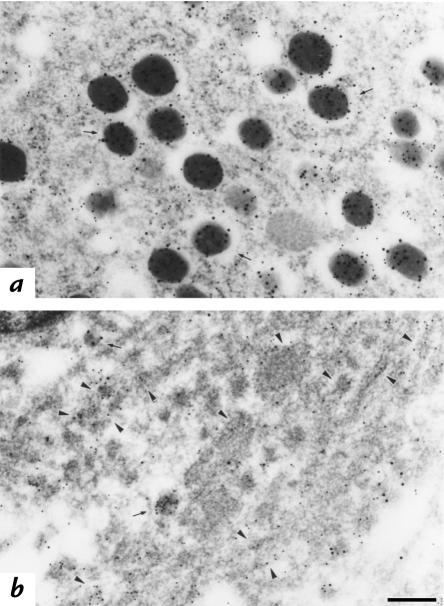
Figure 6.
Immunolocalization of insulin and C-peptide. β cells from either C57BL/6J (a) or Mody (b) mice were double immunolabeled for insulin (large gold particles) and C-peptide (small gold particles). Although insulin and C-peptide signals are profoundly decreased in the secretory granules in Mody mice compared with the control strain (arrows), a significant amount of C-peptide immunoreactivity exists in the ER (arrowheads) in Mody mice. Bar, 0.25 μm.
Insulin processing in the islets of the Mody mice.
Insulin processing was examined using immunoblotting analysis of islet proteins. Because our anti-insulin antibody has a very weak affinity to insulin on the reducing gels that separate the A and B chains (data not shown), the immunoblotting was performed on nonreducing gels. Insulin migrated as a 5.5-kDa protein close to the position of human insulin in the control islets (Fig. 7, lanes 10 and 11). The amount of insulin was dramatically decreased in the islets of Mody mice, consistent with the results of immunostaining (Fig. 7, lane 12). The immunoblotting by anti–C-peptide antibody on reducing gels revealed 8.6-kDa and 7.6-kDa proteins in the normal islets, whereas only the 8.6-kDa protein was detected in the islets of Mody mice (Fig. 7, lanes 7 and 8). The 8.6-kDa protein was considered proinsulin because it comigrated with human proinsulin (Fig. 7, lane 5). The nature of the 7.6-kDa protein remains unknown, but probably represents some form of proinsulin metabolite with a different structure that cannot be attained in the mutants. A notable reduction in proinsulin levels, but less prominent than that of insulin, was again observed in the islets of Mody mice as in the immunostaining study. Immunoblotting using the same antibody was also performed on nonreducing gels. Proinsulin was detected around the position corresponding to that found on reducing gels in the control (Fig. 7, lane 3). In addition, bands whose molecular weights are approximately twice that of monomeric proinsulin were evident. If they represent dimers, it suggests that proinsulin dimers are physiologically formed during folding through intermolecular disulfide bonds. In addition to these discrete bands, there was a high level of immunoreactivity at the boundary between the stacking and separating gels. This immunoreactivity is clearly specific because it was not detected by the same antibody on reducing gels (Fig. 7, lanes 7 and 8) nor was it detected by different antibodies on nonreducing gels (Fig. 7, lanes 11 and 12). It likely corresponds to complexes with other proteins or aggregates of proinsulin molecules. Alternatively, sodium dodecyl sulfate (SDS) may cause artificial dimers or aggregates of proinsulin in nonreducing gels. Similar protein that cannot enter a resolving gel has been identified as aggregates of expanded polyglutamine-containing ataxin-3 protein that causes Mashado-Joseph disease (19). This high-molecular-weight form was also present in the islets of Mody mice (Fig. 7, lane 4). No other discrete bands immunoreactive to anti–C-peptide antibodies were detected in mutant mice. These findings indicate that little proinsulin is processed to insulin but is accumulated as high molecular weight complexes or aggregates in the β cells of Mody mice.
Molecular chaperones and enzymes involved in proinsulin folding.
The existence of high molecular-weight forms of proinsulin prompted us to examine the possible proteins involved in the folding of proinsulin. There are at least two classes of proteins involved in this process: enzymes that catalyze specific isomerization steps and chaperones that stabilize unfolded or partially folded structures and prevent the formation of inappropriate intra- or interchain interactions (20). The first class of protein includes PDI. PDI, an ER-resident enzyme, catalyzes the isomerization of protein disulfide bonds and thereby facilitates formation of the correct set of disulfide bonds (20). Because the Mody mutation changes a cysteine residue that normally forms an intramolecular disulfide bond of insulin, possible stimulation of PDI expression was examined. PDI was overexpressed in the islets of Mody mice (Fig. 7, lanes 13 and 14).
One of the representative ER chaperones that directly determine the folding of the polypeptide is BiP (21). BiP synthesis is induced by the accumulation of secretory precursors or mutated proteins in the ER or by a number of different stress conditions that increase aberrant protein folding (20). BiP was expressed weakly both in exocrine and endocrine cells in the control pancreas, although some islet cells had focal overexpression (Fig. 4e). In contrast, it was distinguishably overexpressed in the islets of Mody mice (Fig. 4f). Double immunostaining with anti–C-peptide antibodies revealed that BiP overexpression occurred specifically in the β cells (data not shown). Immunoelectron microscopy revealed that BiP was confined to the ER of the control islets and also detected in the abnormally enlarged lumen of the ER in β cells of Mody mice (data not shown). Immunoblotting study confirmed that BiP was overexpressed in the islets of mutant mice (Fig. 7, lanes 15–18). A portion of BiP showed a slower mobility on nonreducing gels, suggesting a formation of complexes with proinsulin (Fig. 7, lanes 15 and 16). These results indicate that PDI and BiP, thought to be involved in the folding of proinsulin, are overexpressed in the islets of Mody mice.
Analysis in CHO cells expressing wild-type and mutant insulin 2.
To examine the intracellular metabolism of proinsulin in details, CHO cell lines were established that constitutively express either wild-type (CHO-Ins2wt) or mutant insulin 2 (CHO-Ins2Mody). There are two major advantages to the use of these cell lines. First, they overcome the difficulty of using a limited number of murine islets. Second, in contrast to the islets of Mody mice, it is not necessary to consider coexisting wild-type insulin 2 or insulin 1 molecules. Among several clones established, CHO-Ins2wt (clone w9) and CHO-Ins2Mody (clone a7) were analyzed in detail. Northern blot analysis revealed that CHO-Ins2Mody cells expressed approximately two times the insulin message than those of CHO-Ins2wt cells (Fig. 8a).
Next, the amount of insulin secreted into the media and the insulin content in cells were determined using anti-insulin antibodies (Fig. 8b). In CHO-Ins2wt cells, a significant amount of IRI was found in the cell extracts, and the amount secreted in the media was increased with time. In contrast, IRI was not detected in either CHO-Ins2Mody cell extracts or their media. As shown in Fig. 9, little proinsulin was processed to insulin in CHO cells, probably because non-endocrine CHO cells lack hormone-specific conversion endopeptidases such as PC1/PC3 or PC2. Although anti-insulin antibodies we used could not detect proinsulin on immunoblots (Fig. 7), they have a significant affinity to human proinsulin in solution in the immunoassay (data not shown). Therefore, most IRI found in these cells should represent proinsulin. The lack of IRI in CHO-Ins2Mody cells indicates either loss of the immunoreactivity or absence of mutant proinsulin. To discriminate these possibilities, immunoblotting analysis of these cell extracts and the media was performed using anti–C-peptide antibodies. Proinsulin was detected in several CHO-Ins2Mody cell lines independently isolated (Fig. 8c, lanes 5–7), and the protein levels in cells were proportional to the RNA levels (data not shown). This finding indicates an actual synthesis and intracellular accumulation of mutant proinsulin in CHO-Ins2Mody cells. It also indicates that mutant proinsulin lost the immunoreactivity to anti-insulin antibodies probably because of the conformational change but preserved that to anti–C-peptide antibodies. Although the media of CHO-Ins2wt cells contained a significant amount of proinsulin (Fig. 8c, lanes 8 and 9), there was no detectable proinsulin in those of CHO-Ins2Mody cells (Fig. 8c, lanes 10–12). These results indicate that mutant proinsulin is not efficiently secreted in the media, but is degraded intracellularly. Proinsulin detected in and secreted from these CHO cell lines comigrated with the 8.6-kDa protein, but not the 7.6-kDa protein, of control islets (Fig. 8c), suggesting that the 7.6 kDa protein possibly corresponds to a conversion intermediate specific to the β cells. The high-molecular-weight form of proinsulin was not found in the extracts of these CHO cell lines on nonreducing gels (data not shown).
To examine how mutant proinsulin is metabolized in these cells, pulse-chase experiments were performed. Following the pulse-labeling of cells with [35S]methionine for 30 minutes and chasing for the indicated period, immunoprecipitation was performed using a mixture of anti-insulin and anti–C-peptide antibodies (Fig. 9). The amount of proinsulin initially synthesized in CHO-Ins2Mody cells was approximately two times that in CHO-Ins2wt cells, consistent with their insulin mRNA levels. Virtually no wild-type or mutant proinsulin was converted to insulin in CHO cells during the 24-hour period. Following the chase, both wild-type and mutant proinsulin disappeared from cells at similar rates. Although the amounts secreted in the media during the first two hours were similar in wild-type and mutant proinsulin (approximately 8% of the initial proinsulin), the secretion rate of proinsulin differed thereafter. Whereas wild-type proinsulin was gradually secreted and accumulated in the media, the amount of mutant proinsulin in the media decreased as time passed. The lower amounts recovered at the later time points in CHO-Ins2wt cells is probably due to the competitive inhibition for immunoprecipitation by accumulated unlabeled proinsulin secreted in the media, as shown in Fig. 8, b and c. This competitive inhibition should not occur in CHO-Ins2Mody cells because there was little proinsulin in the media (Fig. 8c). The decreased secretion of proinsulin into the media of CHO-Ins2Mody cells suggests no further secretion of synthesized mutant proinsulin after two hours. Thus, some portions of mutant proinsulin could be secreted in a relatively short time, but once trapped in the ER, they are destined to be degraded intracellularly. Immunoblotting analysis also confirmed that the amounts of proinsulin secreted in the media during two hours were comparable in these two cell lines (data not shown), whereas those during 24 hours were far different (Fig. 8c).
To obtain further evidence for the role of BiP in the folding of proinsulin, interaction of proinsulin and BiP was directly examined in the CHO cell lines described above. Proinsulin was first immunoprecipitated with a mixture of anti-insulin and anti–C-peptide antibodies, and then the immunoprecipitates were immunoblotted using anti-BiP antibodies. In CHO-Ins2wt cells, BiP formed a complex with wild-type proinsulin (Fig. 10, lane 2). Moreover, increased amounts of the complex were found between BiP and mutant proinsulin in CHO-Ins2Mody cells (Fig. 10, lane 3).
Discussion
This study demonstrates that a mutation of the Ins2 gene on one of the two alleles is responsible for the diabetic phenotype of Mody mice. Although Ins2 is located in the region of the Mody locus identified by the genetic analyses reported previously (5, 6), mutation of Ins2 had not initially been expected to produce the phenotype of Akita mice because it is very different from the symptoms of human subjects with mutations in the insulin gene (22). In humans, several point mutations have been found that result in amino acid substitutions within the proinsulin molecules. All the subjects have been heterozygous for the defective gene. They exhibit a high incidence of glucose intolerance, which is inherited in an autosomal dominant manner. In spite of the very low biologic activity of these mutants, human subjects usually show mild and late-onset symptoms, sometimes even normal glucose tolerance, because of coexisting wild-type insulin (22). On the other hand, diabetes in Mody mice is 100% penetrative and very severe. Another common feature of the human syndrome is hyperinsulinemia or hyperproinsulinemia, depending on the effect of mutations for the processing of proinsulin to insulin. Hyperinsulinemia reflects the reduced clearance rate of mutant insulin molecules through insulin receptor–mediated uptake and degradation because of its very low binding potency to the receptor. Hyperproinsulinemia reflects decreased cleavage or altered subcellular sorting of the mutant proinsulin. In any case, significant amounts of both wild-type and mutant (pro)insulin are obviously secreted from the islets, although the precise efficiency for the secretion is unknown. This feature is in striking contrast to that of Mody mice, which have hypoinsulinemia and a marked reduction of insulin secretion as determined by a perifusion study (5). This decrease cannot be quantitatively explained by a simple loss of the immunoreactivity of the mutant insulin, and indicates that secretion of coexisting wild-type insulin is also impaired in Mody mice.
Mice have two functional insulin genes, Ins1 and Ins2. Therefore, heterozygous Mody mice should have three functional alleles that code wild-type insulin (one Ins2 and two Ins1 alleles). There was no gross alteration in the transcription levels of each allele. Protein levels of insulin and proinsulin, however, were dramatically reduced in the islets of Mody mice. Moreover, only a small number of secretory granules were formed in the β cells, suggesting that their cargo, insulin, is not supplied from the proximal compartments of the secretory pathway. These findings indicate that neither mutant nor wild-type proinsulin are transported to secretory granules. Instead, they are probably degraded intracellularly. This unique feature must arise from the nature of the mutation in this mouse. Ins2Mody codes insulin in which tyrosine has been substituted for cysteine at A7. This cysteine residue normally forms an intramolecular disulfide bond with cysteine at B7. Disruption of the disulfide bonds is expected to induce a drastic conformational change of the molecule. The introduction of the bulky aromatic tyrosine residue may also disrupt the conformation. This would activate enzymes or proteins that are involved in the folding of proinsulin. In fact, we found that PDI and BiP were overexpressed in the islets of Mody mice. Both proteins reside in the ER, where disulfide formation and folding of secreted protein occur.
Disulfide formation can be formed in the wrong temporal order and even randomly in some proteins (23). PDI corrects errors in disulfide pairing by catalyzing isomerization of the bonds. An immunoblotting analysis of the islet protein from normal mice detected dimer-like immunoreactive proinsulin molecules in a nonreducing SDS gel, suggesting at least a transient physiologic formation of intermolecular disulfide bonds. In this case, lack of cysteine at A7 in the mutant proinsulin will leave cysteine at B7 free and thereby increase the possibility that the latter cysteine forms incorrect disulfide bonds with other molecules. This would affect the disulfide bond formation of coexisting wild-type proinsulin and result in an aggregation in Mody mice.
The mechanism to monitor protein folding in the ER is called “quality control” (24). It ensures that nascent proteins that fail to fold correctly are not deployed to distal compartments. The ER luminal chaperone BiP, which is also a subunit of the import machinery, is involved in retrograde protein export of mutated yeast carboxypeptidase yscY (CPY*) to the cytosol for ER-associated degradation (25). Our observation that BiP formed complexes with wild-type proinsulin expressed in CHO cells suggests that this association physiologically occurs in pancreatic β cells. However, further studies are required to examine the specificity and normal functional role of this interaction. In CHO cells expressing mutant proinsulin, there are larger amounts of the complexes. In these cells, most of mutant proinsulin molecules were not secreted efficiently, but degraded intracellularly, although some mutant proinsulin appeared to escape from the ER and was secreted. Thus, the decreased amounts of proinsulin and insulin in the β cells of Mody mice are at least partly the result of intracellular degradation of mutant proinsulin. Prolonged retention of proinsulin in the ER due to persistent binding to ER chaperone such as BiP might increase its degradation. Alternatively, malfolded protein might accumulate in the ER because incorrectly folded intermediates tend to form insoluble aggregates that are often resistant to proteolysis. Once insoluble aggregates are formed, they can then act as seeds, facilitating normal protein to adopt the abnormal conformation, as suggested in some neurodegenerative diseases (19). Because almost all the islet protein immunoreactive to anti–C-peptide antibodies was a high molecular-weight form on nonreducing gels, proinsulin might accumulate as either complexes with molecular chaperones or aggregates in the ER in the β cells of Mody mice. Electron microscopy revealed that the lumen of the ER was prominently enlarged and appeared to be filled with electron-dense materials. Immunogold-electron microscopy of the β cells in mutant mice revealed that most of the C-peptide immunoreactivity resided in the enlarged ER, where C-peptide is still connected to the insulin moiety as a part of proinsulin. In contrast, the C-peptide immunoreactivity in control β cells was mainly localized in the secretory granules.
The findings from Mody mice are instructive for the molecular pathogenesis of diabetes associated with mutation in the insulin gene in humans. Although it is almost certain that the presence of a defective insulin allele can be a significant predisposing factor in the development of diabetes in families, the question of why diabetes develops in people with these mutations has not yet been answered clearly because normal insulin can be produced by one normal allele. The mutant insulin of Mody mice probably exhibits the most dramatic conformational change among the mutants previously described in human cases. It is noteworthy, however, that all mutants causing hyperinsulinemia possess greatly reduced insulin-receptor–binding potency (<5%) due to their conformational changes (22). Furthermore, a significant fraction of mutant proinsulin that causes hyperproinsulinemia is degraded in the β cells of transgenic mice (26). Mutant proinsulin characterized previously might be secreted efficiently once it reaches the Golgi apparatus, but there have been no studies on the effect of mutations on the fraction to be degraded or accumulated in the ER. Considering the findings from Mody mice described in this study, we propose that these mutant proinsulins with modest effects described in humans also accelerate the rate of degradation and accumulation in the ER, which would lead to dysfunction of the β cells. We also predict the existence of mutations in the insulin gene in humans exhibiting decreased levels of IRI. The accumulation of abnormally folded proinsulin might be a hallmark of aging of β cells. This could explain the mode of dominant inheritance and late-onset of this disease. The heterogeneous insulin immunostaining pattern found in β cells of Mody mice might reflect the age of each β cell: the younger cells preserve their function and insulin content, whereas the older cells lose them due to accumulation of denatured proinsulin.
In summary, we demonstrate here that diabetes in Mody mice is caused by a mutation of insulin that replaces a cysteine residue forming an intramolecular disulfide bond. Mutant proinsulin trapped in the ER was either degraded or accumulated intracellularly. The futile synthesis and degradation of proinsulin and accumulation of malfolded proinsulin appeared to induce overall dysfunction of β cells, which could account for a dominant phenotype of this mutation. Through the analysis of this mouse bearing insulin molecules with an extreme conformational change, we underline the importance of the ER in both physiologic and pathologic insulin metabolism in β cells.
Acknowledgments
We wish to thank K. Wakabayashi and H. Kobayashi for guinea pig anti–porcine insulin antibodies. This work was supported by a grant-in-aid for scientific research from the Ministry of Education, Science, and Culture of Japan, and by grants from Mitsukoshi Fund of Medicine, Japan Diabetes Foundation, Uehara Memorial Foundation, and Tanabe Medical Frontier Conference to T. Izumi.
References
- 1.King H, Rewers M. WHO Ad Hoc Diabetes Reporting Group. Global estimates for prevalence of diabetes mellitus and impaired glucose tolerance in adults. Diabetes Care. 1993;16:157–177. doi: 10.2337/diacare.16.1.157. [DOI] [PubMed] [Google Scholar]
- 2.Fajans SS. Scope and heterogeneous nature of MODY. Diabetes Care. 1990;13:49–64. doi: 10.2337/diacare.13.1.49. [DOI] [PubMed] [Google Scholar]
- 3.Froguel P, Vaxillaire M, Velho G. Genetic and metabolic heterogeneity of maturity-onset diabetes of the young. Diabetes Rev. 1997;5:123–130. [Google Scholar]
- 4.Leiter EH, Herberg L. The polygenetics of diabesity in mice. Diabetes Rev. 1997;5:131–148. [Google Scholar]
- 5.Yoshioka M, Kayo T, Ikeda T, Koizumi A. A novel locus, Mody4, distal to D7Mit189 on chromosome 7 determines early-onset NIDDM in nonobese C57BL/6 (Akita) mutant mice. Diabetes. 1997;46:887–894. doi: 10.2337/diab.46.5.887. [DOI] [PubMed] [Google Scholar]
- 6.Kayo T, Koizumi A. Mapping of murine diabetogenic gene Mody on chromosome 7 at D7Mit258 and its involvement in pancreatic islet and β cell development during the perinatal period. J Clin Invest. 1998;101:2112–2118. doi: 10.1172/JCI1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gotoh M, Maki T, Kiyoizumi T, Satomi S, Monaco AP. An improved method for isolation of mouse pancreatic islets. Transplantation. 1985;40:437–438. doi: 10.1097/00007890-198510000-00018. [DOI] [PubMed] [Google Scholar]
- 8.Wentworth BM, Schaefer IM, Villa-Komaroff L, Chirgwin JM. Characterization of the two nonalleic genes encoding mouse preproinsulin. J Mol Evol. 1986;23:305–312. doi: 10.1007/BF02100639. [DOI] [PubMed] [Google Scholar]
- 9.Walker MD, Edlund T, Boulet AM, Rutter WJ. Cell-specific expression controlled by the 5′-flanking region of insulin and chymotrypsin genes. Nature. 1983;306:557–561. doi: 10.1038/306557a0. [DOI] [PubMed] [Google Scholar]
- 10.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 11.Mashima H, et al. Betacellulin and activin A coordinately convert amylase-secreting pancreatic AR42J cells into insulin-secreting cells. J Clin Invest. 1996;97:1647–1654. doi: 10.1172/JCI118591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 13.Kayo T, et al. Developmental expression of proprotein-processing endoprotease furin in rat pancreatic islets. Endocrinology. 1996;137:5126–5134. doi: 10.1210/endo.137.11.8895387. [DOI] [PubMed] [Google Scholar]
- 14.Tanaka S, Yora T, Nakayama K, Inoue K, Kurosumi K. Proteolytic processing of pro-opiomelanocortin occurs in acidifying secretory granules of AtT-20 cells. J Histochem Cytochem. 1997;45:425–436. doi: 10.1177/002215549704500310. [DOI] [PubMed] [Google Scholar]
- 15.Roth J, Taatjes DJ, Tokuyasu KT. Contrasting of Lowicryl K4M thin sections. Histochemistry. 1990;95:123–136. doi: 10.1007/BF00266584. [DOI] [PubMed] [Google Scholar]
- 16.Laemmli UK. Cleavage of structural proteins during the assembly by the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Gilliland, G., Perrin, S., and Bunn, H.F. 1990. Competitive PCR for quantification of mRNA. In PCR protocols: a guide to methods and applications. M.A. Innis, D.H. Gelfand, J.J. Sninsky, and T.J. White, editors. Academic Press. San Diego. 60–69.
- 18.Orci L, et al. Direct identification of prohormone conversion site in insulin-secreting cells. Cell. 1985;42:671–681. doi: 10.1016/0092-8674(85)90124-2. [DOI] [PubMed] [Google Scholar]
- 19.Paulson HL, et al. Intranuclear inclusions of expanded polyglutamine protein in spinocerebellar ataxia type 3. Neuron. 1997;19:333–344. doi: 10.1016/s0896-6273(00)80943-5. [DOI] [PubMed] [Google Scholar]
- 20.Gething M-J, Sambrook J. Protein folding in the cell. Nature. 1992;355:33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- 21.Munro S, Pelham HRB. An Hsp70-like protein in the ER: identity with the 78 kd glucose-regulated protein and immunoglobulin heavy chain binding protein. Cell. 1986;46:291–300. doi: 10.1016/0092-8674(86)90746-4. [DOI] [PubMed] [Google Scholar]
- 22.Steiner DF, et al. Lessons learned from molecular biology of insulin-gene mutations. Diabetes Care. 1990;13:600–609. doi: 10.2337/diacare.13.6.600. [DOI] [PubMed] [Google Scholar]
- 23.Gilbert HF. Protein disulfide isomerase and assisted protein folding. J Biol Chem. 1997;272:29399–29402. doi: 10.1074/jbc.272.47.29399. [DOI] [PubMed] [Google Scholar]
- 24.Kopito RR. ER quality control: the cytoplasmic connection. Cell. 1997;88:427–430. doi: 10.1016/s0092-8674(00)81881-4. [DOI] [PubMed] [Google Scholar]
- 25.Plemper RK, Böhmler S, Bordallo J, Sommer T, Wolf DH. Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature. 1997;388:891–895. doi: 10.1038/42276. [DOI] [PubMed] [Google Scholar]
- 26.Carroll RJ, et al. A mutant human proinsulin is secreted from islets of Langerhans in increased amounts via an unregulated pathway. Proc Natl Acad Sci USA. 1988;85:8943–8947. doi: 10.1073/pnas.85.23.8943. [DOI] [PMC free article] [PubMed] [Google Scholar]