Abstract
Objective:
Gram-negative bacilli are the most important cause of nosocomial urinary tract infections (UTIs). The production of extended-spectrum β-lactamase (ESBL) enzymes is a common mechanism of resistance among these bacteria. The aim of this study was to determine the rate of ESBL producing Gram-negative bacteria causing nosocomial UTI in a referral hospital as well as their susceptibility pattern to the most commonly used antibiotics.
Methods:
In a prospective cross-sectional study performed over a 6-month period, urinary specimens obtained from hospitalized patients with documented culture-proved nosocomial UTI (age range of 1-87 years). Isolated aerobic Gram-negative bacteria underwent further microbiologic tests for detection of ESBL, as well as antimicrobial susceptibility test using Kirby-Bauer (disk diffusion) and E-test methods.
Findings:
During the study period, 213 urine samples were detected to have growth of Gram-negative organism. Escherichia coli was the most frequently isolated organism (61%). ESBL was detected in 102 isolates including 38.5% of E. coli, 39.5% of Klebsiella pneumonia, 88.5% of Pseudomonas aeruginosa, and 100% of Acinetobacter baumannii strains. Imipenem and meropenem were the most effective antibiotics on E. coli and K. pneumoniae strains. P. aeruginosa and A. baumannii strains showed high resistance to all tested antibiotics.
Conclusion:
Large numbers of Gram-negative bacteria causing nosocomial UTIs produce ESBL with most being multidrug-resistant. Therefore, routine ESBL detection testing and subsequent antibiogram with disk diffusion method could be useful to determine the best treatment options for UTI.
Keywords: Antimicrobial, extended-spectrum β-lactamase-producing bacteria, Gram-negative, nosocomial urinary tract infection, susceptibility
INTRODUCTION
Nosocomial infection is a significant complication of hospitalization. Urinary tract infections (UTIs) are the most common type of nosocomial infections.[1] Gram-negative bacilli are the most important cause of these infections.[2] These bacteria are showing rising rates of resistance to current therapies. The production of extended-spectrum β-lactamase (ESBL) enzymes is a common mechanism of resistance. ESBLs are enzymes that confer resistance to most beta-lactam antibiotics including penicillins, cephalosporins, and the monobactam aztreonam.[3] These enzymes have been found exclusively in Gram-negative organisms.[4] Although the prevalence of ESBL-producing Escherichia coli (E. coli) can vary from country to country, resistance rates to many commonly used therapies have increased throughout the world.[5,6,7,8,9] E. coli is the most common cause of UTIs.[10] Cases of UTI caused by ESBL-producing E. coli and Klebsiella pneumonia as well as Pseudomonas aeruginosa, including multidrug-resistant (MDR) strains, are increasing.[11]
Detection of the microbial etiology of infections, including nosocomial UTIs, provides important information in day-to-day decision-making in individual hospitals regarding potential outbreaks, unusual pathogens, antimicrobial resistance, and local trends in the etiology of infections.[12] On the other hand, selection of an appropriate antibiotic for treatment of these infections is dependent to knowledge of both causative pathogens, including drug-resistant organisms, and their antimicrobial susceptibility pattern.
The aim of this study was to determine the rate of ESBL producing Gram-negative bacteria causing nosocomial UTI in our referral teaching hospital as well as their susceptibility pattern to the most commonly used antimicrobials to identify the most appropriate antibiotic treatments for these infections.
METHODS
This was a prospective cross-sectional study performed over a 6-month period from January to July 2013 at Alzahra Hospital, a 950-bed Referral University Hospital in Isfahan, Iran. Urinary specimens were obtained from hospitalized patients (age range of 1-87 years) with documented nosocomial UTI diagnosed using the CDC/NHSN (Centers for Disease Control and Prevention/National Healthcare Safety Network) definition of healthcare-associated infections.[13] The urine samples, collected by midstream clean-catch method, were sent to the hospital's microbiology laboratory and cultured; if microbial growth occurred, differential cultures and tests were performed to identify different bacterial strains. Bacterial species were identified by different microbiologic tests, including growth in eosin-methylene-blue and MacConkey media, Gram stain, urease production, H2S production in sulfur-indole motility media, indole production, motility, methyl red test, citrate utilization, and decarboxylase production. Isolated aerobic Gram-negative bacteria underwent further microbiologic tests for detection of ESBL as well as antimicrobial susceptibility test.
ESBL detection
ESBL detection was performed using the criteria suggested by Clinical and Laboratory Standards Institute (CLSI).[14] Mueller-Hinton agar culture medium (Himedia, India) was inoculated by a direct saline suspension of isolated colonies with turbidity of 0.5 McFarland. Then antibiotic disks (Mast, UK), cefotaxime (30 μg) and ceftazidime (30 μg) were placed on the agar surface at a distance of 30 mm (center to center) from each other. After 16-18 h of incubation at 37°C, results were interpreted by measurement of inhibition zone diameter around each disk. According to CLSI criteria, an inhibition zone of ≤27 mm for cefotaxime and ≤22 mm for ceftazidime indicated that the strain probably produced ESBL.
Confirmatory tests for ESBL detection
Combined disk test was done for strains that showed ESBL production by the previous test.[15] Mueller-Hinton agar was used as culture medium. Antibiotic disks containing cefotaxime (30 μg), cefotaxime/clavulanic acid (30 μg/10 μg), ceftazidime (30 μg) and ceftazidime/clavulanic acid (30 μg/10 μg) were used. If the inhibition zone diameter of each antibiotic combined with clavulanic acid was ≥5 mm larger than that of antibiotic alone, the production of ESBL by the strain was confirmed.
For isolated P. aeruginosa and Acinetobacter baumannii strains, in addition to the above method, double-disk synergy test (DDST) was also used.[16,17] Both tests were done on cloxacillin (250 μg/ml)-containing Mueller-Hinton agar plates to inhibit cephalosporinase activity and enhance the ability of DDST for detection of ESBL.[16] DDST was performed by placing disks of ceftazidime, cefotaxime, aztreonam, and cefepime (30 μg, each) at a distance of 20 mm (center to center) from a disk of amoxicillin/clavulanic acid (20 μg/10 μg). ESBL production was confirmed when the cephalosporin inhibition zone was expanded by the clavulanate.
Antimicrobial susceptibility tests
Isolated ESBL-producing Gram-negative bacteria underwent antimicrobial susceptibility test using Kirby–Bauer (disk diffusion) method followed by E-test method for isolates that showed either intermediate sensitivity or resistance in the first test to determine the reliability of its results. Both tests were performed according to CLSI guidelines and results were interpreted using CLSI breakpoints.[14] The tested antibiotic disks (Mast, UK) included: Ampicillin (10 μg), ampicillin/sulbactam (10 μg/10 μg), ceftazidime (30 μg), cefotaxime (30 μg), cefepime (30 μg), piperacillin/tazobactam (100 μg/10 μg), imipenem (10 μg), meropenem (10 μg), amikacin (30 μg), ciprofloxacin (5 μg), levofloxacin (5 μg), and nitrofurantoin (300 μg). The E-test method was done using the E-test strips (Liofilchem, Italy) of the following antibiotics: Ceftazidime, cefepime, imipenem, amikacin, and ciprofloxacin.
RESULTS
During the study period, 213 urine samples had growth of Gram-negative organism. E. coli was the most frequently isolated organism (n = 130, 61%), followed by K. pneumoniae (n = 38, 17.8%), P. aeruginosa (n = 26, 12.2%), and A. baumannii (n = 9, 4.2%). The remaining 10 isolates (4.7%) were from other species.
Of 213 isolated Gram-negative bacteria, 102 isolates were ESBL-positive which included 50 isolates of E. coli (38.5%), 15 isolates of K. pneumoniae (39.5%), 23 isolates of P. aeruginosa (88.5%), all isolates of A. baumannii (100%), and 5 isolates of other species (50%).
Table 1 shows the susceptibility pattern of isolated ESBL-producing Gram-negative bacteria determined by disk diffusion test. As shown, imipenem and meropenem was the most effective antibiotics on E. coli and K. pneumoniae strains. On the other hand, P. aeruginosa and A. baumannii strains showed high resistance to all tested antibiotics.
Table 1.
Antimicrobial susceptibility pattern of isolated ESBL-producing Gram-negative bacteria determined by disk diffusion test
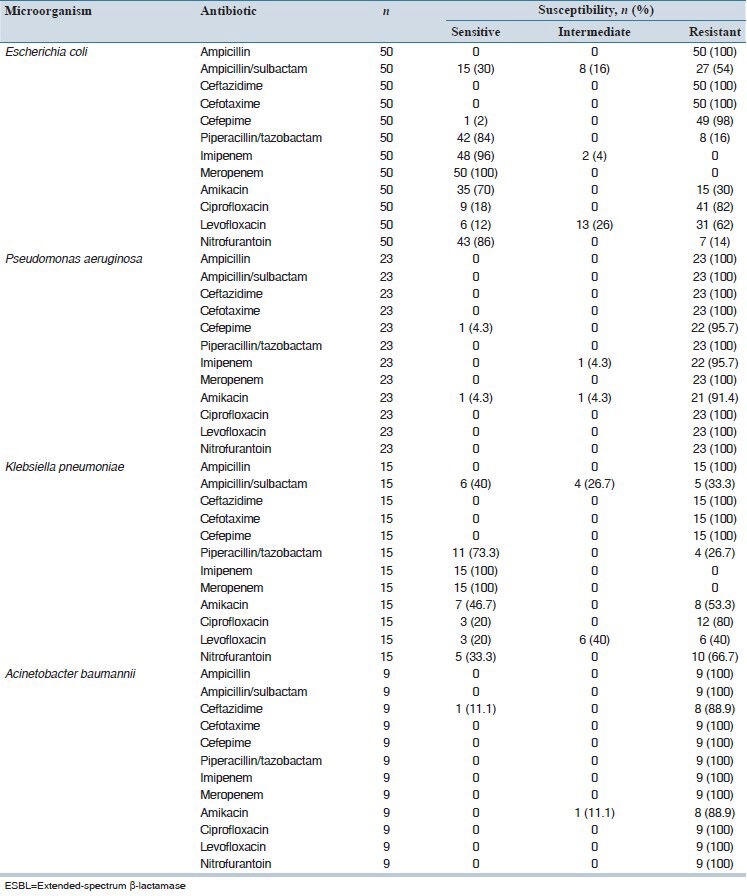
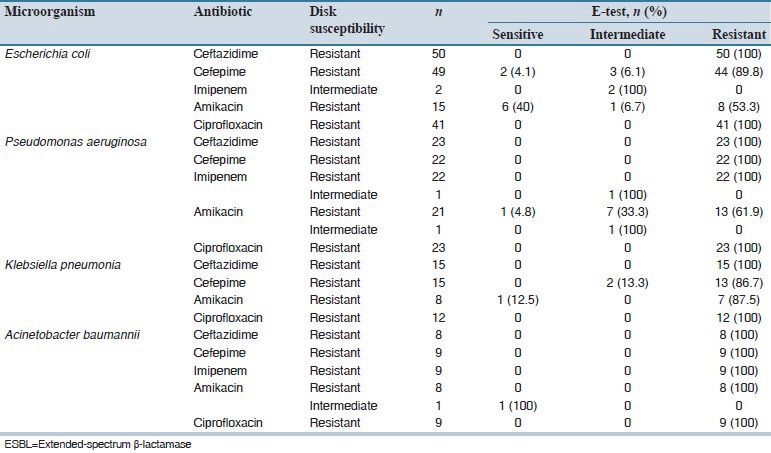
Table 2 presents the results of E-test susceptibility testing performed for five agents against isolates resistant/intermediately resistant to antibiotic disks. As shown, good consistency was seen for results of the two methods. The most frequent inconsistency of results was observed for amikacin against the isolates of E. coli and P. aeruginosa.
Table 2.
Results of E-test method for isolated ESBL-producing Gram.negative bacteria resistant/intermediately resistant to antibiotics in disk diffusion test
DISCUSSION
This study was conducted to detect the most frequent Gram-negative bacterial pathogens including ESBL producers causing nosocomial UTIs in our referral hospital and to determine their antibiotic susceptibility pattern. In our research, E. coli was the most frequently isolated bacteria followed by K. pneumoniae, P. aeruginosa, and A. baumannii. This is consistent with the results of several other studies in Iran[18,19] and other countries[2,20,21] which reported either E. coli or Klebsiella spp. as the most frequent pathogens causing nosocomial UTI. However, the prevalence of other microorganisms is different in various centers. In a study on nosocomial UTIs in 228 hospitals from 29 European countries, the six most commonly isolated microorganisms were, in decreasing order: E. coli (35.6%), Enterococci (15.8%), Candida (9.4%), Klebsiella (8.3%), Proteus (7.9%), and P. aeruginosa (6.9%).[20] However, we did not evaluate the organisms other than Gram-negative bacteria such as Enterococci.
In our study, 47.9% (n = 102) of isolated Gram-negative bacilli were ESBL-positive with E. coli strains being the most frequent agents followed by Pseudomonas, Klebsiella, and Acinetobacter. Similarly, in a study performed in India, 48.3% of isolated uropathogens were found to be ESBL producers.[21] In contrast to our results, in the study of Hosain Zadegan et al. in an Iranian hospital, 23.5% of isolated Gram-negative microorganisms (53 of 222 isolates) were ESBL producers with the most frequent isolates being K. pneumoniae (8.9%), E. coli (4.4%), and P. aeruginosa (4.4%); also, of nine isolated Acinetobacter spp. strains, 2 (0.9%) were ESBL-positive.[22] In another Iranian study conducted by Irajian et al. on different clinical specimens, ESBL was detected among 18.1% of all isolated E. coli and K. pneumoniae strains. Frequency of ESBL production was 17.45% and 19.6% for these two organisms, respectively.[23] These values are very lower than the rates in our study. This may be due to the fact that our study was performed only on urine samples as in the above-mentioned works, the most ESBL producing organisms were found in urine samples (39.6% and 88.4%, respectively).[22,23] Also, difference in the origin of isolated pathogens (nosocomial versus nonnosocomial) may be another contributing factor. Other studies have reported higher rates of ESBL production in K. pneumoniae isolates.[24,25]
Our study showed very high levels of resistance to antibiotics among ESBL-producing Gram-negative bacteria causing nosocomial UTIs. Carbapenems (imipenem and meropenem) and piperacillin/tazobactam were the most effective agents against ESBL-positive E. coli and K. pneumoniae strains. Furthermore, amikacin and nitrofurantoin had acceptable activity only on E. coli isolates. Our results show that high numbers of ESBL-producing Gram-negative bacilli are resistant not only to extended-spectrum cephalosporins including cefepime, but also to aminoglycosides (e.g., amikacin) and quinolones (e.g., ciprofloxacin and levofloxacin). The susceptibility pattern of these bacteria to various antimicrobials depends on ESBL genotype.[26] Consistent with our results, in the study of Tankhiwale et al. in India, multi-drug resistance was found to be significantly more in ESBL producing isolates (90.5%) than non ESBL-producers (68.9%).[21]
Finally, our study showed good consistency between the results of disk diffusion and E-test methods for antimicrobial susceptibility testing of ESBL-producing Gram-negative bacilli. Most inconsistencies were observed for amikacin against E. coli and P. aeruginosa strains. Therefore, as also shown in similar comparative studies,[27,28,29,30,31] it seems that the agreement level for these two methods depends on both antibiotic and microorganism tested.
Our study showed that large numbers of Gram-negative bacteria causing nosocomial UTIs produce ESBL with most being multi-drug resistant (MDR). Therefore, routine ESBL detection testing and subsequent antibiogram with disk diffusion method could be useful to determine the best treatments for UTI.
AUTHORS’ CONTRIBUTION
All authors contributed to the research idea and study design. Rasool Soltani prepared the manuscript.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared.
REFERENCES
- 1.Sharifi Y, Hasani A, Ghotaslou R, Naghili B, Aghazadeh M, Milani M, et al. Virulence and antimicrobial resistance in Enterococci isolated from urinary tract infections. Adv Pharm Bull. 2013;3:197–201. doi: 10.5681/apb.2013.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaynes R, Edwards JR National Nosocomial Infections Surveillance System. Overview of nosocomial infections caused by gram-negative bacilli. Clin Infect Dis. 2005;41:848–54. doi: 10.1086/432803. [DOI] [PubMed] [Google Scholar]
- 3.Pitout JD, Laupland KB. Extended-spectrum beta-lactamase-producing Enterobacteriaceae: An emerging public-health concern. Lancet Infect Dis. 2008;8:159–66. doi: 10.1016/S1473-3099(08)70041-0. [DOI] [PubMed] [Google Scholar]
- 4.Jain A, Roy I, Gupta MK, Kumar M, Agarwal SK. Prevalence of extended-spectrum beta-lactamase-producing Gram-negative bacteria in septicaemic neonates in a tertiary care hospital. J Med Microbiol. 2003;52:421–5. doi: 10.1099/jmm.0.04966-0. [DOI] [PubMed] [Google Scholar]
- 5.Livermore DM. Current epidemiology and growing resistance of gram-negative pathogens. Korean J Intern Med. 2012;27:128–42. doi: 10.3904/kjim.2012.27.2.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hawser SP, Badal RE, Bouchillon SK, Hoban DJ. Trending eight years of in vitro activity of ertapenem and comparators against Escherichia coli from intra-abdominal infections in North America - SMART 2002-2009. J Chemother. 2011;23:266–72. doi: 10.1179/joc.2011.23.5.266. [DOI] [PubMed] [Google Scholar]
- 7.Nakamura T, Komatsu M, Yamasaki K, Fukuda S, Miyamoto Y, Higuchi T, et al. Epidemiology of Escherichia coli, Klebsiella species, and Proteus mirabilis strains producing extended-spectrum β-lactamases from clinical samples in the Kinki Region of Japan. Am J Clin Pathol. 2012;137:620–6. doi: 10.1309/AJCP48PDVKWQOXEZ. [DOI] [PubMed] [Google Scholar]
- 8.Hawser SP, Badal RE, Bouchillon SK, Hoban DJ, Biedenbach DJ, Cantón R, et al. Monitoring the global in vitro activity of ertapenem against Escherichia coli from intra-abdominal infections: SMART 2002-2010. Int J Antimicrob Agents. 2013;41:224–8. doi: 10.1016/j.ijantimicag.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 9.Pitout JD. Infections with extended-spectrum beta-lactamase-producing enterobacteriaceae: Changing epidemiology and drug treatment choices. Drugs. 2010;70:313–33. doi: 10.2165/11533040-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 10.Sievert DM, Ricks P, Edwards JR, Schneider A, Patel J, Srinivasan A, et al. Antimicrobial-resistant pathogens associated with healthcare-associated infections: Summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009-2010. Infect Control Hosp Epidemiol. 2013;34:1–14. doi: 10.1086/668770. [DOI] [PubMed] [Google Scholar]
- 11.Zilberberg MD, Shorr AF. Secular trends in gram-negative resistance among urinary tract infection hospitalizations in the United States, 2000-2009. Infect Control Hosp Epidemiol. 2013;34:940–6. doi: 10.1086/671740. [DOI] [PubMed] [Google Scholar]
- 12.Schaberg DR, Culver DH, Gaynes RP. Major trends in the microbial etiology of nosocomial infection. Am J Med. 1991;91:72S–5S. doi: 10.1016/0002-9343(91)90346-y. [DOI] [PubMed] [Google Scholar]
- 13.Horan TC, Andrus M, Dudeck MA. CDC/NHSN surveillance definition of health care-associated infection and criteria for specific types of infections in the acute care setting. Am J Infect Control. 2008;36:309–32. doi: 10.1016/j.ajic.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 14.No. 1. Vol. 34. PA, USA: CLSI; 2009. Clinical and Laboratory Standards Institute. Performance Standard for Antimicrobial Susceptibility Testing. Document M100-S24. [Google Scholar]
- 15.Livermore DM, Brown DF. Detection of β-lactamase mediated resistance. J Antimicrob Chemother. 2001;35:281–94. doi: 10.1093/jac/48.suppl_1.59. [DOI] [PubMed] [Google Scholar]
- 16.Jiang X, Zhang Z, Li M, Zhou D, Ruan F, Lu Y. Detection of extended-spectrum beta-lactamases in clinical isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2006;50:2990–5. doi: 10.1128/AAC.01511-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tzelepi E, Giakkoupi P, Sofianou D, Loukova V, Kemeroglou A, Tsakris A. Detection of extended-spectrum beta-lactamases in clinical isolates of Enterobacter cloacae and Enterobacter aerogenes. J Clin Microbiol. 2000;38:542–6. doi: 10.1128/jcm.38.2.542-546.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Talebi Taher M, Golestanpour A. Symptomatic nosocomial urinary tract infection in ICU patients: Identification of antimicrobial resistance pattern. Iran J Clin Infect Dis. 2009;4:25–9. [Google Scholar]
- 19.Saffar MJ, Enayti AA, Abdolla IA, Razai MS, Saffar H. Antibacterial susceptibility of uropathogens in 3 hospitals, Sari, Islamic Republic of Iran, 2002-2003. East Mediterr Health J. 2008;14:556–63. [PubMed] [Google Scholar]
- 20.Bouza E, San Juan R, Muñoz P, Voss A, Kluytmans J Co-operative Group of the European Study Group on Nosocomial Infections. A European perspective on nosocomial urinary tract infections I. Report on the microbiology workload, etiology and antimicrobial susceptibility (ESGNI-003 study). European Study Group on Nosocomial Infections. Clin Microbiol Infect. 2001;7:523–31. doi: 10.1046/j.1198-743x.2001.00326.x. [DOI] [PubMed] [Google Scholar]
- 21.Tankhiwale SS, Jalgaonkar SV, Ahamad S, Hassani U. Evaluation of extended spectrum beta lactamase in urinary isolates. Indian J Med Res. 2004;120:553–6. [PubMed] [Google Scholar]
- 22.Hosain Zadegan H, Ramazanzadeh R, Hasany A. Cross-sectional study of extended spectrum beta-lactamase producing gram-negative bacilli from clinical cases in Khorramabad, Iran. Iran J Microbiol. 2009;1:16–9. [Google Scholar]
- 23.Irajian G, Jazayeri-Moghadas A, Beheshti A. Prevalence of extended-spectrum beta-lactamase positive and multidrug resistance pattern of Escherichia coli and Klebsiella pneumonia isolates, Semnan, Iran. Iran J Microbiol. 2009;1:49–53. [Google Scholar]
- 24.Shahid M, Malik A, Akram M, Agrawal LM, Khan AU, Agrawal M. Prevalent phenotypes and antibiotic resistance in Escherichia coli and Klebsiella pneumoniae at an Indian tertiary care hospital: Plasmid-mediated cefoxitin resistance. Int J Infect Dis. 2008;12:256–64. doi: 10.1016/j.ijid.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 25.Durmaz R, Durmaz B, Koroglu M, Tekerekoglu MS. Detection and typing of extended-spectrum beta-lactamases in clinical isolates of the family Enterobacteriaceae in a medical center in Turkey. Microb Drug Resist. 2001;7:171–5. doi: 10.1089/10766290152045048. [DOI] [PubMed] [Google Scholar]
- 26.Lagacé-Wiens PR, Nichol KA, Nicolle LE, Decorby MR, McCracken M, Alfa MJ, et al. ESBL genotypes in fluoroquinolone-resistant and fluoroquinolone-susceptible ESBL-producing Escherichia coli urinary isolates in Manitoba. Can J Infect Dis Med Microbiol. 2007;18:133–7. doi: 10.1155/2007/848194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khalili H, Soltani R, Negahban S, Abdollahi A, Gholami K. Reliability of disk diffusion test results for the antimicrobial susceptibility testing of nosocomial gram-positive microorganisms: Is E-test method better? Iran J Pharm Res. 2012;11:559–63. [PMC free article] [PubMed] [Google Scholar]
- 28.Katz OT, Peled N, Yagupsky P. Evaluation of the current National Committee for Clinical Laboratory standards guidelines for screening and confirming extended-spectrum beta-lactamase production in isolates of Escherichia coli and Klebsiella species from bacteremic patients. Eur J Clin Microbiol Infect Dis. 2004;23:813–7. doi: 10.1007/s10096-004-1223-4. [DOI] [PubMed] [Google Scholar]
- 29.Rahbar M, Yaghoobi M, Fattahi A. Comparison of different laboratory methods for detection of methicillin resistant Staphylococcus aureus. Pak J Med Sci. 2006;22:442–5. [Google Scholar]
- 30.Hsueh PR, Chang JC, Teng LJ, Yang PC, Ho SW, Hsieh WC, et al. Comparison of Etest and agar dilution method for antimicrobial susceptibility testing of Flavobacterium isolates. J Clin Microbiol. 1997;35:1021–3. doi: 10.1128/jcm.35.4.1021-1023.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Erfani Y, Rasti A, Mirsalehian A, Mirafshar SM, Ownegh V. E-test versus disk diffusion method in determining multidrug resistant strains of Escherichia coli in urinary tract infection. Afr J Microbiol Res. 2011;5:608–11. [Google Scholar]


