Abstract
Endophenotypes are measurable biomarkers that are correlated with an illness, at least in part, because of shared underlying genetic influences. Endophenotypes may improve our power to detect genes influencing risk of illness by being genetically simpler, closer to the level of gene action, and with larger genetic effect sizes or by providing added statistical power through their ability to quantitatively rank people within diagnostic categories. Furthermore, they also provide insight into the mechanisms underlying illness and will be valuable in developing biologically-based nosologies, through efforts such as RDoC, that seek to explain both the heterogeneity within current diagnostic categories and the overlapping clinical features between them. While neuroimaging, electrophysiological, and cognitive measures are currently most used in psychiatric genetic studies, researchers currently are attempting to identify candidate endophenotypes that are less genetically complex and potentially closer to the level of gene action, such as transcriptomic and proteomic phenotypes. Sifting through tens of thousands of such measures requires automated, high-throughput ways of assessing and ranking potential endophenotypes, such as the Endophenotype Ranking Value. However, despite the potential utility of endophenotypes for gene characterization and discovery, there is considerable resistance to endophenotypic approaches in psychiatry. In this review, we address and clarify some of the common issues associated with the usage of endophenotypes in the psychiatric genetics community.
Keywords: endophenotype, psychiatric genetics, schizophrenia, bipolar disorder, depression
Introduction
Recently, a number of quantitative trait loci (QTL) have been localized for mental illness through large-scale genome wide association studies of unrelated individuals [Ferreira and others 2008; O’Donovan and others 2008; Purcell and others 2009; Sklar and others 2011] or through family-based linkage studies [Breen and others 2011]. However, these findings explain only a small portion of the genetic variance predisposing to mental illnesses like schizophrenia, bipolar disorder, autism, major depression and addictive disorders [So and others 2011]. Furthermore, QTL localizations from either linkage or GWA do not reflect true gene identifications, as the causal functional variants have not been identified. Rather, a significant QTL indicates that a gene/variant of interest is present in a potentially large chromosomal region. For linkage, which has the ability to detect rare functional variation, this chromosomal region is typically ~10cM (~10Mb of physical sequence). For genome wide association, which is typically limited to common variants, the genomic area of interest is reduced to ~500kb (~250kb on either side of the associated SNP depending on linkage disequilibrium). Once a mental illness risk gene is identified, it must be validated and functionally characterized to further understand its influence on illness liability.
There are two different conceptualizations of what is meant by functional characterization in genetics. The first is focused on the relationship between an identified variant and gene action through direct effects on protein expression or indirect regulatory or epigenetic effects. Typically, this type of genetic functional characterization requires some level of in vitro assay to establish molecular mechanism. The other type of genetic functional characterization involves understanding how a particular gene or its expressed protein produces the cascade of biological changes that ultimately result in increased risk for the clinical symptoms of mental illness. For psychiatry, this second conceptualization requires linking gene action to behavior, a process that is largely unexplored in genetics, with the possible exception of transgenic animals or optogenetic methodologies. In this context, traits that are genetically related to the disorder (behavior) can provide important “trail markers” for traversing the chasm between genotype and behavioral phenotype. Such traits are ideally suited to facilitate the functional characterization of risk genes and should play a major roll when moving beyond a simplistic genotype-phenotype association to delineating the molecular, cellular and system-level mechanisms that give rise to a psychiatric assessment.
The dominant paradigm in psychiatric genetic studies focuses on a specific phenomenologically defined disease entity (i.e. a DSM-diagnosis). However, as with most disease states, this endpoint is relatively distant from the causal disruptions of brain anatomy/physiology associated with mental illness. An alternate strategy involves using endophenotypes, either alone or in conjunction with diagnosis, to identify mental illness risk genes [Blangero 2004; Gottesman and Gould 2003; Gottesman and Shields 1972]. Endophenotypes are heritable risk factors genetically correlated with disease liability, measureable in both affected and unaffected individuals, that are capable of providing greater power to localize and identify disease-related genes than affection status alone [Almasy and Blangero 2001; Blangero 2004; Gottesman and Gould 2003] Although there has been limited use of endophenotypes to identify risk genes in psychiatry [Glahn and others 2012], such an approach has been fruitful in the study of other complex diseases such as heart disease [Kathiresan and others 2009; Sing and others 2003], obesity [Comuzzie and others 1997; Willer and others 2009], diabetes [Mitchell and others 2000] and osteoporosis [Kammerer and others 2003; Kiel and others 2007]. Given the success of the endophenotypic or allied phenotype approach for identifying risk genes in other areas of medicine, it seems imperative to fully explore this potential path to discovering the genetic architecture of mental disorders. Unfortunately, most large-scale studies designed to identify mental illness genetic loci have focused exclusively on diagnosis.
Endophenotypes are critical for the functional characterization of risk genes discovered in traditional psychiatric genetic experiments and, if applied in large-scale studies, could identify novel risk genes, given our lack of understanding of pathophysiology and gene regulation. However, despite the potential utility of endophenotypes for gene characterization and discovery, there is considerable resistance to endophenotypic approaches in psychiatry. In the sections that follow, we address and clarify some of the common issues associated with the usage of endophenotypes in the psychiatric genetics community.
What Are Endophenotypes?
The term “endophenotype” was first coined in insect biology, to describe “microscopic and internal” traits as opposed to “exophenotypes”, being the “obvious and external” (i.e. behavior or physical appearance) [John and Lewis 1966]. In the field of psychiatric genetics, Gottesman & Shields (1972) first mentioned endophenotypes as internal phenotypes discoverable by a “biochemical test or microscopic examination.” More recently, the term was conceptualized in more detail as a measurable trait that is not observable by the unaided eye and that lies more proximal to the underlying genetics of a disorder than the clinical phenotype [Gottesman and Gould 2003]. Over time, specific, testable criteria were developed to aid the objective identification of endophenotypes in psychiatry. According to these criteria, an endophenotype must: (1) be heritable; (2) be associated with the illness; (3) be independent of clinical state (at times requiring a challenge e.g glucose tolerance test); and (4) impairment must co-segregate with the illness within a family (family members that do not meet diagnostic criteria show impairment relative to the general population); and (5) represent reproducible measurements [Gershon and Goldin 1986; Gottesman and Gould 2003; Leboyer and others 1998; Lenox and others 2002].
Conceptual and semantic issues related to the terms endophenotype, intermediate phenotype, and biomarker were fully explicated by Lenzenweger (2013). Critically, heritability and co-segregation requirements differentiate an endophenotype from a biomarker, which can be any biological measure that is influenced by health, illness, or an exogenous factor [Gould and Gottesman 2006]. Thus, endophenotypes are that subset of biomarkers that are influenced by the same genetic factors that confer risk for the illness [Gould and Gottesman 2006; Glahn and others 2012]. This requirement of pleiotropy implies that endophenotypes are directly comparable to allied phenotypes discussed in other areas of complex disease genetics [Almasy and Blangero 2001; Glahn and others 2012]. The term ‘intermediate phenotype’ has recently been used to describe a measure indexing biological risk for a mental illness that is intermediate between gene expression and disease (symptom) presentation [Meyer-Lindenberg and Weinberger, 2006]. However, this term was initially used to describe partial dominance, making its current use in psychiatric genetics somewhat ambiguous [Lenzenweger, 2013].
What Kind of Traits Can Be Endophenotypes?
Neuroimaging, electrophysiological and cognitive variables are the most oft-cited endophenotypes in the psychiatric literature [Bramon and others 2005; Glahn and others 2003; Glahn and others 2007b; Hasler and others 2006; Hasler and others 2004; Lenzenweger 2013; McDonald and others 2004; Miller and Rockstroh 2013; Slaats-Willemse and others 2003; Snitz and others 2006]. For example, deficits in spatial working memory, where working memory refers to the online manipulation of task-relevant information [Goldman-Rakic 1995], have been observed in patients with schizophrenia [Lee and Park 2005; Park and Holzman 1992; Reichenberg and Harvey 2007] and also in their healthy first-degree relatives [Glahn and others 2003; Park and others 1995]. Similarly, when asked to complete working-memory tasks in the scanner both schizophrenia patients [Glahn and others 2005; Minzenberg and others 2009] and their healthy relatives [MacDonald and others 2009] exhibit aberrant activation patterns in those regions thought to govern working-memory ability. As both working-memory ability (h2 = 0.66) and the brain activation associated with completing working-memory tasks (h2 = 0.40–0.65) are highly heritable [Blokland and others 2008; Knowles and others 2014], working memory and its related brain activity appear to be reasonable candidate endophenotypes for schizophrenia. However, while working memory is a theoretically appealing endophenotype for the illness, it is only one of many potential schizophrenia endophenotypes. Indeed, working memory, like other cognitive and imaging measures may be less effective endophenotypes than other potential biomarkers.
Although behavioral, cognitive, electrophysiological and neuroimaging based measures have received the most attention when examining putative endophenotypes for mental illness, these traits tend to have complex genetic architectures that could potentially limit their utility for gene discovery and characterization. As postulated by Gottesman and Gould (2003), one of the primary assumptions of the endophenotypic approach is that the underlying genetic architecture of the endophenotype itself is relatively simpler than that of the disease and also relatively closer to the action of the gene. While many cognitive and imaging measures are strongly influenced by genetics, it is possible that these high heritability estimates reflect the overall cumulative genetic effect on the trait but do not reveal the subtle complexities therein or the specific composition and architecture of the underlying causal genes [Almasy 2003]. That is, it is not necessarily true that behavioral, electrophysiological, and imaging candidate endophenotypes have genetic architectures any less complex than that of their associated mental illnesses. One solution is to identify endophenotypes for mental illness that are closer to gene action.
The expression of a gene depends on the process of transcription. Template DNA within the nucleus is copied, or transcribed, from the beginning to the end of a gene by RNA polymerase which gives rise to mRNA. A spliced version of the mRNA is then transported to the ribosomes for translation into a protein. Gene expression techniques, for example hybridization, can be used to measure the level of a transcript, or the RNA of a given gene, which in turn gives an indication of how active a gene is in an individual. Transcript-based endophenotypes are clearly a more direct index of gene action than the traditional behavioral or imaging phenotypes. Many factors may influence gene expression including the tissue sampled, age, sex and the time of day [Borovecki and others 2005; Radich and others 2004; Whitney and others 2003]. However, there is also substantial genetic influence on gene expression as evidenced by numerous eQTL and eGWAS studies (for a comprehensive review see [Ertekin-Taner 2011; Zou and others 2010]). Thus, gene expression may make for excellent endophenotypes of complex disease where variation in cis-regulatory polymorphisms mediates disease-risk by influencing expression level [Goring and others 2007; Rockman and Wray 2002; Wray 2007]. Unfortunately, relatively few studies have employed transcripts as endophenotypes for mental illnesses (e.g. Glahn et al., 2012), leaving this area of study quite unexplored.
While transcripts and other blood-based measures could be important endophenotypes for gene discovery and molecular characterization, the relationship between these biomarkers and the signs and symptoms that denote mental illnesses are often unclear. It is possible that the most effective approach for understanding liability for mental illness will involve combining more traditional endophenotypes with potentially complex genetic underpinnings with less genetically complex endophenotypes like cis-regulated transcripts. Although relatively few transcriptomic and proteomic endophenotypes for psychiatric disorders have been identified to date, we anticipate that this will change as more high throughput transcriptomic and proteomic studies are undertaken.
Choosing Between Endophenotypes
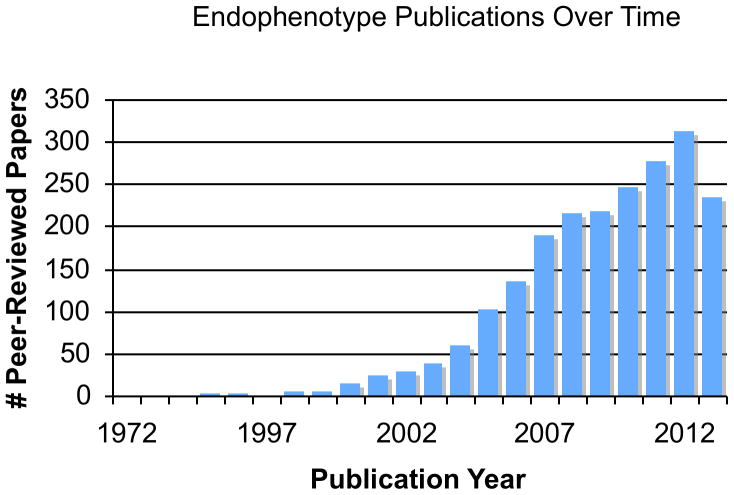
The literature is replete with putative endophenotypes from mental illnesses. Indeed, a simple PubMed search of the term “endophenotype” returns 2111 publications between 1972 and August of 2013. The term “intermediate phenotype” returns an additional 700. Scopus, which has a larger database than PubMed provides 2788 citations for the term “endophenotype.” The rate of endophenotype papers being published annually appears to be exponentially increasing (see Figure 1), with over 300 papers published in 2012. Indeed, Gottesman and Gould’s 2003 review has been referenced over 3000 times. In such an active area of research, the challenge becomes choosing among candidate endophenotypes for subsequent studies. As most endophenotypes are selected based upon a necessarily incomplete understanding of mental illness biology, comparing the utility of one endophenotype over another in terms of their ability to aid in gene discovery or genetic characterization is impossible. Recently, Glahn and colleagues (2012) developed the Endophenotype Ranking Value (ERV), an empirical metric for ranking endophenotypes based upon their genetic similarity to the studied illness. This method involves estimating the standardized genetic covariance between each putative endophenotype and a particular illness (conceptually similar to the co-heritability between traits) and then ranking endophenotypes based on their genetic covariance. Formally, the ERV statistic is defined as the absolute value of the square-root of the heritability of the illness multiplied by the square-root of the heritability of the endophenotype multiplied by the genetic correlation between the endophenotype and the illness: ERVie = |√hi2√he2 ρg|. ERV values vary between 0 and 1, with higher values indicate stronger shared genetic influence between the endophenotype and the illness. This method allows for very large numbers of endophenotypes to be efficiently assessed. Indeed, in their initial paper on the subject, Glahn and colleagues (2012) ranked over 12,000 putative endophenotypes for recurrent major depression (see below for a more involved discussion). It should be noted that the ERV method is applicable to any heritable disease and any set of potentially relevant traits.
Figure 1.
The number of peer-reviewed citations, per year, that include the term endophenotype from 1972 until 2013.
The Role of Unaffected Relatives when Defining Endophenotypes
To test the endophenotype criteria of Gottesman and Gould (2003), family-based studies need to be conducted [Glahn and Blangero 2011]. These must be sufficiently powerful, with large sample sizes and optimal designs, since the effects in unaffected relatives are expected to be subtle. Furthermore, power considerations in these studies must allow for multiple testing corrections for consideration of numerous candidate endophenotypes, for example across brain voxels, transcriptomes, or neuropsychological test batteries. Several family-based designs can be used to define endophenotypes, for example twin studies [Narr and others 2002; van Haren and others 2012], nuclear families [Greenwood and others 2007], or extended pedigrees [Glahn and others 2010; Glahn and others 2007a]. Such family-based studies can test heritability, association with illness and co-segregation within families at once. Furthermore, using genetic correlations the presence of common genetic factors between endophenotype candidate and diagnosis can be inferred. The state-independent criterion of Gottesman and Gould (2003) is often not explicitly tested in these studies since it requires repeated measurements. Instead, it can usually be derived from the literature, and to some degree also assumed when the trait is present in unaffected relatives, and when the heritability is high.
While studies of related individuals are undoubtedly the most powerful and reliable way to investigate endophenotype candidates, the data can be difficult to collect, and require specialized software. Designs using unrelated individuals cannot fully establish endophenotypes, but can nevertheless contribute to the credibility or initial nomination of endophenotype candidates. For example, comparing unrelated unaffected relatives of patients to unrelated controls without family history can indicate familiality of the trait [Johnstone and others 2002; Sprooten and others in press; Sprooten and others 2011], despite not including any related individuals per se. More recently, novel techniques have been developed to empirically derive the additive genetic variance of a trait from genomic data, in unrelated samples from the general population [Lee and others 2012; Yang and others 2011]. Given the cost and effort of obtaining large genetic or imaging datasets, and the abundance and vastness of existing data in unrelated individuals, such novel analysis methods can be extremely valuable.
Utility of Endophenotypes for a Biological Based Psychiatric Nosology
Endophenotypes can help define psychiatric nosology, as they have the potential to provide measures that are sensitive to multiple diagnostic constructs [Glahn and others 2010; Glahn and others 2007a], suggesting shared genetic pathways between diagnostically distinct disorders (e.g. major depressive and bipolar disorders [Ripke and others 2013]), or measures that are specific to a single diagnosis (i.e. analogous to plasma glucose test for diabetes or McDonald criteria for multiple sclerosis [Polman and others 2011]), suggesting unique genetic factors influence illnesses. The NIMH recently proposed the Research Domain Criteria (RDoC) strategy, which encourages researchers to focus their efforts on developing new ways of classifying psychopathology by developing a biologically valid and dimension-based taxonomy of functioning that encompasses behavior, neuroscience and genetics [Cuthbert and Insel 2010; Insel and others 2010]. In essence the RDoC strategy is designed to override the traditional psychiatric approach whereby mental illness is categorized using clusters of symptoms while simultaneously discounting the overlapping pathophysiology of those symptoms, whether that be behavioral neuroscience or genetics [Kapur and others 2012]. It is expected that the dimensions developed using the RDoC strategy will largely dismiss traditional psychiatric diagnoses and that each dimension may cross one or more diagnostic categories that have been treated previously as separate entities [Adam 2013]. Thus RDoC aims to bring psychiatry in line with the rest of the medical field where quantitative, biologically valid tests are used to guide diagnoses. Not only is the endophenotype approach ideally placed to provide solutions to the problems outlined in the RDoC strategy but researchers whose focus has been on endophenotypes for many years are ahead of the game. The aim of RDoC, to deconstruct heterogeneity associated with psychiatric diagnoses using multiple behavioral and neuroscientific measures, is at its core the same as that of the endophenotype approach [Gottesman and Gould 2003]. Endophenotypes represent a key tool for the development of a more refined and biologically-based psychiatric classification system. Perhaps, with the advent of RDoC the focus in the field of psychiatry research will shift from binary classifications of mental illness to quantitative conceptualizations of interrelated disease entities that encompass behavior and biology.
Understanding Heritability: What if the heritability of the endophenotypes is lower than that of the illness?
At the simplest conceptual level, heritability can be thought of as a sort of genetic signal-to-noise ratio. It indicates the relative strength of the overall, unspecified, genetic effects on a trait. More technically, heritability is the proportion of the overall variance in a phenotype that is attributable to the effects of genes [Falconer and Mackay 1996]. It varies from zero to one where higher heritabilities representing stronger genetic effects. Classically, heritability has been estimated from correlations among family members. Mendelian genetics predicts that to the extent that a phenotype is genetically influenced, close relatives, such as siblings, should be more phenotypically correlated than more distant relatives, such as cousins, and the correlation should fall off in a predictable manner with the degree of family relationship. Heritability estimated from twin samples is ‘broad sense’ heritability and includes dominance and epistatic components. Heritability estimated from non-twin family samples is generally ‘narrow sense’ or additive genetic heritability. Generally gene mapping studies use additive genetic models and so it is the additive component that is most useful for assessing potential for gene mapping. But the broad sense/narrow sense distinction is important to keep in mind as a potential explanatory factor if one is comparing heritabilities from different types of study as heritabilities from twin studies are generally larger than heritabilities from family studies. Recently, with the advent of increasingly dense genotype and sequence data in large epidemiological studies, techniques have been developed to estimate heritability using observed genomic sharing at genotyped markers rather than predicted sharing based on pedigree relationship; this can be done in samples of unrelated individuals [Blangero and others 2013; Speed and others 2012; Yang and others 2011]. Heritabilities estimated in this way are generally additive genetic, narrow sense, heritabilities. Another important concept to note is that heritabilities are time- and population-specific. Although the underlying genetic basis of a phenotype may not change much over time or from one population to another, environmental influences can vary substantially. As heritability is the ratio of genetic variance to total variance, increasing or decreasing the environmental sources of variation can change heritability estimates for a phenotype [Turkheimer and others 2003].
Heritability is an indicator of the overall strength of genetic effects on a phenotype and good practice has generally required documenting that a phenotype is heritable before embarking on gene localization studies. One criticism that has been raised of the endophenotype approach in general is that traits that are put forward as potentially promising endophenotypes often have heritabilities that are no better than the dichotomous diagnostic phenotype with which they are correlated. This is presumed to support the argument that the endophenotype provides no better prospect for gene mapping than the yes/no diagnostic trait. However, endophenotypes offer a number of advantages for gene mapping studies even when their heritabilities are equal to or lower than that of the illness itself.
An important feature of heritabilities is that they represent the aggregate effects of an unknown number of genes and genetic variants. As such, a phenotype with a higher heritability may be more difficult to map genes for than a phenotype with a lower heritability whose genetic architecture is simpler. A classic example of this is height, which has a heritability of 0.8 or higher [Crow and Kimura 1970; Silventoinen and others 2003] but many loci contributing to its variation [Willer and others 2009]. Of course, this line of argument raises the question of whether the genetic models behind endophenotypes are any simpler than those behind diagnostic endpoints, particularly when the endophenotypes in question are complex measures of personality, behavior, or cognition. However, there are classes of phenotypes, and therefore potential endophenotypes, that are demonstrably closer to the level of gene action and generally accepted to be influenced by fewer loci some of which have quite large effects. In particular, transcriptomic and proteomic measures that are strongly influenced by specific genes fall into this category (see above).
Assuming equally complex underlying genetic models, endophenotypes still offer advantages for gene mapping as compared to yes/no diagnostic outcomes through the greater inherent power for genetic studies provided by quantitative versus dichotomous phenotypes. It can be shown that unless one assumes a high level of imprecision in a quantitative measure, power for gene mapping is always better with a quantitatively measured phenotype, such as body mass index, than with a direct dichotomization of that phenotype, such as obesity [Blangero and others 2003; Williams and Blangero 2004]. Typically, an endophenotype is not a direct measure of a quantitative trait used as part of the diagnosis, but a related phenotype that is correlated with diagnosis. In this case, the improvement in power of the quantitative phenotype comes in being able to discriminate among affected individuals of greater or lesser severity and among unaffected individuals at higher and lower risk by ordering individuals within each diagnostic category. However, even greater analytic power can be obtained through joint analyses of the dichotomous diagnostic phenotype and the quantitative correlated endophenotype [Glahn and others 2012; Liu and others 2009; Williams and others 1999; Yuan and Diao]. At heart, arguments comparing the heritability of endophenotype and illness introduce a false dichotomy in which studies of illness and endophenotype are assumed to be mutually exclusive. A well-designed genetic study can maximize power by utilizing the complementary information in both endophenotypes and diagnoses.
Conclusions
Endophenotypes are measurable biomarkers that are correlated with an illness, at least in part, because of shared underlying genetic influences. They are heritable, are independent of clinical state, co-segregate with illness within a family, and are impaired in non-affected close relatives of patients as compared to a random population sample. In general, family studies are required to formally test pleiotropy and establish that a candidate endophenotype and the illness of interest share overlapping genetic influences. While neuroimaging, electrophysiological, and cognitive measures are currently most used in psychiatric genetic studies, researchers currently are attempting to identify candidate endophenotypes that are less genetically complex and potentially closer to the level of gene action, such as transcriptomic and proteomic phenotypes. Sifting through tens of thousands of such measures requires automated, high-throughput ways of assessing and ranking potential endophenotypes, such as the Endophenotype Ranking Value.
Endophenotypes may improve our power to detect genes influencing risk of illness by being genetically simpler, closer to the level of gene action, and with larger genetic effect sizes or by providing added statistical power through their ability to quantitatively rank people within diagnostic category. They also provide insight into the mechanisms underlying illness and will be valuable in developing biologically-based nosologies, through efforts such as RDoC, that seek to explain both the heterogeneity within current diagnostic categories and the overlapping clinical features between them. Endophenotypes have been successfully used in genetic studies of schizophrenia and depression, contributing to gene finding and to understanding of potential mechanisms.
Quantitative risk factors have a long and productive history in other areas of complex disease genetics and recent technological advancements have made it both logistically and economically feasible to collect tens or hundreds of thousands of measures of transcriptomic, proteomic, neuroanatomical, neurological, cognitive, and behavioral phenotypes. These potential endophenotypes compliment traditional case/control genetic studies and offer many advantages for advancing discovery and understanding in psychiatric genetics.
Acknowledgments
Financial support for this study was provided by the following National Institute of Mental Health Grants: MH0708143 (Principal Investigator [PI]: DCG), MH078111 (PI: JB), MH083824 (PI: DCG & JB), HL11323, MH097940 (PI: LA, HR & DG), MH061622 (PI: LA), MH093740 (PI: LA), AA08403, and MH080912 (PI DCG). SOLAR is supported by National Institute of Mental Health Grant MH59490 (PI: JB).
Footnotes
Disclosure of Financial Relationships: Authors have no conflicts of interest to disclose.
Authors declare no competing financial interests in relation to the described work.
References
- Adam D. Mental health: On the spectrum. Nature. 2013;496(7446):416–418. doi: 10.1038/496416a. [DOI] [PubMed] [Google Scholar]
- Almasy L. Quantitative risk factors as indices of alcoholism susceptibility. Ann Med. 2003;35(5):337–343. doi: 10.1080/07853890310004903. [DOI] [PubMed] [Google Scholar]
- Almasy L, Blangero JC. Endophenotypes as quantitative risk factors for psychiatric disease: rationale and study design. American Journal of Medical Genetics. 2001;105(1):42–44. [PubMed] [Google Scholar]
- Blangero J. Localization and identification of human quantitative trait loci: king harvest has surely come. Curr Opin Genet Dev. 2004;14(3):233–240. doi: 10.1016/j.gde.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Blangero J, Diego VP, Dyer TD, Almeida M, Peralta J, Kent JW, Williams JT, Almasy L, Göring HH. A kernel of truth: statistical advances in polygenic variance component models for complex human pedigrees. Adv Genet. 2013;81:1–31. doi: 10.1016/B978-0-12-407677-8.00001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blangero J, Williams JT, Almasy L. Novel family-based approaches to genetic risk in thrombosis. J Thromb Haemost. 2003;1(7):1391–1397. doi: 10.1046/j.1538-7836.2003.00310.x. [DOI] [PubMed] [Google Scholar]
- Blokland G, McMahon K, Hoffman J, Zhu G, Meredith M, Martin N, Thompson P, de Zubicaray G, Wright M. Quantifying the heritability of task-related brain activation and performance during the N-back working memory task: a twin fMRI study. Biol Psychol. 2008;79(1):70–79. doi: 10.1016/j.biopsycho.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borovecki F, Lovrecic L, Zhou J, Jeong H, Then F, Rosas HD, Hersch SM, Hogarth P, Bouzou B, Jensen RV, Krainc D. Genome-wide expression profiling of human blood reveals biomarkers for Huntington’s disease. Proc Natl Acad Sci U S A. 2005;102(31):11023–11028. doi: 10.1073/pnas.0504921102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramon E, McDonald C, Croft RJ, Landau S, Filbey F, Gruzelier JH, Sham PC, Frangou S, Murray RM. Is the P300 wave an endophenotype for schizophrenia? A meta-analysis and a family study. Neuroimage. 2005;27(4):960–968. doi: 10.1016/j.neuroimage.2005.05.022. [DOI] [PubMed] [Google Scholar]
- Breen G, Webb BT, Butler AW, van den Oord EJ, Tozzi F, Craddock N, Gill M, Korszun A, Maier W, Middleton L, Mors O, Owen MJ, Cohen-Woods S, Perry J, Galwey NW, Upmanyu R, Craig I, Lewis CM, Ng M, Brewster S, Preisig M, Rietschel M, Jones L, Knight J, Rice J, Muglia P, Farmer AE, McGuffin P. A Genome-Wide Significant Linkage for Severe Depression on Chromosome 3: The Depression Network Study. Am J Psychiatry. 2011 doi: 10.1176/appi.ajp.2011.10091342. [DOI] [PubMed] [Google Scholar]
- Comuzzie A, Hixson J, Almasy L, Mitchell B, Mahaney M, Dyer T, Stern M, MacCluer J, Blangero J. A major quantitative trait locus determining serum leptin levels and fat mass is located on human chromosome 2. Nature Genetics. 1997;15(3):273–276. doi: 10.1038/ng0397-273. [DOI] [PubMed] [Google Scholar]
- Crow JF, Kimura M. An introduction to population genetics theory. New York: Harper & Row; 1970. p. xiv.p. 591. [Google Scholar]
- Cuthbert BN, Insel TR. Toward new approaches to psychotic disorders: the NIMH Research Domain Criteria project. Schizophr Bull. 2010;36(6):1061–1062. doi: 10.1093/schbul/sbq108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertekin-Taner N. Gene expression endophenotypes: a novel approach for gene discovery in Alzheimer’s disease. Mol Neurodegener. 2011;6:31. doi: 10.1186/1750-1326-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer DS, Mackay TFC. Introduction to quantitative genetics. Harlow, England; New York: Prentice Hall; 1996. p. xv.p. 464. [Google Scholar]
- Ferreira M, O’Donovan M, Meng Y, Jones I, Ruderfer D, Jones L, Fan J, Kirov G, Perlis R, Green E, Smoller J, Grozeva D, Stone J, Nikolov I, Chambert K, Hamshere M, Nimgaonkar V, Moskvina V, Thase M, Caesar S, Sachs G, Franklin J, Gordon-Smith K, Ardlie K, Gabriel S, Fraser C, Blumenstiel B, Defelice M, Breen G, Gill M, Morris D, Elkin A, Muir W, McGhee K, Williamson R, Macintyre D, Maclean A, StClair D, Robinson M, VanBeck M, Pereira A, Kandaswamy R, McQuillin A, Collier D, Bass N, Young A, Lawrence J, Nicol Ferrier I, Anjorin A, Farmer A, Curtis D, Scolnick E, McGuffin P, Daly M, Corvin A, Holmans P, Blackwood D, Gurling H, Owen M, Purcell S, Sklar P, Craddock N. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008 doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershon E, Goldin L. Clinical methods in psychiatric genetics. I. Robustness of genetic marker investigative strategies. Acta Psychiatrica Scandinavica. 1986;74:113–118. doi: 10.1111/j.1600-0447.1986.tb10594.x. [DOI] [PubMed] [Google Scholar]
- Glahn D, Almasy L, Barguil M, Hare E, Peralta J, Kent JJ, Dassori A, Contreras J, Pacheco A, Lanzagorta N, Nicolini H, Raventós H, Escamilla M. Neurocognitive endophenotypes for bipolar disorder identified in multiplex multigenerational families. Arch Gen Psychiatry. 2010;67(2):168–177. doi: 10.1001/archgenpsychiatry.2009.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glahn D, Almasy L, Blangero J, Burk G, Estrada J, Peralta J, Meyenberg N, Castro M, Barrett J, Nicolini H, Raventós H, Escamilla M. Adjudicating neurocognitive endophenotypes for schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2007a;144B(2):242–249. doi: 10.1002/ajmg.b.30446. [DOI] [PubMed] [Google Scholar]
- Glahn D, Blangero J. Why endophenotype development requires families. Chinese Science Bulletin. 2011;56:3382–3384. [Google Scholar]
- Glahn D, Ragland J, Abramoff A, Barrett J, Laird A, Bearden C, Velligan D. Beyond hypofrontality: a quantitative meta-analysis of functional neuroimaging studies of working memory in schizophrenia. Hum Brain Mapp. 2005;25(1):60–69. doi: 10.1002/hbm.20138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glahn D, Therman S, Manninen M, Huttunen M, Kaprio J, Lönnqvist J, Cannon T. Spatial working memory as an endophenotype for schizophrenia. Biol Psychiatry. 2003;53(7):624–626. doi: 10.1016/s0006-3223(02)01641-4. [DOI] [PubMed] [Google Scholar]
- Glahn D, Thompson P, Blangero J. Neuroimaging endophenotypes: strategies for finding genes influencing brain structure and function. Hum Brain Mapp. 2007b;28(6):488–501. doi: 10.1002/hbm.20401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glahn DC, Curran JE, Winkler AM, Carless MA, Kent JW, Charlesworth JC, Johnson MP, Göring HH, Cole SA, Dyer TD, Moses EK, Olvera RL, Kochunov P, Duggirala R, Fox PT, Almasy L, Blangero J. High dimensional endophenotype ranking in the search for major depression risk genes. Biol Psychiatry. 2012;71(1):6–14. doi: 10.1016/j.biopsych.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman-Rakic PS. Cellular basis of working memory. Neuron. 1995;14(3):477–485. doi: 10.1016/0896-6273(95)90304-6. [DOI] [PubMed] [Google Scholar]
- Goring HH, Curran JE, Johnson MP, Dyer TD, Charlesworth J, Cole SA, Jowett JB, Abraham LJ, Rainwater DL, Comuzzie AG, Mahaney MC, Almasy L, Maccluer JW, Kissebah AH, Collier GR, Moses EK, Blangero J. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat Genet. 2007;39(10):1208–1216. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- Gottesman II, Gould TD. The Endophenotype Concept in Psychiatry: Etymology and Strategic Intentions. American Journal of Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- Gottesman II, Shields J. Schizophrenia and genetics: A twin study vantage point. New York: Academic Press; 1972. [Google Scholar]
- Gould T, Gottesman I. Psychiatric endophenotypes and the development of valid animal models. Genes Brain Behav. 2006;5(2):113–119. doi: 10.1111/j.1601-183X.2005.00186.x. [DOI] [PubMed] [Google Scholar]
- Greenwood T, Braff D, Light G, Cadenhead K, Calkins M, Dobie D, Freedman R, Green M, Gur R, Gur R, Mintz J, Nuechterlein K, Olincy A, Radant A, Seidman L, Siever L, Silverman J, Stone W, Swerdlow N, Tsuang D, Tsuang M, Turetsky B, Schork N. Initial heritability analyses of endophenotypic measures for schizophrenia: the consortium on the genetics of schizophrenia. Arch Gen Psychiatry. 2007;64(11):1242–1250. doi: 10.1001/archpsyc.64.11.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasler G, Drevets WC, Gould TD, Gottesman, Manji HK. Toward Constructing an Endophenotype Strategy for Bipolar Disorders. Biol Psychiatry. 2006 Jan 6; doi: 10.1016/j.biopsych.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Hasler G, Drevets WC, Manji HK, Charney DS. Discovering endophenotypes for major depression. Neuropsychopharmacology. 2004;29(10):1765–1781. doi: 10.1038/sj.npp.1300506. [DOI] [PubMed] [Google Scholar]
- Insel T, Cuthbert B, Garvey M, Heinssen R, Pine DS, Quinn K, Sanislow C, Wang P. Research domain criteria (RDoC): toward a new classification framework for research on mental disorders. Am J Psychiatry. 2010;167(7):748–751. doi: 10.1176/appi.ajp.2010.09091379. [DOI] [PubMed] [Google Scholar]
- John B, Lewis KR. Chromosome variability and geographic distribution in insects. Science. 1966;152(3723):711–721. doi: 10.1126/science.152.3723.711. [DOI] [PubMed] [Google Scholar]
- Johnstone EC, Lawrie SM, Cosway R. What does the Edinburgh high-risk study tell us about schizophrenia? Am J Med Genet. 2002;114(8):906–912. doi: 10.1002/ajmg.b.10304. [DOI] [PubMed] [Google Scholar]
- Kammerer CM, Schneider JL, Cole SA, Hixson JE, Samollow PB, O’Connell JR, Perez R, Dyer TD, Almasy L, Blangero J, Bauer RL, Mitchell BD. Quantitative trait loci on chromosomes 2p, 4p, and 13q influence bone mineral density of the forearm and hip in Mexican Americans. J Bone Miner Res. 2003;18(12):2245–2252. doi: 10.1359/jbmr.2003.18.12.2245. [DOI] [PubMed] [Google Scholar]
- Kapur S, Phillips AG, Insel TR. Why has it taken so long for biological psychiatry to develop clinical tests and what to do about it? Mol Psychiatry. 2012;17(12):1174–1179. doi: 10.1038/mp.2012.105. [DOI] [PubMed] [Google Scholar]
- Kathiresan S, Willer C, Peloso G, Demissie S, Musunuru K, Schadt E, Kaplan L, Bennett D, Li Y, Tanaka T, Voight B, Bonnycastle L, Jackson A, Crawford G, Surti A, Guiducci C, Burtt N, Parish S, Clarke R, Zelenika D, Kubalanza K, Morken M, Scott L, Stringham H, Galan P, Swift A, Kuusisto J, Bergman R, Sundvall J, Laakso M, Ferrucci L, Scheet P, Sanna S, Uda M, Yang Q, Lunetta K, Dupuis J, de Bakker P, O’Donnell C, Chambers J, Kooner J, Hercberg S, Meneton P, Lakatta E, Scuteri A, Schlessinger D, Tuomilehto J, Collins F, Groop L, Altshuler D, Collins R, Lathrop G, Melander O, Salomaa V, Peltonen L, Orho-Melander M, Ordovas J, Boehnke M, Abecasis G, Mohlke K, Cupples L. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41(1):56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel D, Demissie S, Dupuis J, Lunetta K, Murabito J, Karasik D. Genome-wide association with bone mass and geometry in the Framingham Heart Study. BMC Med Genet. 2007;8(Suppl 1):S14. doi: 10.1186/1471-2350-8-S1-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles E, MAC, de Almeida M, JEC, DRM, Sprooten E, Dyer T, Göring H, Olvera R, Fox P, LA, Duggirala R, Kent J, Blangero J, Glahn DC. Genome-Wide Significant Localization for Working and Spatial Memory: Identifying Genes for Psychosis Using Models of Cognition. Am J Med Genet B Neuropsychiatr Genet. 2014;165(1):84–95. doi: 10.1002/ajmg.b.32211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leboyer M, Bellivier F, Nosten-Bertrand M, Jouvent R, Pauls D, Mallet J. Psychiatric genetics: search for phenotypes. Trends in Neurosciences. 1998;21(3):102–105. doi: 10.1016/s0166-2236(97)01187-9. [DOI] [PubMed] [Google Scholar]
- Lee J, Park S. Working memory impairments in schizophrenia: a meta-analysis. J Abnorm Psychol. 2005;114(4):599–611. doi: 10.1037/0021-843X.114.4.599. [DOI] [PubMed] [Google Scholar]
- Lee SH, Decandia TR, Ripke S, Yang J, Sullivan PF, Goddard ME, Keller MC, Visscher PM, Wray NR (PGC-SCZ) TSPG-WASC, (ISC) TISC (MGS) TMGoSC. Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat Genet. 2012;44(3):247–250. doi: 10.1038/ng.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenox RH, Gould TD, Manji HK. Endophenotypes in bipolar disorder. American Journal of Medical Genetics. 2002;114(4):391–406. doi: 10.1002/ajmg.10360. [DOI] [PubMed] [Google Scholar]
- Lenzenweger MF. Endophenotype, intermediate phenotype, biomarker: definitions, concept comparisons, clarifications. Depress Anxiety. 2013;30(3):185–189. doi: 10.1002/da.22042. [DOI] [PubMed] [Google Scholar]
- Liu J, Pei Y, Papasian CJ, Deng HW. Bivariate association analyses for the mixture of continuous and binary traits with the use of extended generalized estimating equations. Genet Epidemiol. 2009;33(3):217–227. doi: 10.1002/gepi.20372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald AW, Thermenos HW, Barch DM, Seidman LJ. Imaging genetic liability to schizophrenia: systematic review of FMRI studies of patients’ nonpsychotic relatives. Schizophr Bull. 2009;35(6):1142–1162. doi: 10.1093/schbul/sbn053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald C, Bullmore ET, Sham PC, Chitnis X, Wickham H, Bramon E, Murray RM. Association of genetic risks for schizophrenia and bipolar disorder with specific and generic brain structural endophenotypes. Arch Gen Psychiatry. 2004;61(10):974–984. doi: 10.1001/archpsyc.61.10.974. [DOI] [PubMed] [Google Scholar]
- Meyer-Lindenberg A, Weinberger DR. Intermediate phenotypes and genetic mechanisms of psychiatric disorders. Nat Rev Neurosci. 2006;7(10):818–827. doi: 10.1038/nrn1993. [DOI] [PubMed] [Google Scholar]
- Miller GA, Rockstroh B. Endophenotypes in psychopathology research: where do we stand? Annu Rev Clin Psychol. 2013;9:177–213. doi: 10.1146/annurev-clinpsy-050212-185540. [DOI] [PubMed] [Google Scholar]
- Minzenberg M, Laird A, Thelen S, Carter C, Glahn D. Meta-analysis of 41 functional neuroimaging studies of executive function in schizophrenia. Arch Gen Psychiatry. 2009;66(8):811–822. doi: 10.1001/archgenpsychiatry.2009.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell BD, Cole SA, Bauer RL, Iturria SJ, Rodriguez EA, Blangero J, MacCluer JW, Hixson JE. Genes influencing variation in serum osteocalcin concentrations are linked to markers on chromosomes 16q and 20q. J Clin Endocrinol Metab. 2000;85(4):1362–1366. doi: 10.1210/jcem.85.4.6571. [DOI] [PubMed] [Google Scholar]
- Narr KL, van Erp TG, Cannon TD, Woods RP, Thompson PM, Jang S, Blanton R, Poutanen VP, Huttunen M, Lonnqvist J, Standerksjold-Nordenstam CG, Kaprio J, Mazziotta JC, Toga AW. A twin study of genetic contributions to hippocampal morphology in schizophrenia. Neurobiol Dis. 2002;11(1):83–95. doi: 10.1006/nbdi.2002.0548. [DOI] [PubMed] [Google Scholar]
- O’Donovan M, Craddock N, Norton N, Williams H, Peirce T, Moskvina V, Nikolov I, Hamshere M, Carroll L, Georgieva L, Dwyer S, Holmans P, Marchini J, Spencer C, Howie B, Leung H, Hartmann A, Möller H, Morris D, Shi Y, Feng G, Hoffmann P, Propping P, Vasilescu C, Maier W, Rietschel M, Zammit S, Schumacher J, Quinn E, Schulze T, Williams N, Giegling I, Iwata N, Ikeda M, Darvasi A, Shifman S, He L, Duan J, Sanders A, Levinson D, Gejman P, Cichon S, Nöthen M, Gill M, Corvin A, Rujescu D, Kirov G, Owen M, Buccola N, Mowry B, Freedman R, Amin F, Black D, Silverman J, Byerley W, Cloninger C. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008;40(9):1053–1055. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- Park S, Holzman PS. Schizophrenics show spatial working memory deficits. Archives of General Psychiatry. 1992;49(12):975–982. doi: 10.1001/archpsyc.1992.01820120063009. [DOI] [PubMed] [Google Scholar]
- Park S, Holzman PS, Goldman-Rakic PS. Spatial working memory deficits in the relatives of schizophrenic patients. Arch Gen Psychiatry. 1995;52(10):821–828. doi: 10.1001/archpsyc.1995.03950220031007. [DOI] [PubMed] [Google Scholar]
- Polman CH, Reingold SC, Banwell B, Clanet M, Cohen JA, Filippi M, Fujihara K, Havrdova E, Hutchinson M, Kappos L, Lublin FD, Montalban X, O’Connor P, Sandberg-Wollheim M, Thompson AJ, Waubant E, Weinshenker B, Wolinsky JS. Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann Neurol. 2011;69(2):292–302. doi: 10.1002/ana.22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Wray N, Stone J, Visscher P, O’Donovan M, Sullivan P, Sklar P, Consortium IS. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460(7256):748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radich JP, Mao M, Stepaniants S, Biery M, Castle J, Ward T, Schimmack G, Kobayashi S, Carleton M, Lampe J, Linsley PS. Individual-specific variation of gene expression in peripheral blood leukocytes. Genomics. 2004;83(6):980–988. doi: 10.1016/j.ygeno.2003.12.013. [DOI] [PubMed] [Google Scholar]
- Reichenberg A, Harvey PD. Neuropsychological impairments in schizophrenia: Integration of performance-based and brain imaging findings. Psychol Bull. 2007;133(5):833–858. doi: 10.1037/0033-2909.133.5.833. [DOI] [PubMed] [Google Scholar]
- Ripke S, Wray NR, Lewis CM, Hamilton SP, Weissman MM, Breen G, Byrne EM, Blackwood DH, Boomsma DI, Cichon S, Heath AC, Holsboer F, Lucae S, Madden PA, Martin NG, McGuffin P, Muglia P, Noethen MM, Penninx BP, Pergadia ML, Potash JB, Rietschel M, Lin D, Müller-Myhsok B, Shi J, Steinberg S, Grabe HJ, Lichtenstein P, Magnusson P, Perlis RH, Preisig M, Smoller JW, Stefansson K, Uher R, Kutalik Z, Tansey KE, Teumer A, Viktorin A, Barnes MR, Bettecken T, Binder EB, Breuer R, Castro VM, Churchill SE, Coryell WH, Craddock N, Craig IW, Czamara D, De Geus EJ, Degenhardt F, Farmer AE, Fava M, Frank J, Gainer VS, Gallagher PJ, Gordon SD, Goryachev S, Gross M, Guipponi M, Henders AK, Herms S, Hickie IB, Hoefels S, Hoogendijk W, Hottenga JJ, Iosifescu DV, Ising M, Jones I, Jones L, Jung-Ying T, Knowles JA, Kohane IS, Kohli MA, Korszun A, Landen M, Lawson WB, Lewis G, Macintyre D, Maier W, Mattheisen M, McGrath PJ, McIntosh A, McLean A, Middeldorp CM, Middleton L, Montgomery GM, Murphy SN, Nauck M, Nolen WA, Nyholt DR, O’Donovan M, Oskarsson H, Pedersen N, Scheftner WA, Schulz A, Schulze TG, Shyn SI, Sigurdsson E, Slager SL, Smit JH, Stefansson H, Steffens M, Thorgeirsson T, Tozzi F, Treutlein J, Uhr M, van den Oord EJ, Van Grootheest G, Völzke H, Weilburg JB, Willemsen G, Zitman FG, Neale B, Daly M, Levinson DF, Sullivan PF Consortium MDDWGotPG. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry. 2013;18(4):497–511. doi: 10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman MV, Wray GA. Abundant raw material for cis-regulatory evolution in humans. Mol Biol Evol. 2002;19(11):1991–2004. doi: 10.1093/oxfordjournals.molbev.a004023. [DOI] [PubMed] [Google Scholar]
- Silventoinen K, Sammalisto S, Perola M, Boomsma DI, Cornes BK, Davis C, Dunkel L, De Lange M, Harris JR, Hjelmborg JV, Luciano M, Martin NG, Mortensen J, Nistico L, Pedersen NL, Skytthe A, Spector TD, Stazi MA, Willemsen G, Kaprio J. Heritability of adult body height: a comparative study of twin cohorts in eight countries. Twin Res. 2003;6(5):399–408. doi: 10.1375/136905203770326402. [DOI] [PubMed] [Google Scholar]
- Sing CF, Stengård JH, Kardia SL. Genes, environment, and cardiovascular disease. Arterioscler Thromb Vasc Biol. 2003;23(7):1190–1196. doi: 10.1161/01.ATV.0000075081.51227.86. [DOI] [PubMed] [Google Scholar]
- Sklar P, Ripke S, Scott LJ, Andreassen OA, Cichon S, Craddock N, Edenberg HJ, Nurnberger JI, Rietschel M, Blackwood D, Corvin A, Flickinger M, Guan W, Mattingsdal M, McQuillin A, Kwan P, Wienker TF, Daly M, Dudbridge F, Holmans PA, Lin D, Burmeister M, Greenwood TA, Hamshere ML, Muglia P, Smith EN, Zandi PP, Nievergelt CM, McKinney R, Shilling PD, Schork NJ, Bloss CS, Foroud T, Koller DL, Gershon ES, Liu C, Badner JA, Scheftner WA, Lawson WB, Nwulia EA, Hipolito M, Coryell W, Rice J, Byerley W, McMahon FJ, Schulze TG, Berrettini W, Lohoff FW, Potash JB, Mahon PB, McInnis MG, Zöllner S, Zhang P, Craig DW, Szelinger S, Barrett TB, Breuer R, Meier S, Strohmaier J, Witt SH, Tozzi F, Farmer A, McGuffin P, Strauss J, Xu W, Kennedy JL, Vincent JB, Matthews K, Day R, Ferreira MA, O’Dushlaine C, Perlis R, Raychaudhuri S, Ruderfer D, Hyoun PL, Smoller JW, Li J, Absher D, Thompson RC, Meng FG, Schatzberg AF, Bunney WE, Barchas JD, Jones EG, Watson SJ, Myers RM, Akil H, Boehnke M, Chambert K, Moran J, Scolnick E, Djurovic S, Melle I, Morken G, Gill M, Morris D, Quinn E, Mühleisen TW, Degenhardt FA, Mattheisen M, Schumacher J, Maier W, Steffens M, Propping P, Nöthen MM, Anjorin A, Bass N, Gurling H, Kandaswamy R, Lawrence J, McGhee K, McIntosh A, McLean AW, Muir WJ, Pickard BS, Breen G, St Clair D, Caesar S, Gordon-Smith K, Jones L, Fraser C, Green EK, Grozeva D, Jones IR, Kirov G, Moskvina V, Nikolov I, O’Donovan MC, Owen MJ, Collier DA, Elkin A, Williamson R, Young AH, Ferrier IN, Stefansson K, Stefansson H, Thornorgeirsson T, Steinberg S, Gustafsson O, Bergen SE, Nimgaonkar V, Hultman C, Landén M, Lichtenstein P, Sullivan P, Schalling M, Osby U, Backlund L, Frisén L, Langstrom N, Jamain S, Leboyer M, Etain B, Bellivier F, Petursson H, Sigur Sson E, Müller-Mysok B, Lucae S, Schwarz M, Schofield PR, Martin N, Montgomery GW, Lathrop M, Oskarsson H, Bauer M, Wright A, Mitchell PB, Hautzinger M, Reif A, Kelsoe JR, Purcell SM Group PGCBDW. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43(10):977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaats-Willemse D, Swaab-Barneveld H, de Sonneville L, van der Meulen E, Buitelaar J. Deficient response inhibition as a cognitive endophenotype of ADHD. J Am Acad Child Adolesc Psychiatry. 2003;42(10):1242–1248. doi: 10.1097/00004583-200310000-00016. [DOI] [PubMed] [Google Scholar]
- Snitz BE, Macdonald AW, Carter CS. Cognitive deficits in unaffected first-degree relatives of schizophrenia patients: a meta-analytic review of putative endophenotypes. Schizophr Bull. 2006;32(1):179–194. doi: 10.1093/schbul/sbi048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So HC, Gui AH, Cherny SS, Sham PC. Evaluating the heritability explained by known susceptibility variants: a survey of ten complex diseases. Genet Epidemiol. 2011;35(5):310–317. doi: 10.1002/gepi.20579. [DOI] [PubMed] [Google Scholar]
- Speed D, Hemani G, Johnson MR, Balding DJ. Improved heritability estimation from genome-wide SNPs. Am J Hum Genet. 2012;91(6):1011–1021. doi: 10.1016/j.ajhg.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprooten E, Brumbaugh M, Knowles E, McKay D, Lewis J, Barrett J, Landau S, Cyr L, Kochunov P, Winkler A, Pearlson G, Glahn D. Reduced White-Matter Integrity in Sibling Pairs Discordant for Bipolar Disorder. Am J Psychiatry. doi: 10.1176/appi.ajp.2013.12111462. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprooten E, Sussmann JE, Clugston A, Peel A, McKirdy J, Moorhead TW, Anderson S, Shand AJ, Giles S, Bastin ME, Hall J, Johnstone EC, Lawrie SM, McIntosh AM. White matter integrity in individuals at high genetic risk of bipolar disorder. Biol Psychiatry. 2011;70(4):350–356. doi: 10.1016/j.biopsych.2011.01.021. [DOI] [PubMed] [Google Scholar]
- Turkheimer E, Haley A, Waldron M, D’Onofrio B, Gottesman II. Socioeconomic status modifies heritability of IQ in young children. Psychol Sci. 2003;14(6):623–628. doi: 10.1046/j.0956-7976.2003.psci_1475.x. [DOI] [PubMed] [Google Scholar]
- van Haren NE, Rijsdijk F, Schnack HG, Picchioni MM, Toulopoulou T, Weisbrod M, Sauer H, van Erp TG, Cannon TD, Huttunen MO, Boomsma DI, Hulshoff Pol HE, Murray RM, Kahn RS. The genetic and environmental determinants of the association between brain abnormalities and schizophrenia: the schizophrenia twins and relatives consortium. Biol Psychiatry. 2012;71(10):915–921. doi: 10.1016/j.biopsych.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney AR, Diehn M, Popper SJ, Alizadeh AA, Boldrick JC, Relman DA, Brown PO. Individuality and variation in gene expression patterns in human blood. Proc Natl Acad Sci U S A. 2003;100(4):1896–1901. doi: 10.1073/pnas.252784499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer C, Speliotes E, Loos R, Li S, Lindgren C, Heid I, Berndt S, Elliott A, Jackson A, Lamina C, Lettre G, Lim N, Lyon H, McCarroll S, Papadakis K, Qi L, Randall J, Roccasecca R, Sanna S, Scheet P, Weedon M, Wheeler E, Zhao J, Jacobs L, Prokopenko I, Soranzo N, Tanaka T, Timpson N, Almgren P, Bennett A, Bergman R, Bingham S, Bonnycastle L, Brown M, Burtt N, Chines P, Coin L, Collins F, Connell J, Cooper C, Smith G, Dennison E, Deodhar P, Elliott P, Erdos M, Estrada K, Evans D, Gianniny L, Gieger C, Gillson C, Guiducci C, Hackett R, Hadley D, Hall A, Havulinna A, Hebebrand J, Hofman A, Isomaa B, Jacobs K, Johnson T, Jousilahti P, Jovanovic Z, Khaw K, Kraft P, Kuokkanen M, Kuusisto J, Laitinen J, Lakatta E, Luan J, Luben R, Mangino M, McArdle W, Meitinger T, Mulas A, Munroe P, Narisu N, Ness A, Northstone K, O’Rahilly S, Purmann C, Rees M, Ridderstråle M, Ring S, Rivadeneira F, Ruokonen A, Sandhu M, Saramies J, Scott L, Scuteri A, Silander K, Sims M, Song K, Stephens J, Stevens S, Stringham H, Tung Y, Valle T, Van Duijn C, Vimaleswaran K, Vollenweider P, Waeber G, Wallace C, Watanabe R, Waterworth D, Watkins N, Witteman J, Zeggini E, Zhai G, Zillikens M, Altshuler D, Caulfield M, Chanock S, Farooqi I, Ferrucci L, Guralnik J, Hattersley A, Hu F, Jarvelin M, Laakso M, Mooser V, Ong K, Ouwehand W, Salomaa V, Samani N, Spector T, Tuomi T, Tuomilehto J, Uda M, Uitterlinden A, Wareham N, Deloukas P, Frayling T, Groop L, Hayes R, Hunter D, Mohlke K, Peltonen L, Schlessinger D, Strachan D, Wichmann H, McCarthy M, Boehnke M, Barroso I, Abecasis G, Hirschhorn J. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41(1):25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J, Blangero J. Power of variance component linkage analysis-II. Discrete traits. Ann Hum Genet. 2004;68(Pt 6):620–632. doi: 10.1046/j.1529-8817.2004.00128.x. [DOI] [PubMed] [Google Scholar]
- Williams JT, Van Eerdewegh P, Almasy L, Blangero J. Joint multipoint linkage analysis of multivariate qualitative and quantitative traits. I. Likelihood formulation and simulation results. Am J Hum Genet. 1999;65:1134–1147. doi: 10.1086/302570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8(3):206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- Yang J, Manolio TA, Pasquale LR, Boerwinkle E, Caporaso N, Cunningham JM, de Andrade M, Feenstra B, Feingold E, Hayes MG, Hill WG, Landi MT, Alonso A, Lettre G, Lin P, Ling H, Lowe W, Mathias RA, Melbye M, Pugh E, Cornelis MC, Weir BS, Goddard ME, Visscher PM. Genome partitioning of genetic variation for complex traits using common SNPs. Nat Genet. 2011;43(6):519–525. doi: 10.1038/ng.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Diao G. Joint association analysis of bivariate quantitative and qualitative traits. BMC Proc. 2011;5(Suppl 9):S74. doi: 10.1186/1753-6561-5-S9-S74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou F, Carrasquillo MM, Pankratz VS, Belbin O, Morgan K, Allen M, Wilcox SL, Ma L, Walker LP, Kouri N, Burgess JD, Younkin LH, Younkin SG, Younkin CS, Bisceglio GD, Crook JE, Dickson DW, Petersen RC, Graff-Radford N, Ertekin-Taner N. Gene expression levels as endophenotypes in genome-wide association studies of Alzheimer disease. Neurology. 2010;74(6):480–486. doi: 10.1212/WNL.0b013e3181d07654. [DOI] [PMC free article] [PubMed] [Google Scholar]