Summary
Although prokaryotes ordinarily undergo binary fission to produce two identical daughter cells, some are able to undergo alternative developmental pathways that produce daughter cells of distinct cell morphology and fate. One such example is a developmental program called sporulation in the bacterium Bacillus subtilis, which occurs under conditions of environmental stress. Sporulation has long been used as a model system to help elucidate basic processes of developmental biology including transcription regulation, intercellular signaling, membrane remodeling, protein localization, and cell fate determination. This review highlights some of the recent work that has been done to further understand prokaryotic cell differentiation during sporulation and its potential applications.
Introduction
Understanding the mechanisms that drive cell differentiation and morphogenesis are essential in answering the question of how organisms develop. However, elucidating these mechanisms can be difficult due to the complex and intertwined processes that occur during development (Sasai, 2013; Hartwell and Weinert, 1989). One approach to this problem has been to study the relatively simple developmental program of endospore formation (“sporulation”) in the bacterium Bacillus subtilis. During sporulation a single rod-shaped cell divides asymmetrically, resulting in two genetically identical daughter cells that undergo different cell fates (Stragier and Losick, 1996; Errington, 2003; Piggot and Hilbert, 2004). Sporulation in B. subtilis is a particularly attractive system to look at cell differentiation and morphogenesis not only because of the relative simplicity of the sporulation developmental program, but also because of the genetic tractability of the system. B. subtilis is naturally competent and genes necessary for sporulation are often non-essential for normal growth, both of which facilitate the identification of novel factors that participate in this developmental process. As a result, sporulation studies have provided significant insights into basic biological processes such as differential gene expression, membrane remodeling, intercellular communication, subcellular protein localization, and morphogenesis.
B. subtilis is ubiquitous in nature and can successfully adapt to various changes in the environment. Under stressful conditions B. subtilis is able to initiate many survival mechanisms such as motility, uptake of exogenous DNA, biofilm formation and sporulation (Vlamakis et al., 2013; Rao et al., 2008; Burton and Dubnau, 2010). The purpose of sporulation is to produce a largely metabolically inactive dormant cell type called an “endospore” (hereafter, referred to simply as a “spore”) that is able to survive harsh environmental conditions until favorable growth conditions are restored (Paredes-Sabja et al., 2011). Bacterial spores are one of nature’s most resilient cell types and are able to survive under controlled laboratory conditions for several decades, and perhaps even longer in the environment (Jacotot and Virat, 1954; Cano and Borucki, 1995; Setlow, 2007; Vreeland et al., 2000). When the spore senses environmental conditions are conducive to growth, it is able to germinate and resume its vegetative cell cycle.
Sporulation initiates when the rod-shaped B. subtilis divides asymmetrically, elaborating a “polar septum” that results in two genetically identical but morphologically distinct compartments: a larger “mother cell” and a smaller “forespore”, each of which will ultimately experience different cell fates (Fig. 1). Both compartments briefly remain side-by-side, held together by the external cell wall. The initially flat polar septum then begins to curve as the mother cell swallows the forespore (a process called “engulfment”), producing a forespore that resides as a double membrane-bound, roughly spherical, organelle inside the mother cell cytosol. The forespore eventually matures into a partially dehydrated, dormant cell that is released into the environment when the mother cell undergoes programmed cell lysis.
Figure 1.
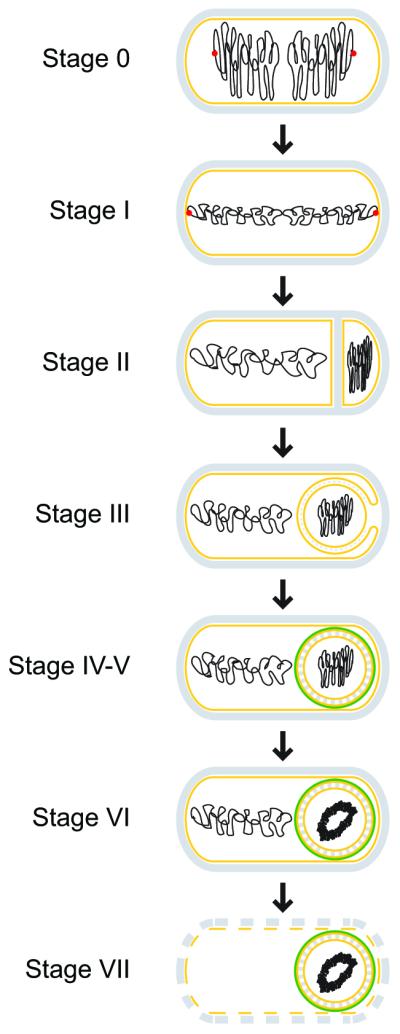
Schematic representation of morphological changes that occur during sporulation in Bacillus subtilis. Distinct stages of sporulation are denoted with a Roman numeral, according to the numbering scheme proposed by Ryter (Ryter, 1965). Peptidoglycan is depicted in gray, membranes are depicted in yellow, DNA is depicted in black, the position of the origin of replication of the chromosomes is shown as a red dot at stage 0 and I, and the spore coat is depicted in green. At stage 0, chromosomes are replicated, but no obvious morphological landmarks of sporulation are yet present. Stage I is defined by chromosome condensation and the anchoring of the origins of replication to the extreme poles of the cell. In stage II, the polar septum is elaborated, followed by engulfment of the forespore in stage III. Stage IV and V represent cortex and coat assembly, respectively. Stage VI refers to “spore maturation”; a particularly obvious morphological feature elaborated at this stage is the tightly condensed, toroidal structure of the forespore chromosome. In stage VII, the mother cell lyses, releasing the mature, largely dormant spore into the environment.
Here, we will provide a brief overview of the molecular mechanisms underlying the sporulation process from the decision of a cell to enter the sporulation program to how the cell undergoes the significant morphological changes in order to become a distinct cell type that is resistant to various environmental insults, with an attempt to highlight primary literature that has been published recently. As a roadmap, we will present the sporulation program in separate sections in which so-called “stages” of sporulation are numbered “0” to “V”, reflecting a classical nomenclature that was based on various morphological landmarks as viewed by electron microscopy (Schaeffer et al., 1963; Piggot and Coote, 1976). It is important to note, though, that as techniques have advanced, it has become clear that sporulation is not composed of distinct stages that occur sequentially. Instead, these stages actually lie along a continuum in which there may be significant overlap in terms of when a “stage” begins and ends.
Stage 0: The decision to sporulate
As with many other developmental programs, the entry into sporulation is closely regulated and relies on a series of feedback and feed-forward loops. In the bacterial population sporulation does not occur homogenously, but rather occurs in subpopulations. Presumably, this is a bet-hedging strategy that allows the cell to absolutely confirm the need to sporulate prior to engaging in this highly energy-consuming and, once committed, irreversible developmental program (Veening et al., 2008).
The first bet-hedging strategy to delay entry into sporulation is the “cannibalistic” behavior displayed by a subpopulation of cells that are the first to detect the onset of starvation conditions. During cannibalism, this subpopulation of cells kills neighboring isogenic siblings that have not yet detected the onset of such conditions (Gonzalez-Pastor et al., 2003). Cannibalism is reliant on two secreted killing factors, Skf and Sdp (Liu et al., 2010; Perez Morales et al., 2013), which are produced by cells in a biofilm that produce the biofilm matrix (Lopez et al., 2009). The death of surrounding cells releases nutrients into the environment to support the growth of the subpopulation that produced the toxins. The toxin producing subpopulation is protected through the concurrent production of a protective factor (Ellermeier et al., 2006). In this way, cannibalism is thought to be a mechanism to delay sporulation and to eliminate cells that are no longer beneficial to the population as it moves toward biofilm formation (Mitri et al., 2011). Consistent with this model, deletion of genes required for cannibalism result in a faster and more homogeneous entry of cells into the sporulation program (Gonzalez-Pastor et al., 2003).
The transition of B. subtilis from vegetative growth to sporulation is largely governed by the transcriptional master regulator Spo0A, which also regulates biofilm formation (Hamon and Lazazzera, 2001). Spo0A transcriptional activity is activated by a ‘phosphorelay’ system that is governed by five autophosphorylating histidine kinases (KinA-KinE) that respond to different environmental stresses. While “limited nutrient availability” is broadly defined as the signal for entry into sporulation, the identification of specific molecular ligands that activate the histidine kinases has remained elusive. This difficulty is largely due to the wide array of environmental inputs sensed by the bacterium and the somewhat redundant functions of the sensor kinases (LeDeaux et al., 1995). A recent strategy to approach the identification of molecular ligands through co-crystallization successfully identified pyruvate as a potential ligand of KinD (Wu et al., 2013). Pyruvate is involved in numerous metabolic pathways and it seems reasonable that its levels in the extracellular environment may serve as an indicator of growth conditions. However, it is still unclear whether pyruvate is the physiological ligand of KinD and what effects this interaction may have on sporulation.
Upon activation and autophosphorylation, the phosphoryl group from the histidine kinases is transferred to Spo0A via the phosphotransferases Spo0F and Spo0B which results in an active phosphorylated Spo0A (referred to as “Spo0A~P”) (Burbulys et al., 1991). Spo0A~P then goes on to directly regulate the expression of approximately 121 genes (Molle et al., 2003) including activation of genes necessary for sporulation. Counter-balancing the production of Spo0A~P are several phosphatases including members of the Rap family of phosphatases (Rap A, B, E and H) and the Spo0E phosphatase (Perego et al., 1994). Regulation of the activity of the kinases and phosphatases determines the levels of Spo0A~P and ultimately whether or not sporulation is initiated. The activity of the Rap phosphatases is regulated by small peptides encoded by phr genes, which are often found in operons with the rap genes (Mueller and Sonenshein, 1992). X-ray analyses have indicated that Phr peptides bind and regulate Rap activity by inducing a conformational change (Baker and Neiditch, 2011; Parashar et al., 2013; Gallego del Sol and Marina, 2013).
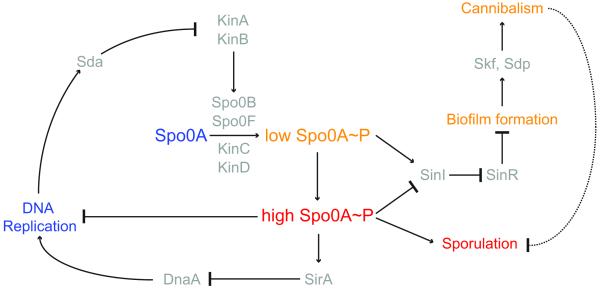
The levels of Spo0A~P are responsible for determining the bacterium’s developmental choices. Lower levels of Spo0A~P result in biofilm formation through promotion of matrix production while higher levels of Spo0A~P promote sporulation (Fig. 2). The mechanism through which Spo0A~P is able to regulate these two distinct cell fates is dependent on its regulation of the levels of the matrix gene repressor SinR and its antirepressor SinI (Chai et al., 2011). The sinI regulatory region has numerous Spo0A~P operator sites that differ in affinity, which allows its expression to be regulated directly by the levels of Spo0A~P. At lower levels of Spo0A~P the high affinity Spo0A~P operator is bound (Fujita and Losick, 2005) and promotes expression of sinI, leading to matrix production and biofilm formation (Fujita et al., 2005). At higher levels of Spo0A~P the lower affinity operators are then able to bind Spo0A~P, which hinders the expression of sinI and promotes expression of sporulation genes that also have low affinity Spo0A~P operators (Chai et al., 2011; Kearns et al., 2005). While high levels of Spo0A~P are important for regulating entry into sporulation, the dynamics through which it achieves high levels of Spo0A~P is also important. The gradual accumulation of Spo0A~P appears to exert a temporal control over the Spo0A regulon, which is necessary for robust sporulation (Vishnoi et al., 2013).
Figure 2.
Genetic circuitry that governs the entry into sporulation. Arrows indicate activation; repression is denoted by a bar. Developmental events are depicted in the color corresponding to Spo0A levels or phosphorylation states that govern that event. Thus, unphosphorylated Spo0A corresponds to active DNA replication; low levels of phosphorylated Spo0A (SpoA~P) leads to biofilm formation and cannabilistic behavior; and high levels of phosphorylated Spo0A drives the entry into sporulation. Proteins other than Spo0A that participate in each activation or repression step are depicted in gray.
Previous studies proposed a model in which there is a threshold level of Spo0A~P, in addition to the phosphorelay components, that must be crossed in order for sporulation initiation to occur (Eswaramoorthy et al., 2010a; Eswaramoorthy et al., 2010b; Fujita and Losick, 2005). However, recent studies have found significant overlap in Spo0A~P levels in sporulating and non-sporulating cells (Levine et al., 2012) indicating there may also be other downstream events that are responsible for the decision to enter sporulation. Consistent with this idea is the observation that entry into sporulation may still be reversible after activation of several Spo0A-regulated spo genes and only becomes irreversible upon activation of σF in the forespore and σE in the mother cell (Dworkin and Losick, 2005). The decision to commit to sporulation instead seems to rely on the ultrasensitive activation of σE (Narula et al., 2012), which occurs after asymmetric septation and σF activation in the forespore.
Stage I: Axial filamentation and ensuring correct chromosome copy number
At the onset of sporulation, the cell harbors two chromosomes: one for the mother cell and one for the forespore. The duplicated chromosomes form a condensed serpentine-like structure called the axial filament (Ryter et al., 1966) that stretches from one pole of the cell to the other. The RacA protein is necessary for anchoring the two chromosomes to the cell poles to promote proper chromosome segregation (Ben-Yehuda et al., 2003). RacA binds to GC-rich inverted repeats located around the origin of replication (Ben-Yehuda et al., 2005) and localizes to the cell poles through its interaction with DivIVA, which in turn localizes to the two poles of the cell through the recognition of highly negatively curved membranes (Lenarcic et al., 2009; Ramamurthi and Losick, 2009). In this way, RacA ensures that each daughter cell receives one origin of replication (and, by extension, one chromosome). As such, RacA chromosomal binding sites are functionally analogous to eukaryotic centromeres, and RacA itself, while not physically driving chromosome movement, provides a function analogous to that of the eukaryotic mitotic spindle in maintaining chromosome integrity during eukaryotic cell division.
In addition to proper chromosome segregation, proper chromosome number is also necessary for robust sporulation and has been found to be tightly regulated via at least three proteins: SirA (sporulation inhibitor of replication A), Sda (suppressor of dnaA1) and Spo0A~P. Transcription of sirA is under the control of Spo0A~P and occurs upon entry into sporulation. SirA interacts directly with DnaA to inhibit its binding to the origin of replication, which prevents the initiation of additional rounds of DNA replication during sporulation (Rahn-Lee et al., 2011). Sda, on the other hand, inhibits entry into sporulation by binding to the major sporulation histidine kinase KinA during active DNA replication and in response to DNA damage and replication defects (Cunningham and Burkholder, 2009). As a result, entry into sporulation is restricted to the period between rounds of DNA replication (Burkholder et al., 2001; Veening et al., 2009). While Spo0A~P plays an indirect role in regulating chromosome number through its transcriptional regulation of sirA, it has also been found to play a more direct role through its ability to bind to sites around the origin of replication. Removal of Spo0A~P binding sites near the origin of replication resulted in an increase in chromosome copy number indicating that Spo0A~P binding to these sites acts as an additional mechanism to inhibit active DNA replication during sporulation (Boonstra et al., 2013).
Stage II: Asymmetric septation
The sporulation program is driven by a cascade of compartment-specific sigma factors that is initiated by asymmetric division of the cell. Understanding the activation of compartment-specific sigma factors during sporulation has revealed conserved mechanisms underlying intercellular signaling and the coupling of transcription with morphological changes in the cell. The transition from a medial septum to an asymmetric septum, which divides the cell into a mother cell and forespore, is a morphological hallmark of sporulation. This switch to a polar septum is dependent on two factors: an increase in levels of the cell division protein FtsZ and the production of the SpoIIE protein, which performs a poorly understood function in deploying FtsZ to polar sites (Carniol et al., 2005). After asymmetric division, the first sporulation-specific sigma factor, forespore-specific σF, is activated. Prior to asymmetric division, σF is produced under the regulation of Spo0A~P, but held in an inactive state by the anti-sigma factor SpoIIAB. After completion of asymmetric division, SpoIIE (which localizes to the polar septum) performs a second function, wherein it dephosphorylates SpoIIAA, which binds and sequesters SpoIIAB, thereby relieving the inhibition of σF (Duncan et al., 1995). Curiously, although SpoIIE is initially produced in both the mother cell and the forespore, activation of σF only occurs in the forespore. Although the biochemical mechanism of σF activation is well understood, the cell biological mechanism that explains the forespore-specific activation of σF is not well known. Part of the answer may be dependent on a preferential localization of SpoIIE on the forespore side of the polar septum (Guberman et al., 2008) and also the temporary genetic asymmetry between the forespore and mother cell that leads to a decrease in the levels of the SpoIIAB anti-sigma factor in the forespore (Dworkin and Losick, 2001).
The temporary genetic asymmetry arises because, at the time of asymmetric septation, the polar septum bisects the axial filament and results in only about a third of the chromosome being harbored in the forespore (Wu and Errington, 1998). The DNA translocase SpoIIIE then pumps the remaining 70% of the chromosome residing in the mother cell into the forespore (Wu and Errington, 1994; Sharp and Pogliano, 2002; Ptacin et al., 2008; Becker and Pogliano, 2007; Burton et al., 2007; Fiche et al., 2013). Prior to translocation of the remaining 70%, the forespore can only express the genes residing on the 30% of the chromosome it harbors initially. This genetic asymmetry has been proposed to play a part in orchestrating the compartment specific activities that occur throughout sporulation (Frandsen et al., 1999). Another example of this is the mother cell compartment specific re-activation of de novo fatty acid synthesis. Many genes necessary for de novo lipid synthesis are located in the portion of the chromosome initially excluded from the forespore, resulting in the forespore’s inability to re-activate lipid synthesis (Pedrido et al., 2013). Re-activation of de novo lipid synthesis is dependent on Spo0A~P and is required for the mother cell-specific activation of σE, the second sporulation-specific sigma factor (Pedrido et al., 2013).
Similar to σF, σE is produced prior to asymmetric division under the control of Spo0A~P and is found in both compartments after septation (Fujita and Losick, 2002). However, σE is produced initially as an inactive pro-σE precursor and is specifically activated only in the mother cell. While it was previously established that spoIIGA is required for processing of pro-σE to its mature form (Jonas et al., 1988; Stragier et al., 1988), it was only more recently that its product SpoIIGA was identified as a novel type of aspartic protease (Imamura et al., 2008). Modeling and mutational evidence suggest that SpoIIGA forms dimers similar to the HIV-1 protease and support a model in which SpoIIGA exists in an inactive state that is then activated through a conformational change induced by association with SpoIIR (Imamura et al., 2008; Hofmeister et al., 1995). SpoIIR is produced in the forespore under σF control and is then secreted into the intermembrane space of the septum that separates the mother cell and forespore, where it activates SpoIIGA. SpoIIR’s ability to activate SpoIIGA is dependent on the acylation of its threonine residue (T27) and requires de novo fatty acid synthesis (Diez et al., 2012). It is hypothesized that because de novo fatty acid synthesis only occurs in the mother cell, only SpoIIR molecules localized to the mother cell side of the septum would be able to be acylated and thus, only SpoIIGA molecules at the mother cell membrane are activated leading to the mother cell specific activation of σE (Pedrido et al., 2013). The activation of σE then allows transcription of the σE regulon, which includes genes necessary for engulfment.
Stage III: Engulfment
After asymmetric division, the polar septum curves around the forespore as the mother cell “swallows” the forespore in a process called engulfment. The result is a double-membrane bound forespore in the mother cell cytosol. Although the dramatic membrane remodeling that occurs during engulfment superficially resembles that of phagocytosis in eukaryotes, the two processes utilize distinct proteins and cytoskeletal elements to achieve this goal. During engulfment, a peptidoglycan degradation machinery composed of SpoIID, SpoIIM and SpoIIP is initially needed for septal wall thinning and subsequently for movement of the engulfing membranes (Abanes-De Mello et al., 2002). Interestingly, cryo-electron micrographs have revealed that a thin layer of peptidoglycan remains during the engulfment process indicating the peptidoglycan is not completely removed by the degradation machinery (Tocheva et al., 2013). The residual peptidoglycan may be necessary to serve as a template for subsequent peptidoglycan remodeling events during engulfment and cortex assembly. Recent evidence has indicated that in addition to peptidoglycan degradation, membrane movement during engulfment also relies on active peptidoglycan synthesis (Meyer et al., 2010). This raises the possibility that stiff, newly synthesized, polymers of peptidoglycan may be providing a cytoskeletal role, analogous to that of eukaryotic actin during phagocytosis, to provide the force required for directed membrane movement (Meyer et al., 2010). However, when peptidoglycan is altogether removed by lysozyme treatment, cells are still able to undergo engulfment, albeit at reduced levels. This redundant engulfment mechanism is dependent on a forespore protein, called SpoIIQ, and a mother cell protein, called SpoIIIAH, which reach across the intermembrane space that divides the mother cell and forespore and directly interact, resulting in a “zippering” of both compartments (Blaylock et al., 2004; Doan et al., 2005). In what has been described as a “ratchet-like” mechanism, the engulfing membrane is able to move forward through random thermal motion of the membrane, but reverse movement is restricted, as the engulfing membrane is stapled to the inner forespore by the tight interaction of SpoIIIAH and SpoIIQ (Broder and Pogliano, 2006).
At the end of engulfment the engulfing membranes must undergo membrane fission to pinch off and release the forespore. The identification of specific factors responsible for membrane fission and fusion in prokaryotes has been historically difficult due to the complexity of the often essential processes that occur concurrently. Unlike most other membrane remodeling events, the membrane fission event that occurs at the end of engulfment is not essential for viability. A recent screen for mutants that were unable to undergo membrane fission at the end of engulfment led to the identification of a mother cell protein called FisB that was enriched at the site of membrane fission during engulfment and was necessary for robust membrane fission. FisB does not resemble or behave like well-studied eukaryotic membrane remodeling proteins like dynamin or the SNARE proteins. Rather, FisB appears to utilize a novel mechanism to promote membrane remodeling through a preferential association with the phospholipid cardiolipin, which is thought to be enriched along negatively curved leaflets of membranes. The current model is that FisB interacts in trans with cardiolipin-enriched membranes at the leading edge of the engulfing membrane to cause a destabilization of both membranes, which in turn could lead to membrane scission (Doan et al., 2013).
At the end of engulfment, the forespore is a free floating cell in the mother cell cytosol. At this time the SpoIIIAA-SpoIIIAH proteins produced under the control of σE and the SpoIIQ protein under the control of σF are required for the activation of the forespore-specific σG (as noted above, SpoIIIAH and SpoIIQ interact directly with one another to “zipper” the mother cell and forespore together during engulfment). The gene encoding the forespore-specific σG is under the transcriptional control of σF, which ensures that it is only expressed in the forespore. Interestingly, despite being under σF transcriptional control, which occurs after asymmetric septation, activation of σG occurs only after engulfment has finished. How does σG activation occur to coincide with the completion of engulfment? After engulfment, when the forespore is sealed off from the mother cell, the metabolic capacity of the forespore is diminished (Camp and Losick, 2009; Doan et al., 2009). The SpoIIAA-SpoIIIAH and SpoIIQ proteins form a channel between the mother cell and forespore (Meisner et al., 2008) through which the mother cell is able to nurture the forespore through the transfer of what are likely small molecules that enable the forespore to continue expressing genes necessary for sporulation (Camp and Losick, 2009) The structure of the basal components of this “feeding tube” channel has been found to be similar to that of type III protein secretion systems (Meisner et al., 2012; Levdikov et al., 2012). Thus, the activation of σG is thought to occur simply because it is the only sigma factor produced at that specific time and location and is dependent on the arrival of metabolites delivered from the mother cell. While the feeding tube is necessary for activation of σG, it is still not completely understood how activation of σG is precisely linked to the end of engulfment. Indeed, recent observations have indicated that completion of engulfment is actually linked to the completion of chromosome translocation into the forespore, suggesting that chromosome translocation also contributes to the timing of σG activation (Regan et al., 2012).
Similar to σE, the subsequent mother-cell specific transcription factor σK is produced as an inactive pro-σK protein. Pro-σK is cleaved by SpoIVFB (Lu et al., 1995), which is an intermembrane protease that is initially held inactive in a complex along with SpoIVFA and BofA (Resnekov and Losick, 1998). Activation of SpoIVFB occurs through the action of the σG-regulated SpoIVB processing enzyme. SpoIVB can relieve the inhibition imposed by SpoIVFA and BofA both by cleaving SpoIVFA at multiple sites and by activating the alternate protease CtpB, which can also cleave SpoIVFA (Campo and Rudner, 2006).
Stage IV-V: Cortex and Coat assembly
The mature spore is encased in two distinct concentric shells: an outer shell, called the “coat”, which is composed of roughly seventy different proteins, and an inner shell called the “cortex” made of specialized peptidoglycan (Henriques and Moran, 2007; McKenney et al., 2013). Together, these two shells serve to protect the spore from environmental insults (Setlow, 2006). Among the first, if not the first, coat proteins to localize to the outer forespore surface is a small 26-amino-acid protein (a so-called “sprotein” (Hobbs et al., 2011)), that is exclusively produced in the mother cell, named SpoVM (Levin et al., 1993; van Ooij and Losick, 2003). SpoVM distinguishes the forespore membrane from the mother cell membrane through the recognition of the forespore’s positive membrane curvature (Ramamurthi et al., 2009). SpoVM tethers a soluble morphogenetic protein called SpoIVA (Ramamurthi et al., 2006), which is the structural component of the basement layer of the coat, onto the forespore surface (Roels et al., 1992; Price and Losick, 1999). While dynamic cytoskeletal nucleotide binding proteins like actin and tubulin hydrolyze nucleotides in order to disassemble, SpoIVA, a static cytoskeletal element, instead hydrolyzes ATP to drive its self assembly to form the basement layer of the coat, which acts as a scaffold atop which all other coat proteins are deposited (Ramamurthi and Losick, 2008; Castaing et al., 2013). Curiously, the ATPase domain of SpoIVA resembles that of the TRAFAC class of P-loop GTPases. However, SpoIVA, like the myosin/kinesin family of exceptional ATPases in the TRAFAC GTPase class (Leipe et al., 2002), has evolutionarily lost the ability to bind GTP and binds ATP instead.
The coat has been described as having four distinct layers: the basement layer, inner coat, outer coat, and crust (McKenney et al., 2010). Each layer’s proper assembly is largely dependent on one (or two, in the case of the crust) major morphogenetic protein that defines each layer. For example, deletion of spoIVA results in the mis-assembly of the basement layer (and, by extension, all subsequent layers) (Roels et al., 1992); deletion of safA, cotE, or cotZ and cotY result in the improper assembly of the inner coat, outer coat, and crust respectively (McKenney et al., 2010; Zheng et al., 1988; Costa et al., 2006; Chada et al., 2003; Imamura et al., 2011). Another coat protein, SpoVID, has been shown to drive the “encasement” step of coat morphogenesis where coat proteins completely encapsulate the developing forespore (Driks et al., 1994; Wang et al., 2009; McKenney et al., 2010).
A recent study monitoring the dynamics of spore coat assembly found that proteins in the coat can be divided into six classes based on their localization dynamics. Spore coat proteins initially assemble as a scaffold in a focus on the mother cell side of the forespore. The encasement of the forespore by specific classes of coat proteins then occurs in coordinated waves that are largely driven by transcription (McKenney and Eichenberger, 2012). Coat assembly occurs predominantly in the mother cell where the coat is assembled on the surface of the outer forespore membrane. Interestingly, the discovery that the SpoIIIAH-SpoIIQ complex, which requires the forespore-specific synthesis of SpoIIQ, is necessary for successful encasement suggests that the forespore may also participate in coordinating the assembly of the coat (McKenney and Eichenberger, 2012).
Although the coat is spatially separated from the cortex by the outer forespore membrane, deletion of either spoVM or spoIVA not only abrogates the initiation of coat assembly, but also abolishes cortex assembly (Coote, 1972; Piggot and Coote, 1976; Roels et al., 1992; Levin et al., 1993) indicating that coat and cortex assembly are somehow linked. Many factors participating in assembly of each individual structure have been identified; however the mechanisms that coordinate temporal assembly of both structures have remained largely mysterious. Recently, a sprotein (37-amino-acids long) named “CmpA”, which was encoded by a previously unannotated mother cell-specific sporulation gene, was found to participate with SpoVM in coordinating cortex assembly (Ebmeier et al., 2012). Specifically, a model emerged wherein cortex peptidoglycan assembly (but not vegetative peptidoglycan assembly) is repressed by a hitherto-unspecified inhibitory activity of CmpA. Cells that successfully initiate coat assembly by SpoVM and SpoIVA overcome the inhibition imposed by CmpA by removing the protein, likely by regulated proteolysis, and continue through the sporulation program to initiate cortex assembly (Ebmeier et al., 2012). Thus, coordination of the assembly of these two large structures appears to be mediated by at least two small proteins, highlighting the general importance of sproteins in biological processes (Hobbs et al., 2011).
The cortex is made of a specialized peptidoglycan that protects the spore from heat and desiccation. The peptidoglycan in the spore resides between the two membrane layers surrounding the forespore and consists of two layers: an inner germ cell wall and an outer cortex. The germ cell wall is a thin layer adjacent to the inner forespore membrane that has a structure similar to that of the vegetative cell wall (Tipper and Linnett, 1976). The cortex, on the other hand, differs in structure from the vegetative cell wall mainly due to a decreased frequency of transpeptidation between glycan chains (Popham and Setlow, 1993b) and the presence of muramic lactam (Warth and Strominger, 1972). These structural changes in the cortex are brought about by the activities of the low molecular weight penicillin binding proteins, which often have D,D-carboxypeptidase activity (Popham et al., 1999), and the CwlD and PdaA proteins, which catalyze the production of muramic lactam from muramic acid (Gilmore et al., 2004). During spore germination the functionally redundant cortex-lytic enzymes SleB and CwlJ (Ishikawa et al., 1998) specifically hydrolyze the cortex peptidoglycan through the recognition of muramic lactam. Mutants with cortexes deficient in muramic lactam are unable to germinate, but can be induced to germinate through exogenous lysozyme treatment (Popham et al., 1996).
It is currently unclear what exactly the functional role of the cortex’s unique peptidoglycan structure is. One theory is that the low degree of crosslinking allows the spore to expand and contract in response to environmental changes (pH, ionic strength, or humidity, for example) without germinating (Ou and Marquis, 1970). Other theories suggest that the low degree of crosslinking may allow the spore to contract (Lewis et al., 1960) or expand (Gould and Dring, 1975; Popham et al., 1999) during spore maturation to attain spore dehydration. Interestingly, the degree of crosslinking throughout the cortex is not homogenous, but rather increases progressively towards the outer cortex layers (Meador-Parton and Popham, 2000). Disruption of this crosslinking gradient does not appear to have significant effects on spore core dehydration suggesting a broad range of cortex crosslinking is permissible to attain spore core dehydration (Meador-Parton and Popham, 2000).
Synthesis of cortex peptidoglycan occurs through similar mechanisms as vegetative cell wall synthesis. Peptidoglycan precursors are produced and modified in the cytosol of the mother cell by the Mur proteins which are also responsible for modification of peptidoglcan precursurs during vegetative cell wall synthesis. During sporulation the production of the Mur proteins is upregulated by σK (Vasudevan et al., 2007). Once properly modified, the peptidoglycan precursors are then tethered to the outer forespore membrane through the formation of the lipid intermediates Lipid I and Lipid II, which are then flipped across the membrane via a Lipid II flippase into the intermembrane space between the outer and inner forespore membranes. Although there are homologs of putative Lipid II flippases that are expressed specifically during sporulation, the identity of the Lipid II flippase during sporulation is currently unknown. The E.coli MviN/MurJ protein has been proposed to be a Lipid II flippase (Ruiz, 2008) and SpoVB was identified as its sporulation-specific homolog (Fay and Dworkin, 2009). However, the discovery that the E. coli FtsW protein which is a part of the SEDS (shape, elongation, division and sporulation) family has in vitro flippase activity (Mohammadi et al., 2011) suggests that its sporulation-specific homolog SpoVE (Ikeda et al., 1989) may also be a Lipid II flippase. Thus, both SpoVB and SpoVE are plausible candidates for being sporulation-specific Lipid II flippases. Consistent with this idea, mutations in either spoVB or spoVE abrogate cortex assembly and result in a buildup of peptidoglycan precursors in the mother cell (Vasudevan et al., 2007).
After translocation into the intermembrane space the lipid-linked precursors are assembled into glycan chains via transglycosylation and peptide crosslinks between the glycan strands are formed via transpeptidation. Tranglycosylation and transpeptidation are performed by the high molecular weight penicillin-binding proteins (PBPs) to produce the meshwork of peptidoglycan that constitutes the cortex. The vegetative PBPs (PBP2B and PBP3) are upregulated during sporulation while PBP1, PBP2A, PBP4, and PBP5 are downregulated (Sowell and Buchanan, 1983). PBP2d and PBP2c are expressed in the forespore during sporulation (Pedersen et al., 2000; Popham and Setlow, 1993a) and have been proposed to play partially redundant roles in synthesizing the spore germ cell wall [McPherson, Driks et al., 2001].
Spore resistance
There are many different factors of the spore that make it able to survive in the environment during harsh conditions. In general, the coat protects the spore from enzymatic assaults such as lysozyme and the cortex is required for protection from high temperature. The coat’s ability to protect the spore from enzymatic assaults has proven useful in resisting predation by bacteriophagous organisms like Tetrahymena (Klobutcher et al., 2006; Laaberki and Dworkin, 2008). The cortex is believed to maintain the spore’s partially dehydrated state [Imae and Strominger 1976; Warth 1978; Mallidis and Scholefield 1987; Ulanowski and Ludlow 1993]1 and this low water content is associated with resistance to heat [Beaman, Koshikawa et al., 1984; Beaman and Gerhardt 1986]. Other factors that contribute to heat resistance and reduction in spore water content include mineralization [Slepecky and Foster 1959; Bender and Marquis 1985; Marquis and Bender 1985; Atrih and Foster 2001] and the presence of the small molecule dipicolinic acid (DPA) [Paidhungat et al., 2000; Setlow et al., 2006]. Additionally, the spore’s DNA is bound by small acid soluble proteins (SASPs) that protect the DNA from damage. Spores that lack SASPs are more susceptible to DNA damaging treatments such as exposure to UV irradiation (Setlow and Setlow, 1987) and hydrogen peroxide (Setlow and Setlow, 1993). A more in depth discussion of spore resistance mechanisms can be found in Leggett et al. (Leggett et al., 2012).
Interest in the ability of spores to survive extraterrestrial environments dates back several decades to when Hagen et al. tested the survival of spores in a simulated Martian environment in an effort to determine the feasibility of extant life on other planets (Hagen et al., 1964). Although the search for extant life is still ongoing, there is significant concern about the contamination of extraterrestrial locations by Earth’s organisms carried on spacecrafts (Nicholson et al., 2009). In order to minimize the contamination risk, much research has been done to explore how and under what conditions terrestrial microorganisms may survive and replicate. In particular, understanding how spores, one of Earth’s hardiest cell types, are able to withstand the harsh conditions of space may help the space biological research field to minimize terrestrial contamination. A recent study by Moeller et al., determined that spore survival in a simulated Mars environment is dependent largely on SASPs, the coat, and dipicolinic acid (Moeller et al., 2012) indicating that survival in Martian environments may depend on the spore’s numerous protective factors.
Applications of sporulation studies
Aside from utilizing sporulation as a model system to understand basic biological processes, other applications of studying sporulation derive from the robust nature of the spore. Bacterial cells have been successfully utilized as whole-cell biosensing systems that rely on genetically modified bacteria which are able to express reporter genes in the presence of an analyte of interest in a dose-dependent matter. Advantages of these systems include low-cost, sensitivity, rapid results and the ability to measure the bioavailability of target analytes (Rawson et al., 1989). However, one disadvantage has thus far been a lack of stability of the cells used in whole-cell biosensing systems in the field. A solution to this problem has been to use organisms that are able to undergo sporulation. Once they have formed spores, the biosensors can then be stored easily for extended periods of time until they are ready to be deployed (Date et al., 2010). The harmless nature of B.subtilis and its genetic tractability, which facilitates the introduction of analyte based reporters, makes it particularly attractive for this purpose. Further studies into sporulation may help in the optimization of spore-based biosensors.
The spore’s unique features also make it amenable for a multitude of other applications. The spore’s outermost layer is composed of proteins making it easy to decorate the spore with proteins of interest through the incorporation of fusion proteins into the organism’s genome. Moreover, such engineered proteins are initially synthesized in the mother cell cytosol and are displayed in the extracellular milieu on the surface of the spore only after mother cell lysis, thereby potentially avoiding protein misfolding issues that may arise when using purified proteins that are subsequently conjugated to a surface. Such display systems may have multiple uses, including the utilization of spores as surface display systems for enzymes or for use as vaccination platforms (Hinc et al., 2010; Mauriello et al., 2004; Isticato et al., 2001).
Concluding thoughts
Since the initial descriptions by Ferdinand Cohn about 140 years ago of heat resistant spores formed by Bacillus subtilis (Cohn, 1877), sporulation has been used as a model system to study a “simple” example of cell differentiation and continues to be used to study fundamental cell biological processes. The non-essential nature of many factors involved in sporulation has greatly facilitated progress made in the field. However, there are still many unanswered questions due to the complex interdependencies and redundancies that are inherent to robust developmental programs.
Despite decades of study, the basic question of how a precursor cell may differentiate into two morphologically distinct, but genetically identical, daughter cells that exhibit different cell fates remains. For example, although in eukaryotes, membrane remodeling events, such as those involved in organelle morphogenesis, endocytosis, and protein trafficking, have been extensively studied, the molecular details underpinning how the architecture of the flat polar septum is altered as the mother cell engulfs the forespore remain an active area of research. Regarding morphogenesis, the spore coat has been a model system for understanding how complex, asymmetric structures may be assembled in an orderly fashion. For years, this structure resisted detailed in vitro investigations, since extensive covalent cross-links prevented the extraction of many coat proteins from mature spores. However, advances in bacterial cell biological techniques have revealed detailed interaction networks between the approximately seventy proteins that make up the coat, and an outstanding challenge will be to recreate these networks in vitro in order to ultimately assemble this structure biochemically. At the heart of the study of sporulation remains the differential, but sequential, activation of transcription factors specifically in the mother cell and forespore that can reveal mechanisms by which adjacent cells can communicate with one another. In B. subtilis, the molecular details of how σF, which is activated in the forespore and sets off the cascade of sigma factor activation, have been exquisitely worked out, yet the cell biological mechanisms that can explain how this activation occurs exclusively in the forespore has been largely unclear. A related question is how asymmetric cell division mechanistically arises in the first place. Additionally, the mysterious chemicals that are transferred from the mother cell to the forespore via the “feeding tube” in order to activate σG exclusively in the forespore await discovery and may reveal how activation of a transcription factor may be linked to completion of a morphological event. In total, then, a remarkable developmental process exhibited by a bacterium originally isolated from the soil at the dawn of the era of modern microbiology will likely continue to provide answers to fundamental biological questions. Developing approaches and strategies to unravel these unanswered questions may provide new tools for further understanding other more complex developmental processes as well.
Acknowledgements
We thank P. Eswaramoorthy for comments on the manuscript. This work was funded by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
References
- Abanes-De Mello A, Sun YL, Aung S, Pogliano K. A cytoskeleton-like role for the bacterial cell wall during engulfment of the Bacillus subtilis forespore. Genes Dev. 2002;16:3253–3264. doi: 10.1101/gad.1039902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker MD, Neiditch MB. Structural basis of response regulator inhibition by a bacterial anti-activator protein. PLoS Biol. 2011;9:e1001226. doi: 10.1371/journal.pbio.1001226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker EC, Pogliano K. Cell-specific SpoIIIE assembly and DNA translocation polarity are dictated by chromosome orientation. Mol Microbiol. 2007;66:1066–1079. doi: 10.1111/j.1365-2958.2007.05992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yehuda S, Fujita M, Liu XS, Gorbatyuk B, Skoko D, Yan J, et al. Defining a centromere-like element in Bacillus subtilis by Identifying the binding sites for the chromosome-anchoring protein RacA. Mol Cell. 2005;17:773–782. doi: 10.1016/j.molcel.2005.02.023. [DOI] [PubMed] [Google Scholar]
- Ben-Yehuda S, Rudner DZ, Losick R. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science. 2003;299:532–536. doi: 10.1126/science.1079914. [DOI] [PubMed] [Google Scholar]
- Blaylock B, Jiang X, Rubio A, Moran CP, Jr., Pogliano K. Zipper-like interaction between proteins in adjacent daughter cells mediates protein localization. Genes Dev. 2004;18:2916–2928. doi: 10.1101/gad.1252704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonstra M, de Jong IG, Scholefield G, Murray H, Kuipers OP, Veening JW. Spo0A regulates chromosome copy number during sporulation by directly binding to the origin of replication in Bacillus subtilis. Mol Microbiol. 2013;87:925–938. doi: 10.1111/mmi.12141. [DOI] [PubMed] [Google Scholar]
- Broder DH, Pogliano K. Forespore engulfment mediated by a ratchet-like mechanism. Cell. 2006;126:917–928. doi: 10.1016/j.cell.2006.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbulys D, Trach KA, Hoch JA. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell. 1991;64:545–552. doi: 10.1016/0092-8674(91)90238-t. [DOI] [PubMed] [Google Scholar]
- Burkholder WF, Kurtser I, Grossman AD. Replication initiation proteins regulate a developmental checkpoint in Bacillus subtilis. Cell. 2001;104:269–279. doi: 10.1016/s0092-8674(01)00211-2. [DOI] [PubMed] [Google Scholar]
- Burton B, Dubnau D. Membrane-associated DNA transport machines. Cold Spring Harb Perspect Biol. 2010;2:a000406. doi: 10.1101/cshperspect.a000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton BM, Marquis KA, Sullivan NL, Rapoport TA, Rudner DZ. The ATPase SpoIIIE transports DNA across fused septal membranes during sporulation in Bacillus subtilis. Cell. 2007;131:1301–1312. doi: 10.1016/j.cell.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp AH, Losick R. A feeding tube model for activation of a cell-specific transcription factor during sporulation in Bacillus subtilis. Genes Dev. 2009;23:1014–1024. doi: 10.1101/gad.1781709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campo N, Rudner DZ. A branched pathway governing the activation of a developmental transcription factor by regulated intramembrane proteolysis. Mol Cell. 2006;23:25–35. doi: 10.1016/j.molcel.2006.05.019. [DOI] [PubMed] [Google Scholar]
- Cano RJ, Borucki MK. Revival and identification of bacterial spores in 25- to 40- million-year-old Dominican amber. Science. 1995;268:1060–1064. doi: 10.1126/science.7538699. [DOI] [PubMed] [Google Scholar]
- Carniol K, Ben-Yehuda S, King N, Losick R. Genetic dissection of the sporulation protein SpoIIE and its role in asymmetric division in Bacillus subtilis. J Bacteriol. 2005;187:3511–3520. doi: 10.1128/JB.187.10.3511-3520.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaing JP, Nagy A, Anantharaman V, Aravind L, Ramamurthi KS. ATP hydrolysis by a domain related to translation factor GTPases drives polymerization of a static bacterial morphogenetic protein. Proc Natl Acad Sci U S A. 2013;110:E151–160. doi: 10.1073/pnas.1210554110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chada VG, Sanstad EA, Wang R, Driks A. Morphogenesis of bacillus spore surfaces. J Bacteriol. 2003;185:6255–6261. doi: 10.1128/JB.185.21.6255-6261.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Kolter R, Losick R. Reversal of an epigenetic switch governing cell chaining in Bacillus subtilis by protein instability. Mol Microbiol. 2011;78:218–229. doi: 10.1111/j.1365-2958.2010.07335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn F. Untersuchungen über bacterien. IV. Beiträge zur biologie der Bacillen. Beiträge zur biologie der Pflanzen. 1877;7:249–276. [Google Scholar]
- Coote JG. Sporulation in Bacillus subtilis. Characterization of oligosporogenous mutants and comparison of their phenotypes with those of asporogenous mutants. J Gen Microbiol. 1972;71:1–15. doi: 10.1099/00221287-71-1-1. [DOI] [PubMed] [Google Scholar]
- Costa T, Isidro AL, Moran CP, Jr., Henriques AO. Interaction between coat morphogenetic proteins SafA and SpoVID. J Bacteriol. 2006;188:7731–7741. doi: 10.1128/JB.00761-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham KA, Burkholder WF. The histidine kinase inhibitor Sda binds near the site of autophosphorylation and may sterically hinder autophosphorylation and phosphotransfer to Spo0F. Mol Microbiol. 2009;71:659–677. doi: 10.1111/j.1365-2958.2008.06554.x. [DOI] [PubMed] [Google Scholar]
- Date A, Pasini P, Daunert S. Fluorescent and bioluminescent cell-based sensors: strategies for their preservation. Adv Biochem Eng Biotechnol. 2010;117:57–75. doi: 10.1007/10_2009_22. [DOI] [PubMed] [Google Scholar]
- Diez V, Schujman GE, Gueiros-Filho FJ, de Mendoza D. Vectorial signalling mechanism required for cell-cell communication during sporulation in Bacillus subtilis. Mol Microbiol. 2012;83:261–274. doi: 10.1111/j.1365-2958.2011.07929.x. [DOI] [PubMed] [Google Scholar]
- Doan T, Coleman J, Marquis KA, Meeske AJ, Burton BM, Karatekin E, Rudner DZ. FisB mediates membrane fission during sporulation in Bacillus subtilis. Genes Dev. 2013 doi: 10.1101/gad.209049.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan T, Marquis KA, Rudner DZ. Subcellular localization of a sporulation membrane protein is achieved through a network of interactions along and across the septum. Mol Microbiol. 2005;55:1767–1781. doi: 10.1111/j.1365-2958.2005.04501.x. [DOI] [PubMed] [Google Scholar]
- Doan T, Morlot C, Meisner J, Serrano M, Henriques AO, Moran CP, Jr., Rudner DZ. Novel secretion apparatus maintains spore integrity and developmental gene expression in Bacillus subtilis. PLoS Genet. 2009;5:e1000566. doi: 10.1371/journal.pgen.1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driks A, Roels S, Beall B, Moran CP, Jr., Losick R. Subcellular localization of proteins involved in the assembly of the spore coat of Bacillus subtilis. Genes Dev. 1994;8:234–244. doi: 10.1101/gad.8.2.234. [DOI] [PubMed] [Google Scholar]
- Duncan L, Alper S, Arigoni F, Losick R, Stragier P. Activation of cell-specific transcription by a serine phosphatase at the site of asymmetric division. Science. 1995;270:641–644. doi: 10.1126/science.270.5236.641. [DOI] [PubMed] [Google Scholar]
- Dworkin J, Losick R. Differential gene expression governed by chromosomal spatial asymmetry. Cell. 2001;107:339–346. doi: 10.1016/s0092-8674(01)00528-1. [DOI] [PubMed] [Google Scholar]
- Dworkin J, Losick R. Developmental commitment in a bacterium. Cell. 2005;121:401–409. doi: 10.1016/j.cell.2005.02.032. [DOI] [PubMed] [Google Scholar]
- Ebmeier SE, Tan IS, Clapham KR, Ramamurthi KS. Small proteins link coat and cortex assembly during sporulation in Bacillus subtilis. Mol Microbiol. 2012;84:682–696. doi: 10.1111/j.1365-2958.2012.08052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellermeier CD, Hobbs EC, Gonzalez-Pastor JE, Losick R. A three-protein signaling pathway governing immunity to a bacterial cannibalism toxin. Cell. 2006;124:549–559. doi: 10.1016/j.cell.2005.11.041. [DOI] [PubMed] [Google Scholar]
- Errington J. Regulation of endospore formation in Bacillus subtilis. Nat Rev Microbiol. 2003;1:117–126. doi: 10.1038/nrmicro750. [DOI] [PubMed] [Google Scholar]
- Eswaramoorthy P, Dinh J, Duan D, Igoshin OA, Fujita M. Single-cell measurement of the levels and distributions of the phosphorelay components in a population of sporulating Bacillus subtilis cells. Microbiology. 2010a;156:2294–2304. doi: 10.1099/mic.0.038497-0. [DOI] [PubMed] [Google Scholar]
- Eswaramoorthy P, Duan D, Dinh J, Dravis A, Devi SN, Fujita M. The threshold level of the sensor histidine kinase KinA governs entry into sporulation in Bacillus subtilis. J Bacteriol. 2010b;192:3870–3882. doi: 10.1128/JB.00466-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay A, Dworkin J. Bacillus subtilis homologs of MviN (MurJ), the putative Escherichia coli lipid II flippase, are not essential for growth. J Bacteriol. 2009;191:6020–6028. doi: 10.1128/JB.00605-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiche JB, Cattoni DI, Diekmann N, Langerak JM, Clerte C, Royer CA, et al. Recruitment, Assembly, and Molecular Architecture of the SpoIIIE DNA Pump Revealed by Superresolution Microscopy. PLoS Biol. 2013;11:e1001557. doi: 10.1371/journal.pbio.1001557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frandsen N, Barak I, Karmazyn-Campelli C, Stragier P. Transient gene asymmetry during sporulation and establishment of cell specificity in Bacillus subtilis. Genes Dev. 1999;13:394–399. doi: 10.1101/gad.13.4.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Gonzalez-Pastor JE, Losick R. High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J Bacteriol. 2005;187:1357–1368. doi: 10.1128/JB.187.4.1357-1368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Losick R. An investigation into the compartmentalization of the sporulation transcription factor sigmaE in Bacillus subtilis. Mol Microbiol. 2002;43:27–38. doi: 10.1046/j.1365-2958.2002.02732.x. [DOI] [PubMed] [Google Scholar]
- Fujita M, Losick R. Evidence that entry into sporulation in Bacillus subtilis is governed by a gradual increase in the level and activity of the master regulator Spo0A. Genes Dev. 2005;19:2236–2244. doi: 10.1101/gad.1335705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego del Sol F, Marina A. Structural basis of Rap phosphatase inhibition by Phr peptides. PLoS Biol. 2013;11:e1001511. doi: 10.1371/journal.pbio.1001511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore ME, Bandyopadhyay D, Dean AM, Linnstaedt SD, Popham DL. Production of muramic delta-lactam in Bacillus subtilis spore peptidoglycan. J Bacteriol. 2004;186:80–89. doi: 10.1128/JB.186.1.80-89.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Pastor JE, Hobbs EC, Losick R. Cannibalism by sporulating bacteria. Science. 2003;301:510–513. doi: 10.1126/science.1086462. [DOI] [PubMed] [Google Scholar]
- Gould GW, Dring GJ. Heat resistance of bacterial endospores and concept of an expanded osmoregulatory cortex. Nature. 1975;258:402–405. doi: 10.1038/258402a0. [DOI] [PubMed] [Google Scholar]
- Guberman JM, Fay A, Dworkin J, Wingreen NS, Gitai Z. PSICIC: noise and asymmetry in bacterial division revealed by computational image analysis at sub-pixel resolution. PLoS Comput Biol. 2008;4:e1000233. doi: 10.1371/journal.pcbi.1000233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen CA, Hawrylewicz EJ, Ehrlich R. Survival of Microorganisms in a Simulated Martian Environment. I. Bacillus Subtilis Var. Globigii. Appl Microbiol. 1964;12:215–218. doi: 10.1128/am.12.3.215-218.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamon MA, Lazazzera BA. The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Mol Microbiol. 2001;42:1199–1209. doi: 10.1046/j.1365-2958.2001.02709.x. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Weinert TA. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- Henriques AO, Moran CP., Jr. Structure, assembly, and function of the spore surface layers. Annu Rev Microbiol. 2007;61:555–588. doi: 10.1146/annurev.micro.61.080706.093224. [DOI] [PubMed] [Google Scholar]
- Hinc K, Isticato R, Dembek M, Karczewska J, Iwanicki A, Peszynska-Sularz G, et al. Expression and display of UreA of Helicobacter acinonychis on the surface of Bacillus subtilis spores. Microb Cell Fact. 2010;9:2. doi: 10.1186/1475-2859-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs EC, Fontaine F, Yin X, Storz G. An expanding universe of small proteins. Curr Opin Microbiol. 2011;14:167–173. doi: 10.1016/j.mib.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmeister AE, Londono-Vallejo A, Harry E, Stragier P, Losick R. Extracellular signal protein triggering the proteolytic activation of a developmental transcription factor in B. subtilis. Cell. 1995;83:219–226. doi: 10.1016/0092-8674(95)90163-9. [DOI] [PubMed] [Google Scholar]
- Ikeda M, Sato T, Wachi M, Jung HK, Ishino F, Kobayashi Y, Matsuhashi M. Structural similarity among Escherichia coli FtsW and RodA proteins and Bacillus subtilis SpoVE protein, which function in cell division, cell elongation, and spore formation, respectively. J Bacteriol. 1989;171:6375–6378. doi: 10.1128/jb.171.11.6375-6378.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura D, Kuwana R, Takamatsu H, Watabe K. Proteins involved in formation of the outermost layer of Bacillus subtilis spores. J Bacteriol. 2011;193:4075–4080. doi: 10.1128/JB.05310-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura D, Zhou R, Feig M, Kroos L. Evidence that the Bacillus subtilis SpoIIGA protein is a novel type of signal-transducing aspartic protease. J Biol Chem. 2008;283:15287–15299. doi: 10.1074/jbc.M708962200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa S, Yamane K, Sekiguchi J. Regulation and characterization of a newly deduced cell wall hydrolase gene (cwlJ) which affects germination of Bacillus subtilis spores. J Bacteriol. 1998;180:1375–1380. doi: 10.1128/jb.180.6.1375-1380.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isticato R, Cangiano G, Tran HT, Ciabattini A, Medaglini D, Oggioni MR, et al. Surface display of recombinant proteins on Bacillus subtilis spores. J Bacteriol. 2001;183:6294–6301. doi: 10.1128/JB.183.21.6294-6301.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacotot H, Virat B. La longevite des spores de B. antbracis (premier vaccin de Pasteur) Annales de l’Institut Pasteur. 1954;87:215–217. [PubMed] [Google Scholar]
- Jonas RM, Weaver EA, Kenney TJ, Moran CP, Jr., Haldenwang WG. The Bacillus subtilis spoIIG operon encodes both sigma E and a gene necessary for sigma E activation. J Bacteriol. 1988;170:507–511. doi: 10.1128/jb.170.2.507-511.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns DB, Chu F, Branda SS, Kolter R, Losick R. A master regulator for biofilm formation by Bacillus subtilis. Mol Microbiol. 2005;55:739–749. doi: 10.1111/j.1365-2958.2004.04440.x. [DOI] [PubMed] [Google Scholar]
- Klobutcher LA, Ragkousi K, Setlow P. The Bacillus subtilis spore coat provides “eat resistance” during phagocytic predation by the protozoan Tetrahymena thermophila. Proc Natl Acad Sci U S A. 2006;103:165–170. doi: 10.1073/pnas.0507121102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laaberki MH, Dworkin J. Role of spore coat proteins in the resistance of Bacillus subtilis spores to Caenorhabditis elegans predation. J Bacteriol. 2008;190:6197–6203. doi: 10.1128/JB.00623-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDeaux JR, Yu N, Grossman AD. Different roles for KinA, KinB, and KinC in the initiation of sporulation in Bacillus subtilis. J Bacteriol. 1995;177:861–863. doi: 10.1128/jb.177.3.861-863.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggett MJ, McDonnell G, Denyer SP, Setlow P, Maillard JY. Bacterial spore structures and their protective role in biocide resistance. J Appl Microbiol. 2012;113:485–498. doi: 10.1111/j.1365-2672.2012.05336.x. [DOI] [PubMed] [Google Scholar]
- Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- Lenarcic R, Halbedel S, Visser L, Shaw M, Wu LJ, Errington J, et al. Localisation of DivIVA by targeting to negatively curved membranes. Embo J. 2009;28:2272–2282. doi: 10.1038/emboj.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levdikov VM, Blagova EV, McFeat A, Fogg MJ, Wilson KS, Wilkinson AJ. Structure of components of an intercellular channel complex in sporulating Bacillus subtilis. Proc Natl Acad Sci U S A. 2012;109:5441–5445. doi: 10.1073/pnas.1120087109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin PA, Fan N, Ricca E, Driks A, Losick R, Cutting S. An unusually small gene required for sporulation by Bacillus subtilis. Mol Microbiol. 1993;9:761–771. doi: 10.1111/j.1365-2958.1993.tb01736.x. [DOI] [PubMed] [Google Scholar]
- Levine JH, Fontes ME, Dworkin J, Elowitz MB. Pulsed feedback defers cellular differentiation. PLoS Biol. 2012;10:e1001252. doi: 10.1371/journal.pbio.1001252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JC, Snell NS, Burr HK. Water Permeability of Bacterial Spores and the Concept of a Contractile Cortex. Science. 1960;132:544–545. doi: 10.1126/science.132.3426.544. [DOI] [PubMed] [Google Scholar]
- Liu WT, Yang YL, Xu Y, Lamsa A, Haste NM, Yang JY, et al. Imaging mass spectrometry of intraspecies metabolic exchange revealed the cannibalistic factors of Bacillus subtilis. Proc Natl Acad Sci U S A. 2010;107:16286–16290. doi: 10.1073/pnas.1008368107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez D, Vlamakis H, Losick R, Kolter R. Cannibalism enhances biofilm development in Bacillus subtilis. Mol Microbiol. 2009;74:609–618. doi: 10.1111/j.1365-2958.2009.06882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Cutting S, Kroos L. Sporulation protein SpoIVFB from Bacillus subtilis enhances processing of the sigma factor precursor Pro-sigma K in the absence of other sporulation gene products. J Bacteriol. 1995;177:1082–1085. doi: 10.1128/jb.177.4.1082-1085.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauriello EM, Ducle H, Isticato R, Cangiano G, Hong HA, De Felice M, et al. Display of heterologous antigens on the Bacillus subtilis spore coat using CotC as a fusion partner. Vaccine. 2004;22:1177–1187. doi: 10.1016/j.vaccine.2003.09.031. [DOI] [PubMed] [Google Scholar]
- McKenney PT, Driks A, Eichenberger P. The Bacillus subtilis endospore: assembly and functions of the multilayered coat. Nat Rev Microbiol. 2013;11:33–44. doi: 10.1038/nrmicro2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney PT, Driks A, Eskandarian HA, Grabowski P, Guberman J, Wang KH, et al. A distance-weighted interaction map reveals a previously uncharacterized layer of the Bacillus subtilis spore coat. Curr Biol. 2010;20:934–938. doi: 10.1016/j.cub.2010.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney PT, Eichenberger P. Dynamics of spore coat morphogenesis in Bacillus subtilis. Mol Microbiol. 2012;83:245–260. doi: 10.1111/j.1365-2958.2011.07936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meador-Parton J, Popham DL. Structural analysis of Bacillus subtilis spore peptidoglycan during sporulation. J Bacteriol. 2000;182:4491–4499. doi: 10.1128/jb.182.16.4491-4499.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisner J, Maehigashi T, Andre I, Dunham CM, Moran CP., Jr. Structure of the basal components of a bacterial transporter. Proc Natl Acad Sci U S A. 2012;109:5446–5451. doi: 10.1073/pnas.1120113109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisner J, Wang X, Serrano M, Henriques AO, Moran CP., Jr. A channel connecting the mother cell and forespore during bacterial endospore formation. Proc Natl Acad Sci U S A. 2008;105:15100–15105. doi: 10.1073/pnas.0806301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer P, Gutierrez J, Pogliano K, Dworkin J. Cell wall synthesis is necessary for membrane dynamics during sporulation of Bacillus subtilis. Mol Microbiol. 2010;76:956–970. doi: 10.1111/j.1365-2958.2010.07155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitri S, Xavier JB, Foster KR. Social evolution in multispecies biofilms. Proc Natl Acad Sci U S A. 2011;108(Suppl 2):10839–10846. doi: 10.1073/pnas.1100292108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller R, Schuerger AC, Reitz G, Nicholson WL. Protective role of spore structural components in determining Bacillus subtilis spore resistance to simulated mars surface conditions. Appl Environ Microbiol. 2012;78:8849–8853. doi: 10.1128/AEM.02527-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi T, van Dam V, Sijbrandi R, Vernet T, Zapun A, Bouhss A, et al. Identification of FtsW as a transporter of lipid-linked cell wall precursors across the membrane. Embo J. 2011;30:1425–1432. doi: 10.1038/emboj.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molle V, Fujita M, Jensen ST, Eichenberger P, Gonzalez-Pastor JE, Liu JS, Losick R. The Spo0A regulon of Bacillus subtilis. Mol Microbiol. 2003;50:1683–1701. doi: 10.1046/j.1365-2958.2003.03818.x. [DOI] [PubMed] [Google Scholar]
- Mueller JP, Sonenshein AL. Role of the Bacillus subtilis gsiA gene in regulation of early sporulation gene expression. J Bacteriol. 1992;174:4374–4383. doi: 10.1128/jb.174.13.4374-4383.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narula J, Devi SN, Fujita M, Igoshin OA. Ultrasensitivity of the Bacillus subtilis sporulation decision. Proc Natl Acad Sci U S A. 2012;109:E3513–3522. doi: 10.1073/pnas.1213974109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson WL, Schuerger AC, Race MS. Migrating microbes and planetary protection. Trends Microbiol. 2009;17:389–392. doi: 10.1016/j.tim.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Ou LT, Marquis RE. Electromechanical interactions in cell walls of gram-positive cocci. J Bacteriol. 1970;101:92–101. doi: 10.1128/jb.101.1.92-101.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parashar V, Jeffrey PD, Neiditch MB. Conformational change-induced repeat domain expansion regulates Rap phosphatase quorum-sensing signal receptors. PLoS Biol. 2013;11:e1001512. doi: 10.1371/journal.pbio.1001512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes-Sabja D, Setlow P, Sarker MR. Germination of spores of Bacillales and Clostridiales species: mechanisms and proteins involved. Trends Microbiol. 2011;19:85–94. doi: 10.1016/j.tim.2010.10.004. [DOI] [PubMed] [Google Scholar]
- Pedersen LB, Ragkousi K, Cammett TJ, Melly E, Sekowska A, Schopick E, et al. Characterization of ywhE, which encodes a putative high-molecular-weight class A penicillin-binding protein in Bacillus subtilis. Gene. 2000;246:187–196. doi: 10.1016/s0378-1119(00)00084-6. [DOI] [PubMed] [Google Scholar]
- Pedrido ME, de Ona P, Ramirez W, Lenini C, Goni A, Grau R. Spo0A links de novo fatty acid synthesis to sporulation and biofilm development in Bacillus subtilis. Mol Microbiol. 2013;87:348–367. doi: 10.1111/mmi.12102. [DOI] [PubMed] [Google Scholar]
- Perego M, Hanstein C, Welsh KM, Djavakhishvili T, Glaser P, Hoch JA. Multiple protein-aspartate phosphatases provide a mechanism for the integration of diverse signals in the control of development in B. subtilis. Cell. 1994;79:1047–1055. doi: 10.1016/0092-8674(94)90035-3. [DOI] [PubMed] [Google Scholar]
- Perez Morales TG, Ho TD, Liu WT, Dorrestein PC, Ellermeier CD. Production of the Cannibalism Toxin SDP Is a Multistep Process That Requires SdpA and SdpB. J Bacteriol. 2013;195:3244–3251. doi: 10.1128/JB.00407-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggot PJ, Coote JG. Genetic aspects of bacterial endospore formation. Bacteriol Rev. 1976;40:908–962. doi: 10.1128/br.40.4.908-962.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggot PJ, Hilbert DW. Sporulation of Bacillus subtilis. Curr Opin Microbiol. 2004;7:579–586. doi: 10.1016/j.mib.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Popham DL, Gilmore ME, Setlow P. Roles of low-molecular-weight penicillin-binding proteins in Bacillus subtilis spore peptidoglycan synthesis and spore properties. J Bacteriol. 1999;181:126–132. doi: 10.1128/jb.181.1.126-132.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popham DL, Helin J, Costello CE, Setlow P. Muramic lactam in peptidoglycan of Bacillus subtilis spores is required for spore outgrowth but not for spore dehydration or heat resistance. Proc Natl Acad Sci U S A. 1996;93:15405–15410. doi: 10.1073/pnas.93.26.15405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popham DL, Setlow P. Cloning, nucleotide sequence, and regulation of the Bacillus subtilis pbpF gene, which codes for a putative class A high-molecular-weight penicillin-binding protein. J Bacteriol. 1993a;175:4870–4876. doi: 10.1128/jb.175.15.4870-4876.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popham DL, Setlow P. The cortical peptidoglycan from spores of Bacillus megaterium and Bacillus subtilis is not highly cross-linked. J Bacteriol. 1993b;175:2767–2769. doi: 10.1128/jb.175.9.2767-2769.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price KD, Losick R. A four-dimensional view of assembly of a morphogenetic protein during sporulation in Bacillus subtilis. J Bacteriol. 1999;181:781–790. doi: 10.1128/jb.181.3.781-790.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptacin JL, Nollmann M, Becker EC, Cozzarelli NR, Pogliano K, Bustamante C. Sequence-directed DNA export guides chromosome translocation during sporulation in Bacillus subtilis. Nat Struct Mol Biol. 2008;15:485–493. doi: 10.1038/nsmb.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahn-Lee L, Merrikh H, Grossman AD, Losick R. The sporulation protein SirA inhibits the binding of DnaA to the origin of replication by contacting a patch of clustered amino acids. J Bacteriol. 2011;193:1302–1307. doi: 10.1128/JB.01390-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthi KS, Clapham KR, Losick R. Peptide anchoring spore coat assembly to the outer forespore membrane in Bacillus subtilis. Mol Microbiol. 2006;62:1547–1557. doi: 10.1111/j.1365-2958.2006.05468.x. [DOI] [PubMed] [Google Scholar]
- Ramamurthi KS, Lecuyer S, Stone HA, Losick R. Geometric cue for protein localization in a bacterium. Science. 2009;323:1354–1357. doi: 10.1126/science.1169218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthi KS, Losick R. ATP-driven self-assembly of a morphogenetic protein in Bacillus subtilis. Mol Cell. 2008;31:406–414. doi: 10.1016/j.molcel.2008.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthi KS, Losick R. Negative membrane curvature as a cue for subcellular localization of a bacterial protein. Proc Natl Acad Sci U S A. 2009;106:13541–13545. doi: 10.1073/pnas.0906851106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao CV, Glekas GD, Ordal GW. The three adaptation systems of Bacillus subtilis chemotaxis. Trends Microbiol. 2008;16:480–487. doi: 10.1016/j.tim.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawson DM, Willmer AJ, Turner AP. Whole-cell biosensors for environmental monitoring. Biosensors. 1989;4:299–311. doi: 10.1016/0265-928x(89)80011-2. [DOI] [PubMed] [Google Scholar]
- Regan G, Itaya M, Piggot PJ. Coupling of sigmaG activation to completion of engulfment during sporulation of Bacillus subtilis survives large perturbations to DNA translocation and replication. J Bacteriol. 2012;194:6264–6271. doi: 10.1128/JB.01470-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnekov O, Losick R. Negative regulation of the proteolytic activation of a developmental transcription factor in Bacillus subtilis. Proc Natl Acad Sci U S A. 1998;95:3162–3167. doi: 10.1073/pnas.95.6.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roels S, Driks A, Losick R. Characterization of spoIVA, a sporulation gene involved in coat morphogenesis in Bacillus subtilis. J Bacteriol. 1992;174:575–585. doi: 10.1128/jb.174.2.575-585.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz N. Bioinformatics identification of MurJ (MviN) as the peptidoglycan lipid II flippase in Escherichia coli. Proc Natl Acad Sci U S A. 2008;105:15553–15557. doi: 10.1073/pnas.0808352105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryter A. Morphologic Study of the Sporulation of Bacillus Subtilis. Ann Inst Pasteur (Paris) 1965;108:40–60. [PubMed] [Google Scholar]
- Ryter A, Schaeffer P, Ionesco H. Cytologic classification, by their blockage stage, of sporulation mutants of Bacillus subtilis Marburg. Ann Inst Pasteur (Paris) 1966;110:305–315. [PubMed] [Google Scholar]
- Sasai Y. Cytosystems dynamics in self-organization of tissue architecture. Nature. 2013;493:318–326. doi: 10.1038/nature11859. [DOI] [PubMed] [Google Scholar]
- Schaeffer P, Ionesco H, Ryter A, Balassa G. Mechanismes de regulation des activites cellulaires chez les microorganisms. Paris: 1963. La sporulation de Bacillus subtilis: etude gendtique et physiologique; pp. 553–563. [Google Scholar]
- Setlow B, Setlow P. Thymine-containing dimers as well as spore photoproducts are found in ultraviolet-irradiated Bacillus subtilis spores that lack small acid-soluble proteins. Proc Natl Acad Sci U S A. 1987;84:421–423. doi: 10.1073/pnas.84.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow B, Setlow P. Binding of small, acid-soluble spore proteins to DNA plays a significant role in the resistance of Bacillus subtilis spores to hydrogen peroxide. Appl Environ Microbiol. 1993;59:3418–3423. doi: 10.1128/aem.59.10.3418-3423.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow P. Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J Appl Microbiol. 2006;101:514–525. doi: 10.1111/j.1365-2672.2005.02736.x. [DOI] [PubMed] [Google Scholar]
- Setlow P. I will survive: DNA protection in bacterial spores. Trends Microbiol. 2007;15:172–180. doi: 10.1016/j.tim.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Sharp MD, Pogliano K. Role of cell-specific SpoIIIE assembly in polarity of DNA transfer. Science. 2002;295:137–139. doi: 10.1126/science.1066274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowell MO, Buchanan CE. Changes in penicillin-binding proteins during sporulation of Bacillus subtilis. J Bacteriol. 1983;153:1331–1337. doi: 10.1128/jb.153.3.1331-1337.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stragier P, Bonamy C, Karmazyn-Campelli C. Processing of a sporulation sigma factor in Bacillus subtilis: how morphological structure could control gene expression. Cell. 1988;52:697–704. doi: 10.1016/0092-8674(88)90407-2. [DOI] [PubMed] [Google Scholar]
- Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet. 1996;30:297–241. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- Tipper DJ, Linnett PE. Distribution of peptidoglycan synthetase activities between sporangia and forespores in sporulating cells of Bacillus sphaericus. J Bacteriol. 1976;126:213–221. doi: 10.1128/jb.126.1.213-221.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tocheva EI, Lopez-Garrido J, Hughes HV, Fredlund J, Kuru E, Vannieuwenhze MS, et al. Peptidoglycan transformations during Bacillus subtilis sporulation. Mol Microbiol. 2013;88:673–686. doi: 10.1111/mmi.12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooij C, Losick R. Subcellular localization of a small sporulation protein in Bacillus subtilis. J Bacteriol. 2003;185:1391–1398. doi: 10.1128/JB.185.4.1391-1398.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan P, Weaver A, Reichert ED, Linnstaedt SD, Popham DL. Spore cortex formation in Bacillus subtilis is regulated by accumulation of peptidoglycan precursors under the control of sigma K. Mol Microbiol. 2007;65:1582–1594. doi: 10.1111/j.1365-2958.2007.05896.x. [DOI] [PubMed] [Google Scholar]
- Veening JW, Murray H, Errington J. A mechanism for cell cycle regulation of sporulation initiation in Bacillus subtilis. Genes Dev. 2009;23:1959–1970. doi: 10.1101/gad.528209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veening JW, Stewart EJ, Berngruber TW, Taddei F, Kuipers OP, Hamoen LW. Bet-hedging and epigenetic inheritance in bacterial cell development. Proc Natl Acad Sci U S A. 2008;105:4393–4398. doi: 10.1073/pnas.0700463105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishnoi M, Narula J, Devi SN, Dao HA, Igoshin OA, Fujita M. Triggering sporulation in Bacillus subtilis with artificial two-component systems reveals the importance of proper Spo0A activation dynamics. Mol Microbiol. 2013;90:181–194. doi: 10.1111/mmi.12357. [DOI] [PubMed] [Google Scholar]
- Vlamakis H, Chai Y, Beauregard P, Losick R, Kolter R. Sticking together: building a biofilm the Bacillus subtilis way. Nat Rev Microbiol. 2013;11:157–168. doi: 10.1038/nrmicro2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vreeland RH, Rosenzweig WD, Powers DW. Isolation of a 250 million-year-old halotolerant bacterium from a primary salt crystal. Nature. 2000;407:897–900. doi: 10.1038/35038060. [DOI] [PubMed] [Google Scholar]
- Wang KH, Isidro AL, Domingues L, Eskandarian HA, McKenney PT, Drew K, et al. The coat morphogenetic protein SpoVID is necessary for spore encasement in Bacillus subtilis. Mol Microbiol. 2009;74:634–649. doi: 10.1111/j.1365-2958.2009.06886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warth AD, Strominger JL. Structure of the peptidoglycan from spores of Bacillus subtilis. Biochemistry. 1972;11:1389–1396. doi: 10.1021/bi00758a010. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Errington J. Bacillus subtilis SpoIIIE protein required for DNA segregation during asymmetric cell division. Science. 1994;264:572–575. doi: 10.1126/science.8160014. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Errington J. Use of asymmetric cell division and spoIIIE mutants to probe chromosome orientation and organization in Bacillus subtilis. Mol Microbiol. 1998;27:777–786. doi: 10.1046/j.1365-2958.1998.00724.x. [DOI] [PubMed] [Google Scholar]
- Wu R, Gu M, Wilton R, Babnigg G, Kim Y, Pokkuluri PR, et al. Insight into the sporulation phosphorelay: crystal structure of the sensor domain of Bacillus subtilis histidine kinase, KinD. Protein Sci. 2013;22:564–576. doi: 10.1002/pro.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng LB, Donovan WP, Fitz-James PC, Losick R. Gene encoding a morphogenic protein required in the assembly of the outer coat of the Bacillus subtilis endospore. Genes Dev. 1988;2:1047–1054. doi: 10.1101/gad.2.8.1047. [DOI] [PubMed] [Google Scholar]