To the Editor:
Elucidation and manipulation of human, animal and plant genomes is key to basic biology research, medical advances and crop improvement. The development of targeted genome editing, particularly the homologous recombination-based gene replacement, is of great value in all organisms. Recent advances in engineered nucleases with programmable DNA binding specificities, such as zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs), have provided valuable means in creating targeted mutations in metazoan and plant genomes with high specificity1–6. However, these technologies demand elaborate design and assembly of individual DNA-binding proteins for each DNA target site1–6. Recently, a simple, versatile and efficient genome engineering technology has been developed based on the bacterial CRISPR (clustered regularly interspaced short palindromic repeats)/CRISPR-associated (Cas) adaptive immune systems7. In type II CRISPR/Cas system from Streptococcus pyogenes, a single Cas9 endonuclease guided by a duplex of mature CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA) could cleave trespassing DNA from bacteriophage or plasmids in a sequence-specific manner7. By reconstitution of the S. pyogenes Cas9 (SpCas9) and an artificial chimera of crRNA and tracrRNA called guide RNA (gRNA) in eukaryotic cells, including yeast, zebrafish, mouse and human, targeted genome editing could be readily achieved via either error-prone non-homologous end joining (NHEJ) or homology-directed repair (HDR) of the intended cleavage site7–14. However, the feasibility and efficacy of the gRNA/Cas9 technology in plants have not been examined.
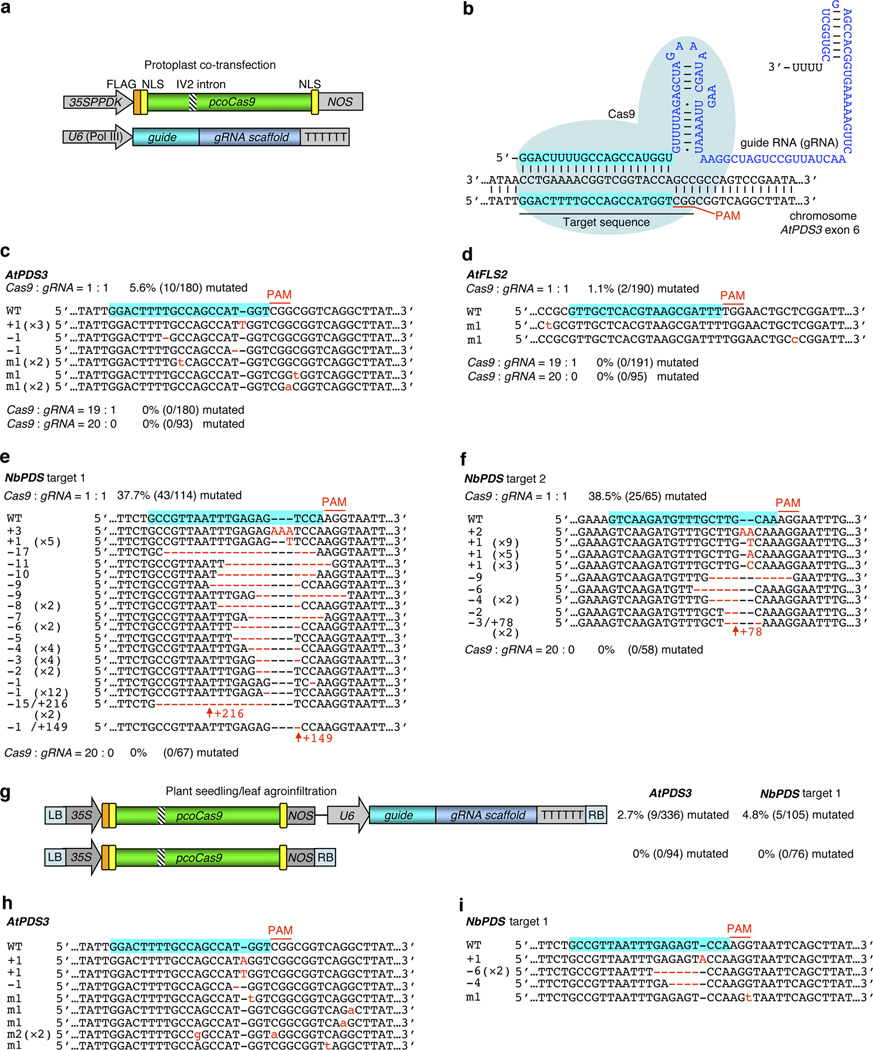
To explore the gRNA/Cas9 technology for plant genome engineering, we first co-expressed the plant codon-optimized SpCas9 (pcoCas9) and a gRNA targeting the Arabidopsis thaliana PDS3 (PHYTOENE DESATURASE) gene (Fig. 1a,b) in Arabidopsis mesophyll protoplasts, which are freshly isolated leaf cells without cell walls. The protoplast transient expression system supports highly efficient DNA co-transfection and protein expression15. The pcoCas9 was expressed under the hybrid constitutive 35SPPDK promoter15, while the gRNA was transcribed from the Arabidopsis U6 polymerase III promoter (Fig. 1a). Notably, pcoCas9 was expressed at a significantly higher level compared to the humanized SpCas99 using the same expression vector in Arabidopsis protoplasts (Supplementary Fig. 1). In addition, pcoCas9 encodes nuclear localization sequences (NLSs) at both protein termini (Fig. 1a) for optimal protein nuclear localization8. A potato IV2 intron (Fig. 1a) was inserted to minimize adverse effects on bacterial growth16 due to potential leaky expression and nuclease activities of pcoCas9 in E. coli during cloning.
Figure 1.
Targeted plant genome editing by gRNA/pcoCas9. (a) Schematic of the gRNA/pcoCas9 constructs for protoplast co-transfection. (b) Diagram of the gRNA/pcoCas9 complex targeting the Arabidopsis AtPDS3 exon 6. (c–f) Targeted genome editing on AtPDS3 (c) and AtFLS2 (d) in Arabidopsis protoplasts and NbPDS (e,f) in tobacco (Nicotiana benthamiana) protoplasts. (g) Binary plasmids for genome editing of AtPDS3 and NbPDS in Arabidopsis and tobacco plants, respectively, via Agrobacterium-mediated transient gene expression. (h,i) Targeted genome editing on AtPDS3 in Arabidopsis seedlings (h) and NbPDS in tobacco leaves (i). The mutation rate in c–g was calculated based on the mutant/total alleles of randomly selected clonal amplicons of the target locus. In c–f, h and i, blue shadow marks the target sequence recognized by cognate gRNA. PAM, the protospacer adjacent motif. DNA insertions, deletions and point mutations are shown in red as upper case letters, dashes and lower case letters, respectively. The upright arrow and number in red indicate a long insertion.
To determine the mutagenesis efficiency of the gRNA/pcoCas9 system in Arabidopsis protoplasts, genomic PCR (gPCR) amplicons using total genomic DNA (gDNA) from transfected protoplasts as templates were cloned and randomly selected for Sanger sequencing. With a DNA ratio of Cas9:gRNA at 1:1 during co-transfection, we detected 10 mutated AtPDS3 target alleles among 180 randomly sequenced amplicons, reaching an approximate mutagenesis frequency of 5.6% (Fig. 1c). Interestingly, a ratio of Cas9:gRNA at 19:1 failed to induce any mutation in 180 sequenced amplicons, and no mutation was detected among 93 sequenced amplicons when pcoCas9 was expressed alone (Fig. 1c). For a second tested gene, AtFLS2 (FLAGELLIN SENSITIVE 2), in Arabidopsis protoplasts, the gRNA/pcoCas9-mediated mutagenesis also only occurred with a DNA ratio of Cas9:gRNA at 1:1 but not at 19:1 (Fig. 1d). In this case, a lower mutagenesis frequency (1.1%) was observed (Fig. 1d). It seemed that gRNA expression is the limiting factor for optimal targeting and mutagenesis in plant cells as in human cells12.
To extend the application of gRNA/pcoCas9-mediated genome editing to other plant systems, we carried out a parallel study using tobacco (Nicotiana benthamiana) protoplasts. Surprisingly, by targeting NbPDS (ortholog of AtPDS3) at two different sites, we obtained substantially higher mutagenesis frequencies than in Arabidopsis, namely 37.7% for the target 1 (Fig. 1e) and 38.5% for the target 2 (Fig. 1f). Interestingly, the gRNA/pcoCas9-induced mutagenesis frequently led to significant DNA deletions or insertions but rare single nucleotide (nt) substitutions in tobacco cells (Fig. 1e,f and Supplementary Fig. 2a,b) as in animal and human cells displaying relatively high mutation rates (e.g., 37.6% in K562 cells and 24.6% in 293T cells)7–14. In contrast, single nt deletions, insertions or substitutions were most frequently detected in Arabidopsis cells with relatively low mutation rates from 1.1 to 5.6% (Fig. 1c,d). Our results clearly demonstrated that the gRNA/pcoCas9 system is effective in plant cells. Whether the different genome mutagenesis frequencies and patterns between Arabidopsis and tobacco by gRNA/pcoCas9 are due to distinct plant genotypes or physiological states requires future investigation.
To validate the occurrence of the gRNA/pcoCas9-induced targeted mutagenesis in the PDS gene in planta, we transiently co-expressed pcoCas9 and AtPDS3- or NbPDS-targeting gRNA via a single binary plasmid (Fig. 1g) in intact leaves of two-week-old Arabidopsis seedlings or five-week-old tobacco plants through Agrobacterium leaf infiltration. Biallelic disruption of PDS in the Arabidopsis or tobacco genome would abolish carotenoid biosynthesis and promote chlorophyll oxidation leading to a photobleached phenotype. We did not observe any visible albino spot on agroinfiltrated leaves from Arabidopsis or tobacco plants 7 days post infiltration. There was either no cell with biallelic disruption of PDS or the population of photobleached cells was too small, as the cell division might have ceased in the infiltrated leaves. Careful screens at the single cell level after the degradation of existing chlorophyll is necessary for further characterization using fluorescent microscopy. Importantly, by sequencing target gPCR amplicons, we did detect precise genomic mutations in the AtPDS3 or NbPDS target sequence in cells from agroinfiltrated leaves with a mutagenesis frequency of 2.7% for AtPDS3 and 4.8% for NbPDS (Fig. 1g). Considering that agroinfiltration exhibits lower efficiency and higher variability in gene transfer than the protoplast transfection15, these mutagenesis frequencies might reflect dilution of the targeted mutations by wild-type gDNA from leaf cells without successful DNA delivery. Notably, the different gRNA/pcoCas9-induced mutagenesis patterns in Arabidopsis and tobacco protoplasts were also observed in corresponding whole plants. While targeted mutations in Arabidopsis seedlings were frequently single nt substitutions (Fig. 1h), those in tobacco plants often involved significant DNA deletions (Fig. 1i). The leaves infiltrated with Agrobacteria expressing pcoCas9 alone did not induce mutations in the target regions (Fig. 1g). These data established that the gRNA/pcoCas9 system is also effective in planta.
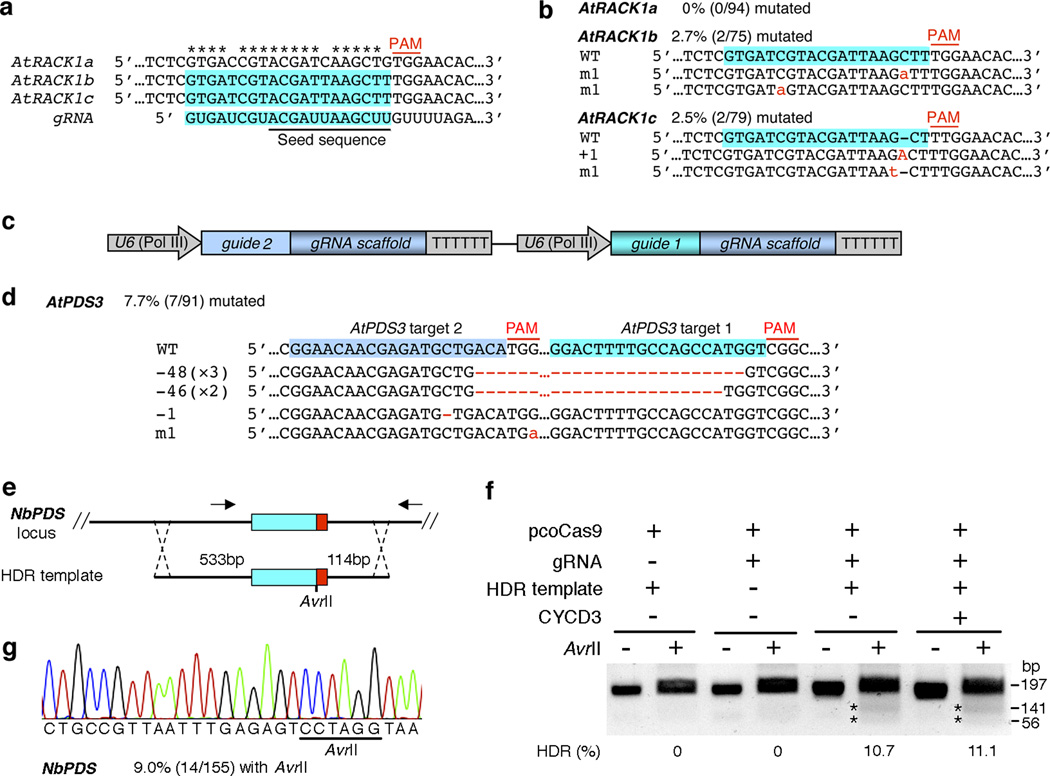
To test whether the gRNA/pcoCas9 system allows multiplex genome editing in Arabidopsis protoplasts, we first identified an identical gRNA target site (target candidate No.2, Supplementary Fig. 3) for both AtRACK1b and AtRACK1c, two members of the Arabidopsis RECEPTOR FOR ACTIVATED C KINASE 1 (RACK1) family (Fig. 2a). By co-expressing pcoCas9 and the cognate gRNA, we observed mutations in both target genes with a comparable mutagenesis frequency (2.5–2.7%, Fig. 2b). Similarly, only single nt substitutions or insertions were detected in these Arabidopsis genes (Fig. 2b). Notably, no mutation was detected in a homologous sequence from AtRACK1a (Fig. 2b), which contains a valid PAM but two mismatches to the 12-bp seed sequence governing the gRNA specificity7–9,13,16,17 (Fig. 2a), illustrating the high specificity of the gRNA/pcoCas9-directed genome editing in plant cells. We further co-expressed pcoCas9 and tandem gRNAs aiming for two juxtaposed targets in AtPDS3 with a 24-bp spacer (Fig. 2c). Interestingly, this simultaneous targeting with two gRNAs led to deletions of up to 48 bp genomic segments between these two target sites by gRNA/pcoCas9 with a mutation frequency of 7.7% (Fig. 2d and Supplementary Fig. 2c). Taken together, these results demonstrated that the gRNA/pcoCas9 system could facilitate multiplex genome editing in plants.
Figure 2.
Multiplex and HDR-mediated genome editing by gRNA/pcoCas9 in plant cells. (a) Diagram of a single gRNA targeting two genes. AtRACK1b and AtRACK1c but not AtRACK1a from the Arabidopsis RACK1 family are gRNA targets. The target sequence recognized by gRNA is shadowed in blue and the gRNA seed sequence is underlined. (b) Targeted mutations induced by gRNA/pcoCas9 in AtRACK1b and AtRACK1c but not AtRACK1a in Arabidopsis protoplasts. (c) Schematic of a tandem gRNA construct. (d) Large genomic deletions are induced by double gRNAs targeting the AtPDS3 locus. In b and d, DNA insertions, deletions and point mutations are shown in red as upper case letters, dashes and lower case letters, respectively. (e) Diagram of the HDR strategy. Successful HDR creates an AvrII site in the target sequence of the NbPDS locus. The arrows represent the primers for gPCR amplification of the target region. (f) AvrII digestion products (marked by asterisks) of NbPDS target amplicons exist upon successful HDR in tobacco protoplasts. Arabidopsis cyclin D-type 3 (CYCD3), a master activator of cell cycle. (g) DNA sequencing evidence of successful HDR in the presence of pcoCas9, gRNA and HDR template.
We next addressed whether the presence of a DNA donor upon gRNA/pcoCas9-mediated generation of a double-strand break would lead to gene replacement by HDR, which could precisely integrate an intended mutation from the DNA donor into the target site. We co-expressed pcoCas9 and the gRNA aiming for the NbPDS target 1 in tobacco protoplasts and concurrently supplied a double-stranded DNA donor that contains a unique AvrII site flanked by a 533-bp left homology arm and a 114-bp right homology arm to the NbPDS locus (Fig. 2e). AvrII digestion of gPCR amplicons spanning the NbPDS target site revealed AvrII incorporation in the target locus with a frequency of 10.7%, and this incorporation strictly relied on both gRNA and the DNA donor (Fig. 2f). Sanger sequencing further verified the anticipated creation of the AvrII site in the target sequence without additional modifications and indicated an HDR-mediated gene replacement at a frequency of 9.0% (Fig. 2g). In addition, we detected NHEJ-mediated targeted mutagenesis at the NbPDS locus with a frequency of 14.2% (Supplementary Fig. 4). As mesophyll protoplasts are isolated from differentiated leaves without active cell division, we tested the possibility of enhancing HDR by triggering ectopic cell division. Co-expression of Arabidopsis CYCD3 (CYCLIN D-TYPE 3), a master activator of cell cycle, hardly promoted the HDR in tobacco protoplasts (Fig. 2f). Exploration of HDR in Arabidopsis protoplasts was unsuccessful, presumably due to intrinsically low efficiency of HDR in Arabidopsis18.
To facilitate genome-wide application of the gRNA/pcoCas9 technology in Arabidopsis, we bioinformatically generated a database for a total of 1,466,718 unique gRNA target sequences in Arabidopsis exons (Supplementary Database), which cover >99% (26,942 out of 27,206) of nuclear protein-encoding genes defined by TAIR10 (The Arabidopsis Information Resource 10). Targeting efficacy and specificity of selected gRNA target candidates from this database need to be experimentally determined each time during future implementation. We also introduced a facile method to manually design a shared gRNA target site specific for multiple homologous target genes by aligning their coding sequences and blasting for off-targets (Supplementary Fig. 3). The gRNA/pcoCas9 technology enables an easy reprogramming of DNA targeting specificity by changing the 20-nt guide sequence in the gRNA without modifying the pcoCas9 protein. We have established a simple and rapid procedure to create a custom gRNA through overlapping PCR (Supplementary Fig. 5). Thus, it is feasible to use a single or tandemly expressed gRNAs (Fig. 2c) to simultaneously target multigene families, which is not easily achievable with ZFNs and TALENs.
We have tested a total of seven target sequences in five target genes in Arabidopsis or tobacco, and obtained targeted mutagenesis in all cases. The variation in mutagenesis efficiency among different genes in Arabidopsis may stem from distinct gRNA binding strength to individual target sequences or distinct chromatin structure and epigenetic state at individual target loci, which requires future investigation. We have demonstrated that plant protoplasts provide an excellent system to rapidly evaluate the efficiency of the gRNA/pcoCas9-mediated genome editing at a specific genomic locus. Our data also suggested that targeting an Arabidopsis gene with multiple gRNAs could improve the success rate of targeted mutagenesis and generate deletions to ensure gene knockout. Significantly, gRNA/pcoCas9 achieved high efficiency of HDR-mediated gene replacement in tobacco protoplasts. The superb potency, simplicity, versatility and specificity of the gRNA/pcoCas9 technology demonstrated in this work promise marker gene-independent and antibiotic selection-free genome engineering with high precisions in diverse plant species to advance basic science and biotechnology.
Supplementary Material
ACKNOWLEDGMENTS
We thank Frederick Ausubel for critical reading of the article, Daniel Voytas for discussion on the HDR strategy, Yan Xiong for the CYCD3 expression plasmid. J.F.L. is supported by the MGH ECOR Postdoctoral Fellowship for Medical Discovery. The Research is supported by the DOE grant DE-FG02-02ER63445 to G.M.C., the NSF grant IOS-0843244 and the NIH grants R01 GM60493 and R01 GM70567 to J.S.
Footnotes
Note: Supplementary information is available in the online version of the paper.
AUTHOR CONTRIBUTIONS
J.F.L. and J.S. designed experiments; J.F.L. and D.Z. performed experiments; J.A., J.E.N., M.M and G.M.C. conducted bioinformatics analyses; J.B. supplied plant materials; J.F.L. and J.S. wrote the manuscript.
REFERENCES
- 1.Zhang F, et al. Proc. Natl. Acad. Sci. USA. 2010;107:12028–12033. doi: 10.1073/pnas.0914991107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Y, et al. Plant Physiol. 2013;161:20–27. doi: 10.1104/pp.112.205179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li T, et al. Nat. Biotechnol. 2012;30:390–392. doi: 10.1038/nbt.2199. [DOI] [PubMed] [Google Scholar]
- 4.Gaj T, et al. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mussolino C, Cathomen T. Nat. Biotechnol. 2013;31:208–209. doi: 10.1038/nbt.2527. [DOI] [PubMed] [Google Scholar]
- 6.Streubel J, et al. Nat. Biotechnol. 2012;30:593–595. doi: 10.1038/nbt.2304. [DOI] [PubMed] [Google Scholar]
- 7.Jinek M, et al. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cong L, et al. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mali P, et al. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hwang WY, et al. Nat. Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho SW, et al. Nat. Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 12.Jinek M, et al. eLIFE. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DiCarlo JE, et al. Nucleic Acids Res. 2013;41:4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H, et al. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoo SD, et al. Nat. Protoc. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- 16.Qi LS, et al. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang W, et al. Nat. Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Pater S, et al. Plant Biotechnol. J. 2013;11:510–515. doi: 10.1111/pbi.12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.