Abstract
The growth of diffraction-quality single crystals is of primary importance in protein X-ray crystallography. Chemical modification of proteins can alter their surface properties and crystallization behavior. The Midwest Center for Structural Genomics (MCSG) has previously reported how reductive methylation of lysine residues in proteins can improve crystallization of unique proteins that initially failed to produce diffraction-quality crystals. Recently, this approach has been expanded to include ethylation and isopropylation in the MCSG protein crystallization pipeline. Applying standard methods, 180 unique proteins were alkylated and screened using standard crystallization procedures. Crystal structures of 12 new proteins were determined, including the first ethylated and the first isopropylated protein structures. In a few cases, the structures of native and methylated or ethylated states were obtained and the impact of reductive alkylation of lysine residues was assessed. Reductive methylation tends to be more efficient and produces the most alkylated protein structures. Structures of methylated proteins typically have higher resolution limits. A number of well-ordered alkylated lysine residues have been identified, which make both intermolecular and intramolecular contacts. The previous report is updated and complemented with the following new data; a description of a detailed alkylation protocol with results, structural features, and roles of alkylated lysine residues in protein crystals. These contribute to improved crystallization properties of some proteins.
Keywords: Chemical modification, Lysine reductive alkylation, Methylation, Ethylation, Isopropylation, Protein crystallization
1 Introduction
X-ray crystallography is the most important method for the elucidation of atomic resolution three-dimensional structures of biological macromolecules. Its success depends essentially on the availability of diffraction-quality single crystals. The generation of protein crystals suitable for structure determination remains a major bottleneck in structural biology. Not all proteins crystallize and an analysis of the data from large-scale structural genomics efforts reveals that at best ~15 % of purified proteins produce a three-dimensional structure. Consequently, there is much interest in exploring salvage approaches to increase the structure determination success rate, specifically by increasing the propensity of proteins for crystallization and improving the diffraction-quality of crystals.
The crystallization of proteins is influenced by many factors associated with the sample itself (impurities, conformational flexibility and local disorder, polydispersity, chemical non-homogeneity, missing interacting partners, insufficient loading with ligands, etc.). Over the years, numerous approaches have been described that address these issues [1–3] but few have been tested vigorously on a large set of protein samples under controlled conditions [4]. Protein surface properties are important for protein crystallization. Modification of the protein surface, either by site-directed mutagenesis [3, 5] or chemical modification [4, 6–9], is a well-established strategy to promote protein crystallization. It is believed that these modifications reduce the surface entropy of the protein [10] and support protein–protein associations [11]. Among protein surface modification strategies, reductive methylation of lysine residues has been successfully applied to obtain good protein samples and high quality crystals for structure determination either in a small sample set or a large sample set in structural genomics centers [4–6, 9, 11, 12]. Some proteins can only be crystallized after methylation [6, 9, 12–14] and often crystals of modified proteins diffract to higher resolution [4, 12, 15]. Besides surface entropy reduction [10] and new surface contact creation [11], it was proposed that N-methyl to oxygen contacts of methylated lysines are important for assisting in the formation of diffraction-quality crystals [4, 14, 16]. Since the potential of other reductive alkylation methods such as ethylation and isopropylation remains unknown, experimental protocols have recently been expanded to include these two in parallel with reductive methylation in the Midwest Center for Structural Genomics (MCSG) pipeline to assess these alternative alkylation approaches.
Reductive alkylation of proteins is a simple, inexpensive method that involves modification of the solvent-exposed ε-amino group of lysine (and under some conditions the N-terminal α-amino group) [17] with reducing agents dimethylamine-borane complex and formaldehyde (for reductive methylation), acetaldehyde (for reductive ethylation) or acetone (for reductive isopropylation). Reductive methylation and ethylation produce both mono-alkyl (mmLys or meLys) and di-alkyl (dmLys or deLys) derivatives. The reaction mechanism for reductive methylation is believed to involve a nucleophilic addition of an unprotonated lysine ε-amino group to formaldehyde to form an N-methylol moiety, which dehydrates and is then hydrogenated to form monomethylated lysine (mmLys). In the second step of the exampled reductive methylation, the secondary amine reacts with a second molecule of formaldehyde, which undergoes hydrogenation leading to dimethylated lysine (dmLys).
R − NH2 +CH2O ↔ R − N = CH2 → R − NH − CH3 → R − N − (CH3)2
The properties of methylated lysines in proteins have been investigated by NMR. The pKa of dmLys measured in calmodulin ranges from 9.29 to 10.23 [17, 18] and is slightly lower than observed for lysine 9.84–10.71 [19]. This is consistent with an observed decrease in the protein isoelectric point after methylation [12].
The chemical modification is fast, specific (only free amino groups are modified), and requires few steps under relatively mild buffer and chemical conditions. Moreover, native and reported methylated proteins show very similar structures and in most cases maintain their biochemical function [6, 9, 15, 17]. In an early effort to assess the efficacy of methylation on a sample set of statistical significance, 370 proteins that have no significant sequence similarity and resisted crystallization efforts in the MCSG during PSI-2 were modified. The results of the evaluation with an improved success rate in protein crystal structures production were reported [4, 16].
Reductive methylation has since continued to be used as an effective salvaging method for proteins that fail in producing diffraction-quality crystals in initial screenings. The proteins tested were biased to those that could be purified in reasonable scale (5–20 mg/ml). Of the 180 proteins that were modified and screened, 12 structures were determined, including the first ethylated and the first isopropylated protein structures (Tables 1 and 2). Together with the previous trial of methylated proteins, 32 alkylated protein structures out of 550 proteins have been determined, a 5.8 % success rate. Considering only ~15 % of proteins purified in their native form result in a crystal structure, the 5.8 % success rate represents a 37 % increase as the proteins targeted in this project derive from a subset of proteins that failed to produce a structure in initial attempts. Therefore, the use of alkylation complements the experiments with native proteins. Reductive alkylation, particularly methylation of protein lysine residues, provides a simple, specific, fast, inexpensive, and efficient method to alter protein surface properties that can improve protein crystallizability and crystalline order and can aid in structure determination. There are very few known side reactions and the method does not require laborious processing of the protein. The method requires a reasonable amount of material and can be applied to several samples in parallel; it does not involve any specialized equipment and therefore can be considered as a good generic approach to salvage projects that failed in the initial crystallization screens. Hence, it fits well into high-throughput approaches for structure determination and suits regular laboratories as well.
Table 1.
Summary of reductive alkylation results for proteins processed in this study
| Alkylation sets |
Number of proteins treated |
Macroscopic crystals harvested |
Diffraction data set collected |
Structure(s) solved |
|---|---|---|---|---|
| Methylation | 180 | 21 | 11 | 10 |
| Ethylation | 74 | 10 | 1 | 1 |
| Isopropylation | 21 | 4 | 1 | 1 |
Table 2.
Structures of alkylated proteins and their properties
| PDB ID | Identifier APC number |
Molecular weight (Da) |
Protein pI | Hydropathy score |
# of Lys in sequence |
Resolution limit (Å) |
|---|---|---|---|---|---|---|
| Methylated | ||||||
| 3R6D | APC100850 | 24,384 | 6.2 | −0.211 | 10 | 1.25 |
| 3OP3 | CDC25CA-c005 | 22,688 | 6.4 | −0.550 | 11 | 2.70 |
| 3QOM | APC100114 | 54,865 | 5.0 | −0.348 | 22 | 2.31 |
| 3PNN | APC100138 | 33,627 | 4.8 | −0.254 | 13 | 1.98 |
| 4DZR | APC100341 | 34,655 | 5.3 | −0.201 | 8 | 2.55 |
| 4EVX | APC101548 | 13,487 | 4.8 | −0.085 | 2 | 1.70 |
| 4H7L | APC100584 | 16,925 | 4.9 | −0.203 | 3 | 2.45 |
| 4I4D | APC109063 | 9,957 | 6.5 | −0.410 | 1 | 2.10 |
| 4IQN | APC101506 | 25,598 | 4.2 | −0.485 | 10 | 1.75 |
| 4JDU | APC105901 | 37,969 | 9.5 | 0.021 | 25 | 1.47 |
| Ethylated | ||||||
| 4IPT | APC100850 | 24,384 | 6.2 | −0.211 | 10 | 1.55 |
| Isopropylated | ||||||
| 4IAG | APC109219 | 14,595 | 4.2 | −0.009 | 1 | 1.90 |
| Native | ||||||
| 4HNGa | APC100850 | 24,384 | 6.2 | −0.211 | 10 | 1.50 |
This native structure is listed for comparison to its pairing methylated and ethylated structures
2 Materials
2.1 Protein Preparation
All proteins were prepared by following the standard procedure developed by the MCSG [20] and Center for Structural Genomics of Infectious Diseases (CSGID). For preparation of protein, details on cloning and protein purification protocols, see the Chapters 5 and 7 in this book. This procedure can also be applied to seleno-methionine labeled proteins. The alkylation protocol requires approximately 5–20 mg of purified proteins at concentrations of 5–10 mg/ml for each sample.
2.2 Reagent Preparation
All reagents are prepared fresh the day of experimentation and all solutions are kept at 4 °C or on ice. They include:
1 M dimethylamine-borane complex (ABC) in deionized water (6 mg of ABC in 100 µl of water).
1 M formaldehyde (for methylation) or 1 M acetaldehyde (for ethylation) or 1 M acetone (for isopropylation) in deionized water.
1 M glycine in deionized water.
1 M dithiothreitol (DTT).
Reaction buffer: 50 mM HEPES pH 8.0, 500 mM NaCl, 5 % (v/v) glycerol, and 10 mM β-mercaptoethanol.
3 Methods
The initial reductive methylation protocol was performed according to Rypniewski et al. [6] and Rayment et al. [9]. The experiment was conducted using sodium borohydride as the reducing agent. However, to reduce foaming and subsequent protein denaturation, the protocol was modified to a more gentle treatment with 1 M dimethylamine-borane complex (ABC) as the reducing agent [12]. More recently, commercial reductive alkylation protocols/kits have also become available (Hampton Research, Inc.). Due to its proven success rate, reductive methylation remains the primary alkylation choice especially when the amount of protein is limited. Otherwise, parallel experiments including reductive ethylation and reductive isopropylation are performed. Experimental protocols for ethylation and isopropylation are similar to the one for methylation except for their second reducing agents as described below. Parallel experiments take advantage of the high-throughput structure determination pipeline in the MCSG and CSGID.
3.1 Reductive Alkylation, Day 1
Add 20 µl of 1 M ABC per 1 ml of protein solution and mix gently.
Immediately add 40 µl of 1 M formaldehyde (for methylation), acetaldehyde (for ethylation) or acetone (for isopropylation) per 1 ml of protein solution, then mix gently.
Incubate the solution at 4 °C for 2 h and repeat the procedure one more time.
At the end of the second incubation, add an additional amount of 10 µl of ABC per 1 ml of protein.
Incubate the solution at 4 °C overnight (12–14 h).
3.2 Reductive Alkylation, Day 2
Add 80 µl of 1 M glycine (to a final concentration of 5 mg/ml) and 6 µl of 1 M DTT (to a final concentration of 5 mM) to quench the reaction.
Leave solution on ice for 2 h.
The modified proteins are either buffer exchanged extensively by dialysis overnight against a large volume of crystallization buffer (20 mM HEPES pH 8.0, 250 mM NaCl, 2 mM DTT) or preferably purified by size exclusion chromatography, which not only removes residual reagents from the reaction, but also separates higher molecular weight protein aggregates, which may have formed during the reaction. In some cases, analysis of the size exclusion chromatography profile can reveal reaction-induced changes in the oligomerization states of the protein.
The modified proteins are then concentrated to the desired concentration for characterization and crystallization (see Note 1).
3.3 Characterization and Crystallization of Modified Proteins
The modified proteins are characterized using SDS PAGE; selected samples are characterized using MALDI-TOF spectrometry (Tecan) or Electrospray Ionization mass spectrometry (ESPI-TOF) with QStar XL (Applied Biosystems Inc.)
The modified proteins are screened for crystallization conditions in sitting drops (Mosquito, TTP Labtech); 0.4 µl of protein is added to 0.4 µl of crystallization solution and equilibrated over a 135 µl well solution. Commercial crystallization formulations available from Hampton Research (Index), Decode Genetics (Emerald Biostructures) (Wizard I & II), and Qiagen (Nextal Biotechnologies) (PEGs II) or the MCSG suite including MCSG-1 to -4 (Microlytic, Inc) are used for the crystal screening. Plates are kept at 4 or 16 °C in Robohotels and imaged with the Minstrel III system (RIGAKU) (see Notes 2 and 3).
Fig. 1.
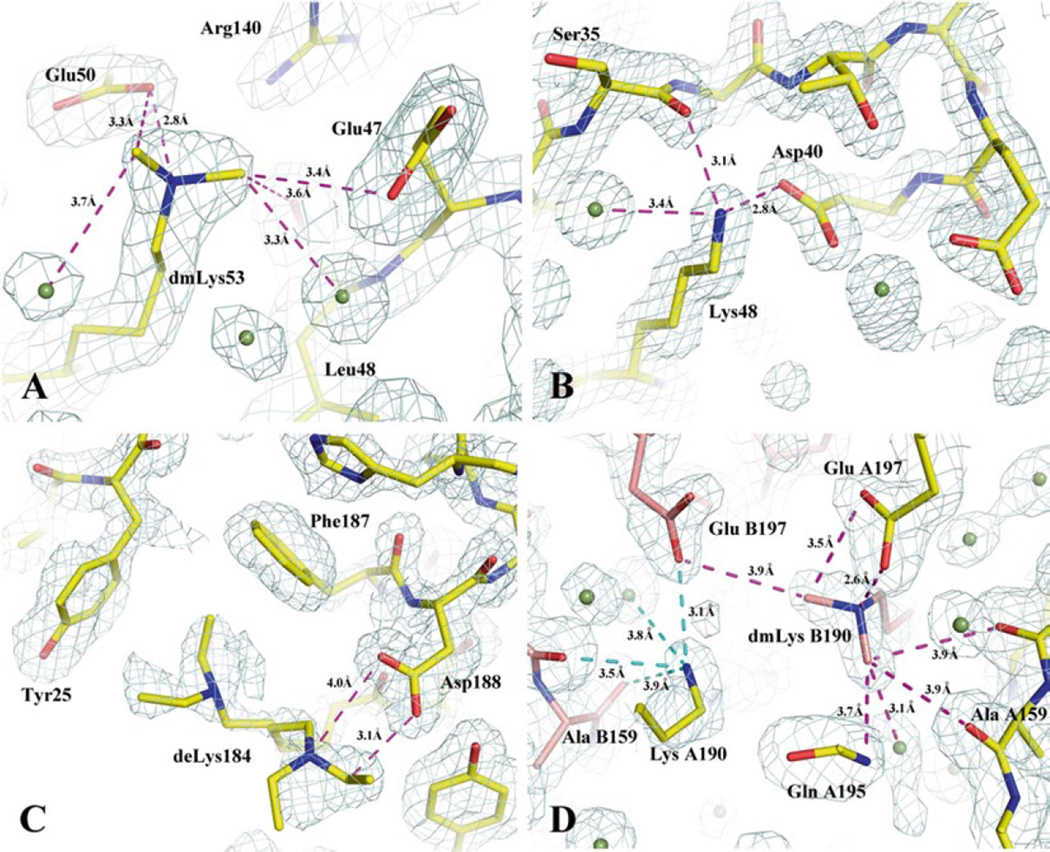
Examples of methylated, unmethylated and ethylated lysines found in alkylated protein structures and the interactions promoted by alkylated lysines. Proteins are drawn in stick format incased in navy mesh of a 2Fo–Fc map contoured at 1σ. (a) A di-methylated lysine 53 (dmLys53) interacts with carboxylates, carbonyl, and water. It is also a part of an extended hydrogen bond network. The figure was prepared based on the structure of a secreted protein from Salmonella enterica subsp. enterica serovar typhimurium str. 14028S (gi: 267994654, PDB code: 4HG1). In this case, the native protein crystals from initial screenings diffracted to about 3.5 Å. After the protein was methylated, the modified protein crystals diffracted to 1.75 Å and its structure determination was straightforward (Table 1, PDB: 4HG1). This was a project in collaboration with the Program for the Characterization of Secreted Effector Proteins (PCSEP) of the Pacific Northwest National Laboratory. (b) An unmethylated lysine (Lys48) involved in multiple interactions (salt bridge and hydrogen bonds) with other protein atoms and one water molecule in a methylated structure (PDB code: 3BED). It is believed that a lysine involved in strong intrachain interactions may prevent the residue from being methylated. The exampled structure is from the mannose/sorbose specific IIA subunit of phosphotransferase system from E. faecalis v583. (c) A diethylated lysine (deLys) in two conformations. Ethylated lysine residues tend to have multiple conformations and be at least partially disordered. In this structure from an ethylated short-chain dehydrogenase/reductase from V. parvula DSM 2008, the deLys184 forms an additional hydrogen bond to Asp188. It also makes hydrophobic contacts to Tyr25 and Phe187. The hydrophobic interaction added from ethylated lysine is believed to be its major feature. (d) In the methylated structure (PDB code: 3BED) as mentioned in (b), the two chains, (A) and (B), make a contact that is nearly twofold symmetric. Across the small interface, methylated lysine 190 from the (B) chain (dmLys B190) forms an extensive interaction to the (A) chain. Carbon atoms from the (A) and (B) chains are colored in yellow and salmon, respectively
Acknowledgments
We wish to thank all members of the Structural Biology Center and Midwest Center for Structural Genomics at Argonne National Laboratory for their help in conducting these experiments. This work was supported by National Institutes of Health Grant number GM GM094585, Contract numbers HHSN272200700058C and HHSN272201200026C and by the US Department of Energy, Office of Biological and Environmental Research, under contract DE-AC02-06CH11357.
Footnotes
Impact of alkylation for protein properties:
A number of studies [12, 16, 17, 21] have indicated that methylation of lysine residues in proteins changes their biochemical properties such as pI, solubility, and hydropathy. After chemical modification, many proteins had noticeably changed properties with the key difference being reduced solubility occasionally associated with protein aggregation and precipitation, as has been reported previously [12].
The alkylation protocol described above requires 5–20 mg protein at a concentration of less than 10 mg/ml. Higher protein concentrations may sometimes produce cross-linked or aggregated protein samples. For about a quarter of the samples, the modified protein became significantly less soluble and could not be recovered after the reaction or displayed significant cross-linking, as revealed by SDS PAGE. In extreme cases, proteins were solidified after a few days of storage at 4 °C. In sporadic cases, proteins were degraded after alkylation, displayed several bands on the SDS PAGE and yielded no crystals. In a third of the cases, the detrimental effect of alkylation on the protein behavior was less pronounced. In approximately another third of the samples, protein properties improved significantly.
The protocol described above is composed of several steps that can be executed in less than 2 days for several proteins in parallel and can be automated using a liquid handling workstation. The completeness of the reaction after the alkylation reaction was assessed using mass spectrometry (data not shown). For those tested that included mostly methylated and some ethylated and isopropylated proteins, the alkylation rates significantly varied. Although the intention was to generate homogeneous fully N-alkylated proteins, the majority of proteins had more than 90 % of lysines modified; however, we observed cases where as little as 50 % of lysines were modified. Our data suggest that methylation is faster and more efficient than ethylation but the actual reaction time of alkylation seems to be protein dependent.
Structural impact of lysine alkylation:
The structural observation of lysine residues after alkylation provides important information about the consequences of the chemical modification.
In alkylated protein structures, the majority of lysine residue side chains are in fact poorly ordered and many could not be observed although they are detected by mass spectrometry. The ordered methylated lysines are mostly in the dmLys form, and are typically involved in interactions with protein and/or solvent (Fig. 1a). These interactions seem to help stabilize the modified side chains. In some low-resolution methylated protein structures, no modified lysine side chain can be reliably built into the model. For example, in the 2.7 Å structure of the human cyclin G associated kinase (GAK) (PDB entry 3OP3), a collaborative project with the Structural Genomics Consortium (SGC, Oxford), no methyl group was built for the six lysines in the structure, though reductive methylation was essential in improving the crystal diffraction limit from 6 to 2.7 Å. However, at a low contour level (0.75σ) in a 2Fo–Fc electron density map, a likely dmLys324 forms a hydrogen bond to the carbonyl group of Val363 in a loop, which may help to stabilize this loop.
In several cases, both alkylated and unalkylated surface lysines were observed in the electron density. Some of these unmethylated lysines are believed to be involved in multiple interactions with neighboring residues (and solvent) and therefore resist alkylation (Fig. 1b). In the structure of the mannose/sorbose specific IIA subunit of the phosphotransferase system from Enterococcus faecalis v583 (PDB entry: 3BED), three of five lysine residues in each of the two protein chains are ordered. Among these, Lys48 remains unmethylated while the other two are clearly modified. There are a few mmLys also observed. Although partial disorder could be a factor, some likely resulted from local geometries where the Nζ atom was involved in a strong hydrogen bond to other protein atom(s) restricting accessibility to reagents.
From limited observations of ethylated and isopropylated protein structures, the ethyl groups or isopropyl groups attached to the Nζ atom of modified lysine tend to have multiple conformations (Fig. 1c). In the ethylated protein structure of a short-chain dehydrogenase/reductase from Veillonella parvula DSM 2008 (PDB entry: 4IPT), three out of ten lysines were modeled as diethylated lysines. No ethyl group on other lysine side chains was observed in electron density maps due to either its absence or disorder. The ordered deLys show multiple conformations and/or partial disorder as well. In the isopropylated protein structure from a zorbamycin binding protein from Streptomyces flavoviridis (PDB entry: 4IAG) the isopropyl group attached to the only lysine of the molecule also has multiple conformations (data not shown).
Roles of alkylated lysines:
The ordered alkylated lysines are engaged in a variety of intramolecular and intermolecular interactions with the protein and solvent. These interactions may help stabilize the protein and create new crystal packing contacts, producing diffraction-quality crystals.
New interactions promoted by alkylated lysine are observed. Besides the interactions that involve contacts between the dmLys methyl groups with carboxylates and carbonyls on the surface of the protein as reported earlier by Kim et al. [4], Shaw et al. [14], and Fan and Joachimiak [16], several additional types of interactions were also observed, such as the interaction with His residues [4]. The deLys shows a new type of hydrophobic interaction (observed for the first time) with the aromatic rings of Tyr and Phe, suggesting that adding a larger hydrophobic alkyl group to lysine changes the nature of the interactions from a hydrogen bond interaction (with methyl group) to a hydrophobic interaction (with ethyl and isopropyl groups) (Fig. 1c). Interestingly, ethylated lysine can still make a hydrogen atom bond through its Nζ nitrogen (Fig. 1c). Experimental observations [4, 11, 12] and quantum mechanical calculations [16] show that the methyl group in dmLys or mmLys is polar and is capable of acting as hydrogen-bond donors in quasi-hydrogen (albeit weaker) bonds formed with oxygen and nitrogen atoms of proteins as well as ordered solvent. In some cases, the interaction is exclusively with a single water molecule, but more often the methylated lysine becomes part of an extensive interaction network on the protein surface (Fig. 1a). It has been proposed that methylation of lysine residues enhances crystal packing by solvent reorganization around methylated lysine side chains, favoring the formation of protein crystals through solvent entropic gain [16]. The distances between methyl carbons in the dmLys (or mmLys) and oxygen or nitrogen atoms range from 3.2 to 3.8 Å, somewhat longer than a typical hydrogen bond distance between two electronegative atoms, as predicted from theoretical calculations (3.28 Å) [16].
Alkylating a lysine expands its interaction radius. Lysine is underrepresented at the protein–protein and crystal packing interfaces [11, 22–24]. Chemical modification to the lysine residue sometimes changes its properties from non-supporting interactions to promoting interactions. For example, methyl groups provide a convenient extension to the ε-amino group of the lysine (i.e., Lys53 in Fig. 1a) allowing a weak, long distance (>4.2 Å) interaction with oxygen or nitrogen (i.e., Glu47 in Fig. 1a) to be replaced with stronger, shorter ε-amine-[N-methyl]—oxygen/nitrogen interactions [16]. Adding methyl groups effectively increases the interaction radius of lysine by 1–1.4 Å. Therefore, modification of Lys, for example, to dmLys may be seen as similar to replacing Lys with a “longer” Arg residue. Arginine has a higher propensity to promote interactions and is found more often on protein–protein [25] and crystal packing interfaces [22–24]. Most interactions with an Arg side chain occur approximately in the plane of the guanidinium moiety. The dimethylamino group, however, is not planar, and interaction with it can occur over a wider range of angles providing a greater interaction surface than Arg, although individual interactions are weaker than those with a guanidium group. Of course, this may also cause “unwanted” effects such as protein aggregation, providing an explanation as to why after methylation some proteins precipitate or change their oligomeric state [12]. Similar mechanisms may happen with lysine ethylation and isopropylation as well.
Alkylated lysine may produce more ordered and tighter packing. Intrachain interactions promoted by alkylated lysine residues can improve protein stability, molecular packing, and help to produce more ordered diffraction-quality crystals (Fig. 1d). Additionally, lysine alkylation alters the surface, changing protein solubility and allowing for the exploration of a different set of protein orientations as they pack to form a crystal. An alkylated protein can explore an “interaction space” that is different from the native protein. This often leads to a new selection of crystal packing contacts with lower lattice disorder as manifested by a reduction in isotropic B-factors. Quantum mechanical calculations showed that methylated lysines attract more ordered solvent molecules [16]. These water molecules are being released to bulk solvent upon protein crystallization leading to a net entropic gain. Additionally, for proteins with both native and alkylated structures, the solvent contents of alkylated crystals are ~4–5 % less than native crystals. Protein crystals with lower solvent content tend to have higher crystalline order and usually diffract to higher resolution [4, 26].
References
- 1.Ferré-D’Amaré AR, Burley SK. Use of dynamic light scattering to assess crystallizability of macromolecules and macromolecular assemblies. Structure. 1994;2:357–359. doi: 10.1016/s0969-2126(00)00037-x. [DOI] [PubMed] [Google Scholar]
- 2.Dong A, Xu X, Edwards AM, et al. In situ proteolysis for protein crystallization and structure determination. Nat Methods. 2007;4:1019–1021. doi: 10.1038/nmeth1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Derewenda ZS. Rational protein crystallization by mutational surface engineering. Structure. 2004;12:529–535. doi: 10.1016/j.str.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 4.Kim Y, Quartey P, Li H, et al. Large-scale evaluation of protein reductive methylation for improving protein crystallization. Nat Methods. 2008;5:853–854. doi: 10.1038/nmeth1008-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D’Arcy A, Stihle M, Kostrewa D, et al. Crystal engineering: a case study using the 24 kDa fragment of the DNA gyrase B subunit from Escherichia coli. Acta Crystallogr D Biol Crystallogr. 1999;55:1623–1625. doi: 10.1107/s0907444999008136. [DOI] [PubMed] [Google Scholar]
- 6.Rypniewski WR, Holden HM, Rayment I. Structural consequences of reductive methylation of lysine residues in hen egg white lysozyme: an X-ray analysis at 1.8-A resolution. Biochemistry. 1993;32:9851–9858. doi: 10.1021/bi00088a041. [DOI] [PubMed] [Google Scholar]
- 7.Means GE, Feeney RE. Chemical modifications of proteins: history and applications. Bioconjug Chem. 1990;1:2–12. doi: 10.1021/bc00001a001. [DOI] [PubMed] [Google Scholar]
- 8.Rayment I. Reductive alkylation of lysine residues to alter crystallization properties of proteins. Methods Enzymol. 1997;276:171–179. [PubMed] [Google Scholar]
- 9.Rayment I, Rypniewski WR, Schmidt-Base K, et al. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 10.Derewenda ZS, Vekilov PG. Entropy and surface engineering in protein crystallization. Acta Crystallogr D Biol Crystallogr. 2006;62:116–124. doi: 10.1107/S0907444905035237. [DOI] [PubMed] [Google Scholar]
- 11.Sledz P, Zheng H, Murzyn K, et al. New surface contacts formed upon reductive lysine methylation: improving the probability of protein crystallization. Protein Sci. 2010;19:1395–1404. doi: 10.1002/pro.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walter TS, Meier C, Assenberg R, et al. Lysine methylation as a routine rescue strategy for protein crystallization. Structure. 2006;14:1617–1622. doi: 10.1016/j.str.2006.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schubot FD, Waugh DS. A pivotal role for reductive methylation in the de novo crystallization of a ternary complex composed of Yersinia pestis virulence factors YopN, SycN and YscB. Acta Crystallogr D Biol Crystallogr. 2004;60:1981–1986. doi: 10.1107/S0907444904023005. [DOI] [PubMed] [Google Scholar]
- 14.Shaw N, Cheng C, Tempel W, et al. (NZ)CH…O contacts assist crystallization of a ParB-like nuclease. BMC Struct Biol. 2007;7:46. doi: 10.1186/1472-6807-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi M, Kubota M, Matsuura Y. Crystallization and improvement of crystal quality for X-ray diffraction of maltooligosyl trehalose synthase by reductive methylation of lysine residues. Acta Crystallogr D Biol Crystallogr. 1999;55:931–933. doi: 10.1107/s0907444999002115. [DOI] [PubMed] [Google Scholar]
- 16.Fan Y, Joachimiak A. Enhanced crystal packing due to solvent reorganization through reductive methylation of lysine residues in oxidoreductase from Streptococcus pneumoniae. J Struct Funct Genomics. 2010;11:101–111. doi: 10.1007/s10969-010-9079-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Means GE. Reductive alkylation of amino groups. Methods Enzymol. 1977;47:469–478. doi: 10.1016/0076-6879(77)47047-2. [DOI] [PubMed] [Google Scholar]
- 18.Zhang M, Thulin E, Vogel HJ. Reductive methylation and pKa determination of the lysine side chains in calbindin D9k. J Protein Chem. 1994;13:527–535. doi: 10.1007/BF01901534. [DOI] [PubMed] [Google Scholar]
- 19.Zhang M, Vogel HJ. Determination of the side chain pKa values of the lysine residues in calmodulin. J Biol Chem. 1993;268:22420–22428. [PubMed] [Google Scholar]
- 20.Kim Y, Dementieva I, Zhou M, et al. Automation of protein purification for structural genomics. J Struct Funct Genomics. 2004;5:111–118. doi: 10.1023/B:JSFG.0000029206.07778.fc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Means GE, Feeney RE. Reductive alkylation of proteins. Anal Biochem. 1995;224:1–16. doi: 10.1006/abio.1995.1001. [DOI] [PubMed] [Google Scholar]
- 22.Anashkina A, Kuznetso E, Esipova N, et al. Comprehensive statistical analysis of residues interaction specificity at protein–protein interfaces. Proteins. 2007;67:1060–1077. doi: 10.1002/prot.21363. [DOI] [PubMed] [Google Scholar]
- 23.Glaser F, Steinberg DM, Vakser IA, et al. Residue frequencies and pairing preferences at protein–protein interfaces. Proteins. 2001;43:89–102. [PubMed] [Google Scholar]
- 24.Juers DH, Matthews BW. Reversible lattice repacking illustrates the temperature dependence of macromolecular interactions. J Mol Biol. 2001;311:851–862. doi: 10.1006/jmbi.2001.4891. [DOI] [PubMed] [Google Scholar]
- 25.Magalhaes A, Maigret B, Hoflack J, et al. Contribution of unusual arginine–arginine short-range interactions to stabilization and recognition in proteins. J Protein Chem. 1994;13:195–215. doi: 10.1007/BF01891978. [DOI] [PubMed] [Google Scholar]
- 26.Wang J, Dauter M, Alkire R, et al. Triclinic lysozyme at 0.65 Å resolution. Acta Crystallogr D Biol Crystallogr. 2007;63:1254–1268. doi: 10.1107/S0907444907054224. [DOI] [PubMed] [Google Scholar]