Abstract
Polycomb group (PcG) proteins play important roles in repressing lineage-specific genes and maintaining the undifferentiated state of mouse embryonic stem cells (mESCs). However, how PcG proteins are recruited to their target genes is largely unknown. Here, we show that the H3K36-specific histone demethylase Kdm2b is highly expressed in mESCs and regulated by the pluripotent factors Oct4 and Sox2 directly. Depletion of Kdm2b in mESCs causes de-repression of lineage-specific genes and induces early differentiation. The function of Kdm2b depends on its CxxC-ZF domain, which mediates its genome-wide binding to CpG islands (CGIs). Kdm2b interacts with the core components of polycomb repressive complex 1 (PRC1) and recruits the complex to the CGIs of early lineage-specific genes. Thus, our study not only reveals an Oct4–Sox2–Kdm2b–PRC1–CGI regulatory axis and its function in maintaining the undifferentiated state of mESCs, but also demonstrates a critical function of Kdm2b in recruiting PRC1 to the CGIs of lineage-specific genes to repress their expression.
mESCs are derived from the inner cell mass of blastocysts. In the presence of leukaemia inhibitory factor (LIF), mESCs can self-renew and maintain their pluripotency in vitro1,2. The pluripotency of mESCs depends on their undifferentiated status being maintained in culture. After undergoing differentiation, they have a reduced capacity for chimaera contribution and lose their germline competence. Thus, the differentiation level of mESCs is inversely correlated with their developmental potential3,4.
Repression of early lineage-specific genes is essential for maintaining the mESC pluripotent state. Previous studies have revealed that both the polycomb group (PcG) proteins and the core pluripotent factors play important roles in repressing early lineage-specific genes in mESCs (refs 5–8). Loss of PcG function results in activation of lineage-specific genes and induces spontaneous differentiation of mESCs (refs 9–11). Consistently, knockout of key PcG genes in mice results in abnormal gastrulation and early embryonic lethality12–15.
Although the function of PcG proteins in repressing differentiation genes in mESCs has been well documented, how they are recruited to the lineage-specific genes is just beginning to be revealed. Several mechanisms that include DNA-binding proteins16, the PRC2-associated JARID2 (refs 17,18) and non-coding RNAs (refs 19–21) have been proposed to recruit PRC2, but whether similar mechanisms are used for PRC1 recruitment is unknown. Previous studies suggested that PRC2-mediated H3K27 methylation can contribute to PRC1 recruitment22,23. However, alternative mechanisms for PRC1 recruitment exist as binding of PRC1 to chromatin can take place independently of PRC2 (refs 24–26). The finding that PRC1- and PRC2-binding sites are associated with CGIs in mESCs suggests that PcG proteins can be potentially recruited to these sites through CGI-binding proteins27. CGIs are short interspersed DNA sequences with multiple CpG dinucleotides and a high GC content that are refractory to DNA methylation. Approximately 70% of the annotated promoters are associated with CGIs (ref. 28). Previous studies suggest that the unique DNA sequence features of CGIs could serve as a signal for the recruitment of PRC1 and PRC2 (ref. 29). However, it is not clear whether CGIs recruit the PcG complexes directly or indirectly through some CGI-binding proteins.
The Kdm2b gene, which encodes a H3K36me2-specific demethylase30, was first identified as a hotspot for proviral insertion in murine tumours31. It contains a CxxC-zinc finger (CxxC-ZF) that has the potential to bind to unmethylated CGIs (ref. 32). Functionally, Kdm2b plays an important role in repressing the Ink4/Arf locus and suppressing premature cellular senescence30,33. Depletion of Kdm2b in leukaemia stem cells blocks leukaemia development in a mouse model34. Recently, Kdm2b was also found to facilitate reprogramming of somatic cells to pluripotent stem cells35,36.
Owing to the functional similarity between Kdm2b and PcG proteins in regulating the Ink4/Arf locus, in somatic cell reprogramming, and its predicted CGI-binding property, we postulate that Kdm2b might have important functions in maintaining mESC pluripotent state by targeting PcG proteins to CGI-containing developmental regulators37–39. Here we present evidence that supports this hypothesis.
RESULTS
Kdm2b is highly expressed in mESCs and is a direct target of Oct4 and Sox2
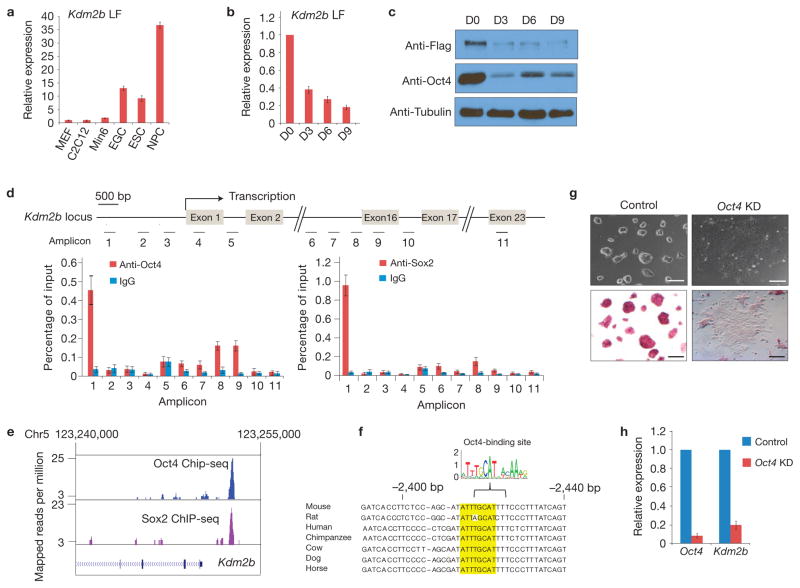
In addition to a JmjC domain responsible for its histone demethylase activity, the long isoform of Kdm2b (Kdm2b LF) harbours a CxxC-ZF, a plant homeodomain (PHD), an F-box and a leucine-rich repeat (LRR). The Kdm2b short isoform (Kdm2b SF), encoded by a transcript variant, lacks the amino-terminal JmjC domain40 (Supplementary Fig. S1A,B). To facilitate characterization of Kdm2b in mESCs, we generated a mESC line with a 3xFlag-tag knocked into the carboxy terminus of the Kdm2b locus (Supplementary Fig. S1C). The correctly targeted mESC clone was validated by genotyping, immunostaining and western blot analyses (Supplementary Fig. S1D–F). Real-time PCR with reverse transcription (RT–qPCR) analyses demonstrated that the Kdm2b LF is highly expressed in mESCs, embryonic germ cells and neural progenitor cells relative to murine embryonic fibroblasts (MEFs), C2C12 myoblasts and pancreatic β cells, whereas the expression of Kdm2b SF exhibited no significant cell type variation (Fig. 1a and Supplementary Fig. S2A). In addition, both the messenger RNA and protein levels of Kdm2b LF are greatly downregulated on embryoid body (EB) differentiation (Fig. 1b,c), whereas the expression of Kdm2b SF is relatively stable during this process (Supplementary Fig. S2B).
Figure 1.
Kdm2b is highly expressed in mESCs and is a direct target of Oct4 and Sox2. (a) Relative expression levels of Kdm2b LF in ESCs, embryonic germ cells (EGCs), neural progenitor cells (NPCs), MEFs, C2C12 myoblasts and pancreatic β cells (Min6) measured by RT–qPCR. Data are mean ± s.d., n = 3. The source data for statistics are provided in Supplementary Table S5. (b) Relative expression levels of Kdm2b LF at different days of embryoid body differentiation measured by RT–qPCR and normalized to Gapdh. The mRNA level of the mESCs (D0) is set as 1. Data are mean ± s.d., n = 3. The source data for statistics are provided in Supplementary Table S5. (c) Western blot analysis of Kdm2b LF and Oct4 protein levels at different days of embryoid body differentiation. Tubulin serves as a loading control. Full scans of blots are shown in Supplementary Fig. S8a. (d) Bottom, ChIP–qPCR analysis demonstrated that Oct4 and Sox2 occupy the Kdm2b gene promoter. Data are presented as percentage of input DNA. The Kdm2b gene locus and the location of the various amplicons are indicated in the diagram at the top of the panel. The TSS and the relevant exons are indicated. Data are mean ± s.d., n = 3. The source data for statistics are provided in Supplementary Table S5. (e) An analysis of published ChIP-seq data45 showing that Oct4 and Sox2 occupancy is enriched at the promoter of the mouse Kdm2b gene. (f) Presence of a consensus Oct4-binding sequence (highlighted) at the promoter region of mammalian Kdm2b genes. (g) Representative images show the morphology (top panels) and alkaline phosphatase activity (bottom panels) of control and Oct4 knockdown mESC cells. Scale bars, 100 μm. (h) Relative expression levels of Oct4 and Kdm2b in the control and Oct4 knockdown mESCs. The relative expression levels are normalized to Gapdh. The level of gene expression in the control knockdown mESCs is set as 1. Data are mean ± s.d., n = 3. The source data for statistics are provided in Supplementary Table S5.
The expression pattern of Kdm2b LF suggests that its expression might be regulated by pluripotent transcription factors. This is supported by chromatin immunoprecipitation and quantitative PCR (ChIP–qPCR) analyses (Fig. 1d). Consistently, analysis of a published data set41 demonstrated that the occupancy of both Oct4 and Sox2 is enriched at the promoter of the Kdm2b LF gene (Fig. 1e), where a consensus mammalian Oct4-DNA-binding sequence is identified (Fig. 1f). Lentiviral shRNA-mediated depletion of Oct4 resulted in downregulation of Kdm2b LF expression (Fig. 1g,h), which further supports a role for Oct4 in regulating Kdm2b LF expression. Collectively, these data demonstrate that Kdm2b LF is highly expressed in mESCs and its expression level is directly controlled by Oct4 and Sox2.
Depletion of Kdm2b induces early differentiation of mESCs
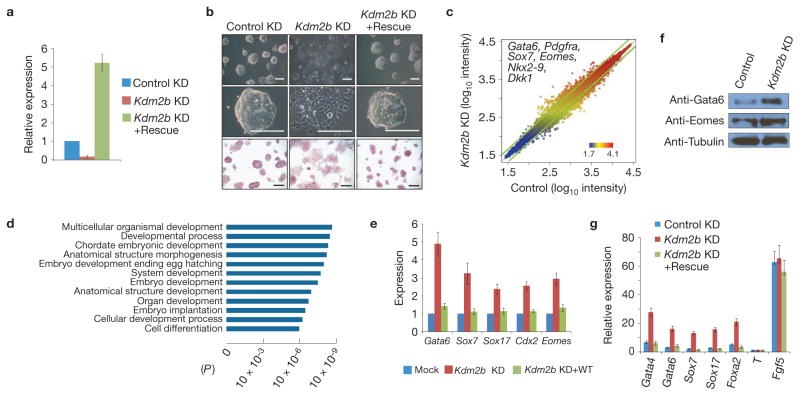
To explore the function of Kdm2b in mESCs, we depleted Kdm2b by a lentiviral shRNA vector targeting both the short and long forms of the Kdm2b transcripts30,34. In parallel, we also established an shRNA-resistant Kdm2b-expression vector for rescue purposes. RT–qPCR analyses demonstrated an 85% knockdown, whereas the rescue vector expressed Kdm2b at about fivefold of the control level (Fig. 2a and Supplementary Fig. S4A). Although the Kdm2b knockdown mESCs still aggregate and are positive for alkaline phosphatase staining, they form flat, loose and morphologically distinctive colonies that resemble typical extra-embryonic endoderm cells (ExEn; Fig. 2b, compare the first two columns). Importantly, these morphological changes can be rescued by expression of Kdm2b (Fig. 2b, compare columns 2 and 3), indicating that the observed morphological change is caused by Kdm2b deficiency.
Figure 2.
Depletion of Kdm2b induces mESC differentiation and causes aberrant gene expression. (a) Relative mRNA levels of Kdm2b in control, knockdown and rescued mESCs measured by RT–qPCR. The relative expression levels are normalized to Gapdh. The level of Kdm2b in the control knockdown mESCs is set as 1. Values represent means ± s.d. from three biological replicates. (b) Representative images show that the morphological changes of Kdm2b knockdown can be rescued by expression of exogenous Kdm2b (upper and middle panels). Bottom panels are images of alkaline phosphatase activity of the various mESCs. Scale bars, 100 μm. (c) Scatter plot shows the upregulated and downregulated genes in response to Kdm2b knockdown in mESCs. The top and bottom boundaries represent a ±1.5-fold change in gene expression. Representative early lineage genes upregulated by Kdm2b knockdown are listed. (d) Gene Ontology analysis of upregulated genes in response to Kdm2b knockdown. (e) Relative expression levels of representative primitive endoderm and trophoectoderm specific genes in control, Kdm2b knockdown and wild-type Kdm2b rescued mESCs. The mRNA levels were measured by RT–qPCR analyses. The mRNA level of Kdm2b in the control mESCs is arbitrarily set as 1. Data are mean ± s.d., n = 3. The source data for statistics are provided in Supplementary Table S5. (f) Western blot analyses show that Kdm2b knockdown causes an increase in Gata6 and Eomes protein levels. Tubulin serves as a loading control. Full scans of blots are shown in Supplementary Fig. S8b. (g) RT–qPCR analyses show the relative expression levels of lineage-specific genes in the control, Kdm2b knockdown and Kdm2b rescued mESCs at day 4 of embryoid body differentiation. The expression levels are normalized to Gapdh. The mRNA level of undifferentiated mESCs is set as 1. Data are mean ± s.d., n = 3. The source data for statistics are provided in Supplementary Table S5.
Interestingly, immunostaining of the pluripotent factors indicated that knockdown of Kdm2b does not significantly alter the protein levels of the core pluripotent factors Oct4, Sox2 and Nanog (Supplementary Fig. S3A–C). Furthermore, Kdm2b knockdown does not affect the kinetics of cell growth or colony-forming capacity (Supplementary Fig. S3D,E). The Kdm2b knockdown mESCs are able to maintain their ExEn-like morphology and propagation capacity when cultured in the presence of LIF (data not shown). These results suggest that depletion of Kdm2b in mESCs induces early differentiation although their self-renewal and colony-formation capacity are maintained.
Depletion of Kdm2b in mESCs induces expression of early lineage-specific genes
To understand the molecular mechanism by which Kdm2b maintains the mESC state, we performed microarray analyses. Compared with the control, knockdown of Kdm2b resulted in upregulation or downregulation (average ≥1.5-fold changes) of 303 and 507 genes, respectively (Fig. 2c and Supplementary Table S1, GEO access number GSE41316). Consistent with the observed morphological changes, early lineage genes such as the ExEn-specific genes (Gata6, Pdgfrα and Sox7) and the trophoectoderm genes (Cdx2 and Eomes) are upregulated in the Kdm2b knockdown mESCs (Supplementary Table S1). A gene ontology (GO) analysis revealed that the upregulated genes are enriched for genes involved in embryonic development and cellular differentiation (P ≤10−6) (Fig. 2d). However, the downregulated genes are enriched for genes related to lysosome, lytic vacuole and lipid metabolic processes (P ≤10−6; data not shown).
RT–qPCR analyses confirmed that the ExEn- (Gata6, Sox7 and Sox17) as well as the trophoectoderm- (Cdx2 and Eomes) specific genes are upregulated on Kdm2b knockdown (Fig. 2e), whereas the expression levels of mesoderm and ectoderm genes are not significantly altered (Supplementary Fig. S4C). Upregulation of Gata6 and Eomes was further confirmed by western blot analysis (Fig. 2f). Importantly, de-repression of these genes can be rescued by enforced expression of wild-type Kdm2b (Fig. 2e). Consistent with the immunostaining results (Supplementary Fig. S3A–C), Kdm2b knockdown does not alter pluripotent gene (Oct4, Sox2 and Nanog) expression. Neither does it alter the expression of PRC1 (Ring1b and Bmi1) or PRC2 (Ezh2) components (Supplementary Fig. S4C). On the basis of these results, we conclude that the phenotypic changes of Kdm2b- depleted mESCs are caused by the expression of early developmental genes.
To further confirm that Kdm2b depletion affects early lineage gene expression, we subjected the Kdm2b-depleted mESCs to embryoid body formation assays. RT–qPCR analyses of 4-day embryoid bodies demonstrated that although all lineage genes are upregulated after embryoid body induction, the endoderm-specific genes (Gata4, Gata6, Sox7, Sox17 and Foxa2) are markedly upregulated in the Kdm2b knockdown cells, which can be rescued by expression of an shRNA-resistant Kdm2b (Fig. 2g). Collectively, these data suggest that Kdm2b is not only required for the repression of early lineage-specific genes in mESCs, but is also required for the regulation of gene expression during early differentiation.
The function of Kdm2b in mESCs depends on its CxxC-ZF domain, but not its H3K36me2 demethylase activity
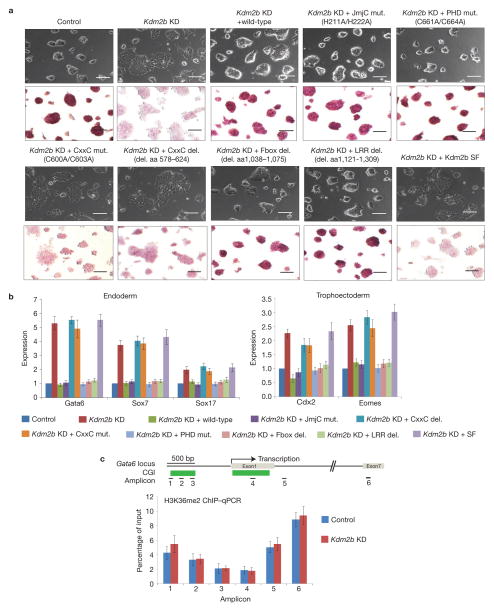
To understand the molecular basis by which Kdm2b represses early lineage-specific genes in mESCs, we attempted to rescue the differentiation phenotypes by expressing wild-type Kdm2b and several Kdm2b mutants. We first confirmed equal expression of wild-type and mutant Kdm2b by RT–qPCR and western blot analyses (Supplementary Fig. S4A,B). Interestingly, with the exception of the short form or the CxxC-ZF deletion (Δaa578–624) or point mutations (C600A/C603A), all of the other mutations in the JmjC domain (H211A/H222A), PHD domain (C661A/C664A), F-Box (Δaa1038–1075) or LRR domain (Δaa1121–1309) can rescue the early differentiation phenotypes induced by Kdm2b knockdown (Fig. 3a). Consistent with these observed phenotypes, RT–qPCR analyses demonstrated that neither the CxxC-ZF mutants, nor Kdm2b SF can repress early lineage-specific genes induced by Kdm2b knockdown (Fig. 3b). These results suggest that Kdm2b-mediated developmental gene repression is independent of its histone demethylase activity. In support of this notion, ChIP–qPCR analyses demonstrated that the H3K36me2 level at the Gata6 gene locus is not significantly altered in the Kdm2b knockdown cells (Fig. 3c).
Figure 3.
Knockdown of Kdm2b in mESCs induces expression of early lineage-specific differentiation genes. (a) Representative images show the morphology (top panels) and alkaline phosphatase activity (bottom panels) of control, Kdm2b knockdown and various rescued cells. Scale bars, 100 μm. (b) Relative expression levels of representative endoderm-and trophoectoderm-specific genes in control, Kdm2b knockdown, and the wild-type and various mutant rescued mESCs. The mRNA levels were measured by RT–qPCR analyses. The mRNA level of Kdm2b in control knockdown mESCs is set as 1. Data are mean ± s.d., n = 3. The source data for statistics are provided in Supplementary Table S5. (c) Bottom, ChIP–qPCR analyses of H3K36me2 at the Gata6 locus in control and Kdm2b knockdown mESCs. Top, schematic representation of the mouse Gata6 genomic structure; associated CGIs as well as the locations of the six amplicons are indicated. Results presented are the percentage of input. Data are mean ± s.d., n = 3. The source data for statistics are provided in Supplementary Table S5.
Kdm2b occupies CGIs in mESCs
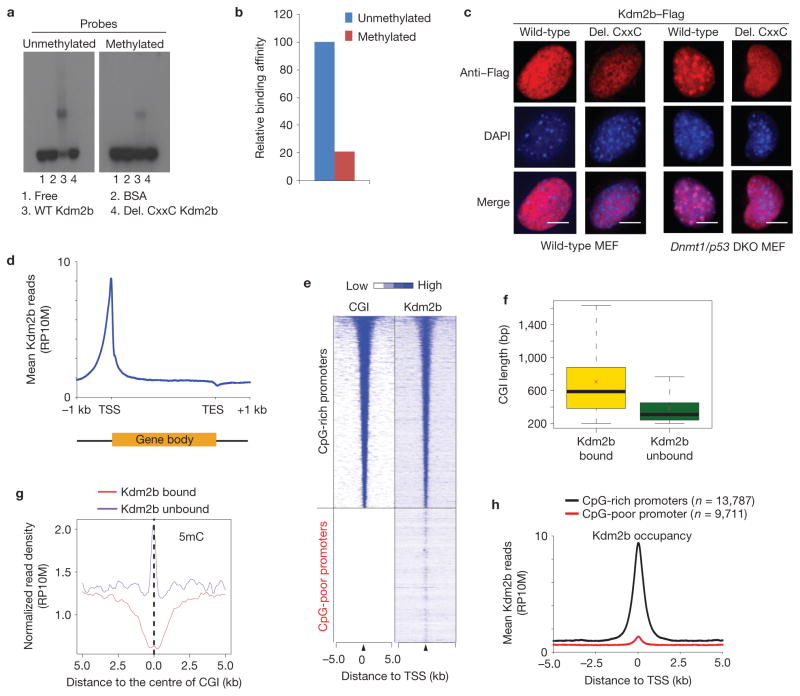
Previous studies have demonstrated that proteins with a CxxC-ZF domain can bind to unmethylated CpGs (refs 42,43), which prompted us to investigate whether Kdm2b has a similar property. Electrophoresis mobility shift assays (EMSAs) demonstrated that the wild-type Kdm2b, but not the CxxC-ZF deletion mutant, preferentially binds to unmethylated CpG-rich DNA (Fig. 4a, compare lanes 3 and 4), with about fivefold higher affinity (Fig. 4b).
Figure 4.
Kdm2b binds to unmethylated CGIs in vitro and in vivo. (a) EMSAs show that Kdm2b preferentially binds to unmethylated CpG-rich probes compared with the methylated probes. The binding activity depends on the CxxC-ZF domain (compare lanes 3 and 4). (b) Quantification of the relative Kdm2b-binding activity. The relative DNA binding activity is calculated as: intensity of shifted upper band/(intensity of upper band + intensity of lower band) ×100. The signal of binding with the unmethylated probe is set as 100. (c) Representative images showing the localization of wild-type and CxxC-ZF deletion mutant Kdm2b in wild-type and Dnmt1/p53 DKO murine embryonic fibroblasts. Note that wild-type Kdm2b, but not the CxxC-ZF deletion mutant, is localized to DAPI-heavy foci in the Dnmt1/p53 MEFs. Scale bars, 5 μm. (d) Global Kdm2b occupancy profile shows Kdm2b is enriched at TSSs but is depleted at TESs. Average Kdm2b signal for all annotated genes is shown along the transcription units from 1 kilobase (kb) upstream of TSS to 1 kb downstream of TES. RP10M, reads per 10 million reads. (e) Heat map representation of CGIs and Kdm2b occupancy at all annotated gene promoters (5 kb flanking TSSs of Refseq genes) in mESCs. The heat map is rank-ordered from genes with longest CGIs to no CGI within 5 kb genomic regions flanking TSSs. (f) The relationship between the lengths of CGIs and Kdm2b-binding states. The whiskers in the boxplot extend up to 1.5 times the interquartile range and data points beyond this range have been omitted for clarity of presentation. The x symbols show the group means. (g) The average 5-methylcytosine (5mC) signals of Kdm2b-bound and Kdm2b-unbound CGIs (ref. 45). (h) Kdm2b-binding profiles centred to TSS for CpG-rich promoters and CpG-poor promoters. The average Kdm2b signal for all annotated genes of the two groups within 5 kb genomic regions flanking TSSs is shown.
To demonstrate that Kdm2b binds to unmethylated CpG-rich DNA in vivo, we expressed a Flag-tagged Kdm2b in both wild-type and Dnmt1/p53 double-knockout MEFs (Dnmt1/p53 DKO MEFs). Depletion of DNA methyltransferase 1 (Dnmt1) leads to loss of DNA methylation at the pericentric heterochromatin marked by DAPI-heavy foci32. Immunostaining revealed that both wild-type and CxxC-ZF deletion mutants distribute evenly in the nuclei of wild-type MEFs (Fig. 4c and Supplementary Fig. S5, first two columns). In contrast, wild-type Kdm2b co-localizes with DAPI-heavy foci in all Dnmt1/p53 DKO MEFs (Fig. 4c and Supplementary Fig. S5, third column). Deletion of the CxxC-ZF domain abolishes this localization pattern (Fig. 4c and Supplementary Fig. S5, last column). These data support that Kdm2b binds to unmethylated CpG-rich sequences in vivo through its CxxC-ZF domain.
The above results suggest that Kdm2b, similar to other CxxC-ZF-containing proteins, binds to unmethylated CGIs in mESCs (refs 32,44). To map its genomic location, we performed ChIP-sequencing (ChIP-seq) analyses in mESCs and identified a total of 29,525 Kdm2b peaks with high confidence (P <10−8, >5-fold enrichment over IgG control; GEO access number: GSE41316). Data analyses revealed that Kdm2b is enriched at promoter, 5′UTR and coding regions (Supplementary Fig. S6). Notably, Kdm2b is highly enriched at transcriptional start sites (TSSs) but depleted at transcriptional end sites (TESs; Fig. 4d). Importantly, Kdm2b occupancy highly overlaps with annotated CGIs (Fig. 4e). Among the total number of 15,963 CGIs analysed, 13,444 (84.2%) are bound by Kdm2b. Compared with the Kdm2b-bound CGIs, the unbound CGIs (15.8%) have a relatively shorter CGI length and a higher level of DNA methylation, suggesting that Kdm2b preferentially binds to the long unmethylated CGIs in cells45 (Fig. 4f,g). Consistently, Kdm2b is enriched at the CpG-rich promoters compared with the CpG-poor promoters (Fig. 4h). Collectively, these results demonstrate that Kdm2b binds to CGIs in mESCs.
Kdm2b and PRC1 components associate and co-occupy the same genomic loci
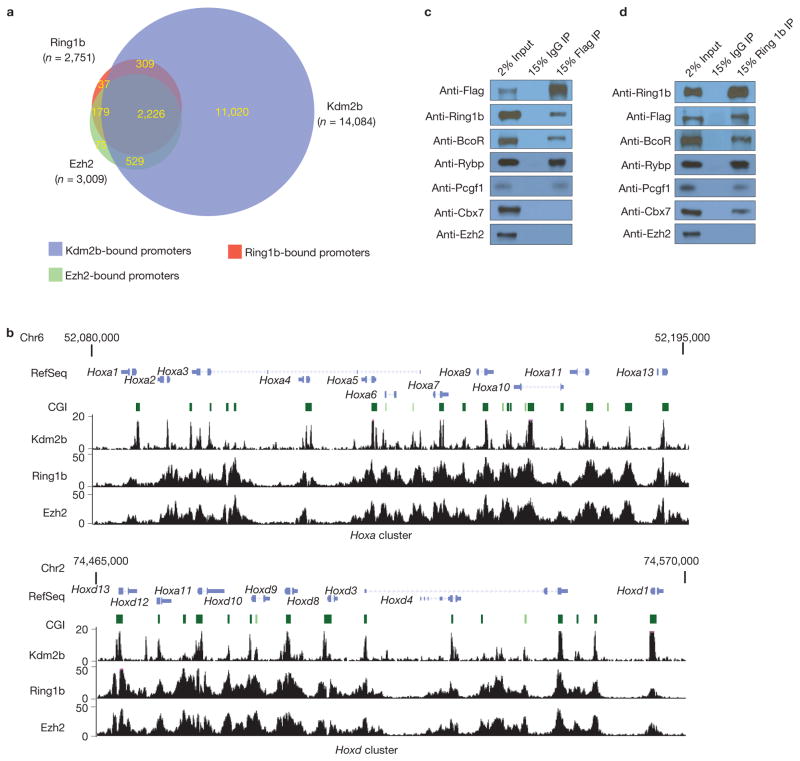
As the function of Kdm2b in repressing developmental genes in mESCs depends on its CxxC-ZF DNA-binding domain but not its histone demethylase activity (Fig. 3), we speculate that its transcriptional regulatory function may be mediated through its associated proteins recruited to the CGIs. Previous studies demonstrated that KDM2B associates with RING1B, RYBP and BCOR in cancer cells46,47 and together they form a PRC1 subcomplex in human HEK293T cells48, suggesting that Kdm2b might be able to recruit PRC1 to CGIs of developmental genes in mESCs. To explore this possibility, we investigated whether Kdm2b and PRC1 components co-occupy the same genomic loci in mESCs. ChIP-seq analyses revealed that of the 23,498 unique promoters analysed, Ring1b and Ezh2 occupy 2,751 and 3,009 gene promoters, respectively. Given that 2,405 promoters are co-occupied by both Ring1b and Ezh2, PRC1 and PRC2 work together in regulating most of their target genes. Notably, most Ring1b-(92%) and Ezh2- (91%) bound promoters are also bound by Kdm2b (Fig. 5a,b and Supplementary Table S2), supporting the notion that Kdm2b recruits PcG proteins to the CGIs in mESCs.
Figure 5.
Kdm2b and PRC1 interact and co-occupy genomic loci (a) A Venn diagram shows the numbers of Ring1b-, Ezh2- and Kdm2b-binding promoters identified by ChIP-seq and their relationship. Co-occupied promoters are indicated by the overlapped areas in the diagram. The numbers of binding sites are indicated in the corresponding area. (b) Bottom, examples of Kdm2b-, Ring1b- and Ezh2-binding peaks across the Hoxa and Hoxd clusters in mESCs. The y axis represents the number of reads in each 10 million total reads. Top, schematic representation of the mouse Hoxa and Hoxd clusters, with the genes and associated CGIs indicated. (c) Co-immunoprecipitation and western blot analysis showing the proteins co-immunoprecipitated with Kdm2b. Full scans of blots are shown in Supplementary Fig. S8c. (d) Co-immunoprecipitation and western blot analysis showing the proteins co-immunoprecipitated with Ring1b. Full scans of blots are shown in Supplementary Fig. S8d.
To demonstrate that Kdm2b can indeed associate with PRC1 in mESCs, we performed immunoprecipitation using protein extracts derived from the Flag–Kdm2b-tagged mESCs (Supplementary Fig. S1). We found that Flag–Kdm2b co-immunoprecipitated with Ring1b, BcoR, Rybp and Pcgf1, but not Cbx7 or Ezh2 (Fig. 5c). Conversely, Ring1b co-immunoprecipitated with Kdm2b, BcoR, Rybp, Pcgf1 and Cbx7 (Fig. 5d). These results suggest that Kdm2b can form a PRC1-like subcomplex in mESCs that includes Ring1b, BcoR, Rybp and Pcgf1, which is consistent with previous findings46–48. We designate this unique Kdm2b-containing PRC1 complex as Kdm2b–PRC1. Notably, Cbx7, which is the predominant Cbx protein in mESCs that mediates PRC1 binding to H3K27me3 (ref. 49), can be pulled-down by Ring1b, but not Kdm2b, indicating that the Kdm2b–PRC1 complex is distinct from the Cbx7-containing PRC1 complex (referred to as Cbx7–PRC1). To determine why Kdm2b SF fails to rescue the Kdm2b-knockdown-induced gene expression change and phenotypes (Fig. 3a,b), we expressed the Flag–Kdm2b SF in mESCs and performed co-immunoprecipitation experiments, which demonstrated that Kdm2b SF failed to co-immunoprecipitate with Ring1b (Supplementary Fig. S7A), suggesting that the N-terminal region of Kdm2b LF is required for its interaction with PRC1. Under our experimental conditions, Ezh2 failed to co-immunoprecipitate with either Kdm2b or Ring1b (Fig. 5c,d), indicating that Ezh2 is recruited to CGIs through alternative mechanisms.
Kdm2b and PcG proteins co-occupy CGI-containing early differentiation genes in mESC
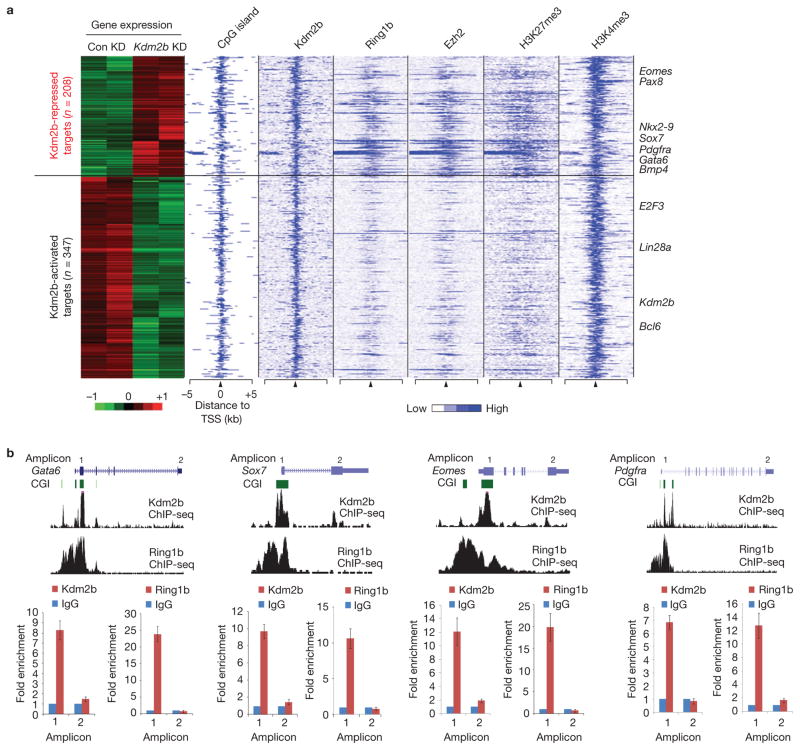
To determine whether the direct target genes of Kdm2b contribute to the Kdm2b knockdown phenotypes, we cross-examined the microarray and ChIP-seq data sets to identify Kdm2b direct targets whose expression is altered on Kdm2b depletion. This analysis identified 208 upregulated and 347 downregulated Kdm2b target genes (Supplementary Table S3). Interestingly, the upregulated gene group is enriched for Ring1b and Ezh2 binding compared with the downregulated gene group (Fig. 6a), suggesting that Kdm2b-repressed genes are preferentially targeted by PcG proteins. Although both groups of genes are associated with H3K4me3, only the upregulated genes are associated with H3K27me3, indicating that the Kdm2b-repressed genes are enriched for bivalent developmental genes (Eomes, Pdgfra, Sox7, Gata6 and so on), which is consistent with the microarray analyses demonstrating that the de-repressed genes are enriched for the GO terms of early embryonic development and cell differentiation (Fig. 2d and Supplementary Table S1).
Figure 6.
Kdm2b and Ring1b co-occupy and repress early differentiation genes. (a) Heat map representation of differentially expressed Kdm2b targets in control and Kdm2b knockdown mESCs. The associated CGIs, the binding profiles of Kdm2b, Ring1b and Ezh2, and the distribution of H3K27me3 and H3K4me3 (ref. 54) within 5 kb genomic regions flanking the TSS of each gene are also shown. Representative differentially expressed genes are listed on the right of the panel. (b) Top, ChIP-seq results of Kdm2b and Ring1b at four representative genes Gata6, Sox7, Eomes and Pdgfra. Schematic illustration of the genomic structure of the gene locus, the associated CGIs and the location of amplicons analysed by ChIP–qPCR are also shown. Bottom, the relative enrichment of Kdm2b and Ring1b occupancy at the location of the amplicons indicated. The enrichment of occupancy was measured by ChIP–qPCR. The enrichment from the IgG control is set as 1. Data are mean ± s.d., n = 3. The source data for statistics are provided in Supplementary Table S5.
To verify the ChIP-seq results, we performed ChIP–qPCR to examine the occupancy of Kdm2b and Ring1b at representative early lineage genes including Gata6, Sox7, Eomes and Pdgfra. Results shown in Fig. 6b demonstrate that both Kdm2b and Ring1b co-occupy the CGIs of these genes, but they are not present in the TESs (Fig. 6b). All of these data suggest that Kdm2b and its associated PRC1 subcomplex target to the developmental genes directly in mESCs.
Depletion of Kdm2b impairs the recruitment of Ring1b to its targets
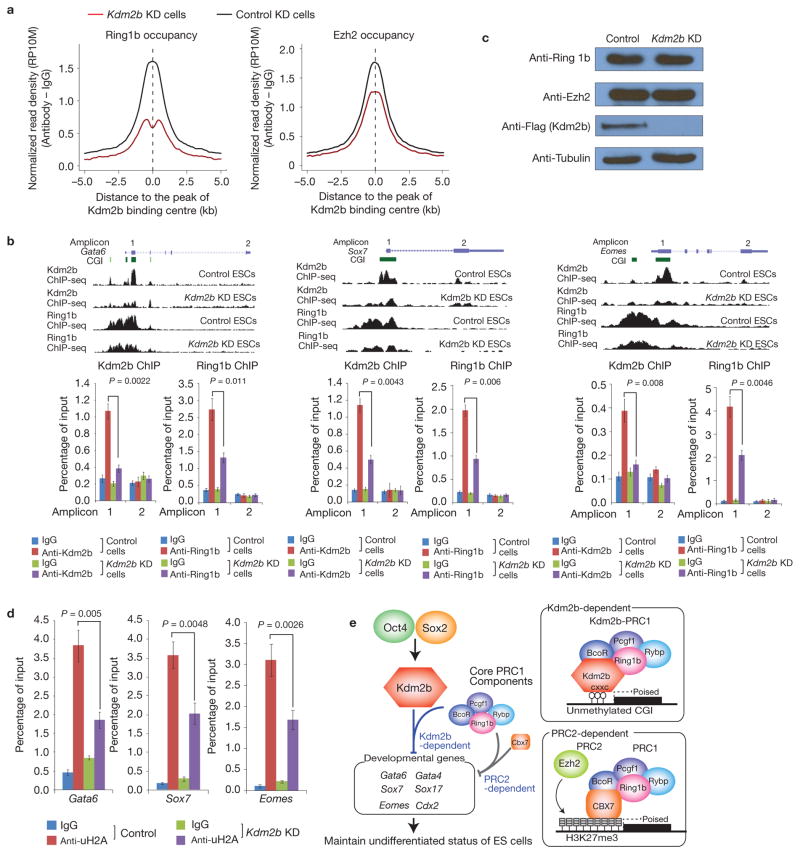
As Kdm2b can directly bind to the CGIs and also physically associate with the PRC1 core components and co-occupy CGI-containing developmental genes in mESCs, it may contribute to the recruitment of PRC1 to the CGIs of these genes. To investigate this possibility, we performed ChIP-seq analyses for Ring1b and Ezh2 under control and Kdm2b-depleted conditions and compared their genomic occupancy. We found that genome-wide Ring1b occupancy at Kdm2b-bound sites is reduced for 63% on Kdm2b knockdown (Fig. 7a). Although genomic occupancy of Ezh2 is also affected, the extent of reduction is less pronounced (22%) compared with that of Ring1b (Fig. 7a), which is consistent with the fact that Kdm2b associates with Ring1b but not Ezh2 (Fig. 5c,d). To further examine whether reduced Ring1b occupancy is caused by activated transcription, we analysed the occupancy of Ring1b and Ezh2 at the Kdm2b, PRC1 and PRC2 co-bound promoters that do not exhibit activation on Kdm2b knockdown. This analysis revealed reduced Ring1b occupancy regardless of whether the genes are activated or not (Supplementary Fig. S7B), indicating that reduced Ring1b occupancy is not a secondary effect of gene activation.
Figure 7.
Depletion of Kdm2b impairs recruitment of Ring1b to the CGIs of target genes. (a) Genome-wide occupancy of Ring1b and Ezh2 at all identified Kdm2b peaks in control (black) and Kdm2b knockdown (red) mESCs. The average signal within 5 kb genomic regions flanking the centre of Kdm2b peaks is shown. (b) Top, ChIP-seq results of Kdm2b and Ring1b at three representative genes Gata6, Sox7 and Eomes in control and Kdm2b knockdown mESCs. Schematic illustration of the genomic structure of the locus, the associated CGIs and the location of amplicons analysed by ChIP–qPCR are also shown. Bottom, the relative enrichment of Kdm2b and Ring1b occupancy at the indicated amplicons. The enrichment of occupancy was measured by ChIP–qPCR. Data are mean ± s.d., n = 3; P values (two-tailed t-test) are listed. The source data for statistics are provided in Supplementary Table S5. (c) Western blot analysis of the Ring1b, Ezh2 and Kdm2b protein levels in control and Kdm2b knockdown cells. Tubulin serves as a loading control. Full scans of blots are shown in Supplementary Fig. S8e. (d) The relative enrichment of H2A ubiquitylation levels at three representative genes (Gata6, Sox7 and Eomes) in control and Kdm2b knockdown mESCs. The enrichment was measured by ChIP–qPCR. Data are mean ± s.d., n = 3; P values (two-tailed t-test) are listed. The source data for statistics are provided in Supplementary Table S5. (e) Schematic representation of the Oct4–Sox2–Kdm2b–PRC1–CGI regulatory axis as well as the Kdm2b-dependent and the PRC2-dependent PRC1 recruitment mechanisms. The two recruitment mechanisms are likely to work in concert to silence early lineage differentiation genes in mESCs.
We further verified the ChIP-seq results by ChIP–qPCR analyses of Kdm2b and Ring1b occupancy at representative early lineage-specific genes Gata6, Sox7 and Eomes that are bound by Kdm2b and de-repressed on Kdm2b knockdown (Figs 3b and 6b). Consistent with ChIP-seq results, we found Kdm2b and Ring1b co-occupy the CGIs near the TSSs (Fig. 7b). On Kdm2b knockdown, binding of Ring1b, but not Ezh2, to the CGIs of these genes is significantly reduced (Fig. 7b and Supplementary Fig S7C). As Kdm2b knockdown does not affect Ring1b or Ezh2 levels (Fig. 7c and Supplementary Fig. S4C), the most reasonable explanation for the decreased genomic occupancy of Ring1b is that Kdm2b contributes to its recruitment. Consistent with the role of PRC1 in H2A ubiquitylation, ChIP–qPCR analyses demonstrated that the histone H2A ubiquitylation levels at its target sites are reduced by approximately 50% in the Kdm2b knockdown cells (Fig. 7d).
To examine whether genomic binding of Kdm2b depends on PRC1, we performed Kdm2b ChIP analysis in Ring1b knockdown cells (Supplementary Fig. S7D). We found that Kdm2b occupancy is not affected by Ring1b knockdown (Supplementary Fig. S7E). We thus conclude that genomic binding of Kdm2b does not require an intact PRC1 complex. Collectively, the above results demonstrate that Kdm2b is required for the recruitment of a PRC1 subcomplex to CGI-containing early lineage genes to silence their expression in mESCs.
DISCUSSION
Kdm2b interacts and recruits PRC1 to CGIs of early differentiation genes in mESCs
In this study, we provide several lines of evidence supporting a critical function of Kdm2b in the recruitment of PRC1 to the CGIs of developmental genes in mESCs. First, we demonstrate that Kdm2b preferentially binds to unmethylated CG-rich DNA through its CxxC-ZF domain (Fig. 4a–c). Second, we demonstrate that Kdm2b is highly enriched at CGIs (Fig. 4d–h), and most PRC1- and PRC2-binding sites are co-occupied by Kdm2b (Fig. 5a,b). Third, Kdm2b can stably associate with PRC1 components to form a PRC1-like complex (Fig. 5c,d). Finally, knockdown of Kdm2b impaired genome-wide as well as locus-specific binding of Ring1b to chromatin (Fig. 7a,b).
A recent study identified Kdm2b as a component of one of six PRC1 subcomplexes in HEK293T cells48. However, our study revealed that over 90% of PRC1-bound sites are co-occupied by Kdm2b in mESCs (Fig. 5a). The difference is probably due to the different cell types (mESCs versus differentiated human cells). Despite the global reduction in chromatin binding of the PRC1 components in the Kdm2b knockdown cells, we note that a certain level of Ring1b binding still remains in Kdm2b knockdown cells (Fig. 7a,b). The remaining Ring1b occupancy may be mediated by an alternative recruitment mechanism such as recognition of the H3K27me3 by the chromodomain of Cbx7 in the Cbx7–PRC1 complex. Similar to our result, a recent study demonstrated that Ring1b binding is reduced, but not completely removed in Eed-null mESCs (ref. 25). Therefore, it is likely that both Kdm2b-mediated CGI binding and Cbx-protein-mediated H3K27me3 recognition contribute to PRC1 recruitment.
We note that, under high stringency, our ChIP-seq analyses revealed that Kdm2b occupies most CGIs, whereas only around 20% of Kdm2b-binding sites are co-bound by the PRC1 complex (Fig. 5a). However, when analysed under reduced stringency, we found that more Kdm2b-bound CGIs are co-occupied by PRC1 at low levels (data not shown), which is consistent with a recent report50. Moreover, of the Kdm2b and Ring1b co-bound genes, only a small group of genes involved in early lineage differentiation are de-repressed on Kdm2b knockdown (Figs 2c, 3b and 6a). Further analysis revealed that genes bound by Kdm2b alone have much higher expression levels than the Kdm2b/Ring1b co-bound genes in mESCs, suggesting that these genes belong to the CGI-associated constitutively expressed genes and genes of the pluripotency network51 (data not shown). As Kdm2b depletion results in a significant reduction of genomic binding of Ring1b (Fig. 7a,b), Kdm2b seems to be necessary for the recruitment of PRC1 to CGIs. On the other hand, it is likely that PRC1 occupancy may not be determined by Kdm2b alone but also by other factors such as local transcriptional activities.
On the basis of previous results and our present study, we propose that at least two mutually non-exclusive mechanisms exist for the recruitment of PRC1 to its target genes in mESCs: a PRC2-dependent recruitment mechanism in which the chromodomain of Cbx7 binds to H3K27me3 to mediate the recruitment of a Cbx7–PRC1 complex; Kdm2b-dependent recruitment in which Kdm2b mediates the recruitment of a Kdm2b–PRC1-like complex, which does not contain Cbx7, to CGIs (Fig. 7e). These two recruitment mechanisms are not mutually exclusive as the Kdm2b-repressed targets are also highly enriched for H3K27me3 (Fig. 6a) and can be targeted by Cbx7. It is likely that multiple recruitment mechanisms work in concert to achieve efficient repression of developmental genes in mESCs.
The Oct4–Sox2–Kdm2b–PRC1–CGI regulatory axis and mESCs maintenance
Previous studies have revealed that both pluripotent factors and PcG proteins are required for maintaining mESCs in an undifferentiated state. However, how pluripotent factors and PcG proteins work together to achieve this is unclear. In this study we provide evidence suggesting that Kdm2b may be the missing link between pluripotent factors and PcG proteins. We first demonstrate that Kdm2b is highly expressed in mESCs and its expression level is greatly reduced on differentiation (Fig. 1a–c). We also demonstrate that Kdm2b is a direct target of the pluripotent factors Oct4 and Sox2 (Fig. 1d–f) and depletion of Oct4 greatly reduced the Kdm2b level in mESCs (Fig. 1g,h). Given that both the pluripotent factors and Kdm2b are required for maintaining mESCs in an undifferentiated state (Figs 2 and 3), the function of Oct4 and Sox2 in mESC maintenance is at least partly mediated by Kdm2b activation. In addition, Kdm2b and Ring1b not only co-occupy a set of CGI-containing differentiation genes (Fig. 6a,b), depletion of Kdm2b and Ring1b in mESCs also exhibited a set of similar phenotypes (Fig. 2)11,15. These data support that Kdm2b and Ring1b are functionally linked. Collectively, our study uncovers a hierarchic regulatory axis including Oct4 and Sox2, Kdm2b, PRC1 and CGIs that plays an important role in silencing lineage-specific genes and maintaining the undifferentiated state of mESCs (Fig. 7e).
Our study not only establishes a critical function for Kdm2b in recruiting PRC1, but also reveals a regulatory axis important in maintaining the undifferentiated state of mESCs. As the embryonic pluripotent factors Oct4 and Sox2 are also found to be expressed in various adult tissue stem cells and cancer stem cells52,53, it will be interesting to investigate whether the same Oct4–Sox2–Kdm2b–PRC1–CGIs regulatory axis exists and functions in tissue stem cells and cancer development.
METHODS
Vector construction and lentivirus production
Stable knockdown was achieved using a lentiviral system obtained from the National Institutes of Health AIDS Research and Reference Reagent Program. The shRNA targeting Kdm2b (5′-GCTCCAACTCAGTTACTGT-3′), Ring1b (5′-GCACTCATCAGCAAGATTTAT-3′) and Oct4 (5′-GCACGAGTGGAAAGCAACT-3′) and the control shRNA (5′-GTTCAGATGTGCGGCGAGT-3′) were cloned into the BbsI/HindIII sites under the U6 promoter. To generate Kdm2b expression vectors, the wild-type and mutated Kdm2b complementary DNAs were PCR amplified, and cloned into the SpeI/EcoRI sites under the EF1α promoter. To generate lentiviral viruses, the transducing vectors pTY, pHP and pHEF1α–VSVG were co-transfected into HEK293T cells30. The supernatant was collected at 24, 36 and 48 h after transfection, filtered through a 0.45 μm membrane and concentrated using a spin column (EMD Millipore).
Cell culture and lentiviral transduction
E14 mESCs were cultured on 0.1%-gelatin-coated plates in DMEM medium (Sigma) supplemented with 15% fetal bovine serum (Sigma), 1 × penicillin/streptomycin, 1× non-essential amino acid, 1 × sodium pyruvate, 1 × GlutaMax, 1 × beta-mercaptoethanol (Invitrogen) and 1000 units ml−1 LIF (ESGRO, EMD Millipore). Around 1× 105 ES cells per well in 24-well plates were infected with lentivirus at a multiplicity of infection (MOI) of 10. At 48 h after transduction, puromycin (2 μg ml−1) was added to the medium to select transduced cells.
Proliferation and colony-formation assays
For proliferation assays, 1 × 105 control, Kdm2b knockdown and Kdm2b rescued mESCs were cultured on 0.1%-gelatin-coated plates and the cell numbers were counted every two days. A growth curve was generated by calculating the average cell numbers of three independent experiments at each time point. For colony-formation assays, around 500 control, Kdm2b knockdown and Kdm2b rescued single mESCs were seeded on gelatin-coated plates. The numbers of colonies were counted 5 days later.
Embryoid body formation assay
mESCs were trypsinized and suspended as single cells. After removal of the medium, the cells were washed 3 times to remove the LIF in the medium. The cells were plated onto 6-well low-attachment plates (Corning Life Sciences) at 1× 105 cells per well in ESC medium without LIF.
Western blot analysis
Total proteins were separated by electrophoresis using 6–8% PAGE gel. The proteins were transferred on nitrocellulose membrane and western blotting was performed using antibodies against Flag (F1804, clone M2, Sigma, 1:5,000), Flag (F7425, Sigma, 1:2,000), Gata6 (sc-9055, Santa Cruz Biotechnology, 1:500), Eomes (4540, Cell Signaling Technology, 1:1,000), BcoR (AP-7359C, Abgent, 1:1,000), Cbx7 (07981, EMD Millipore, 1:1,000), Rybp (ab5976, Abcam, 1:1,000), Pcgf1 (sc-109392, Santa Cruz Biotechnology, 1:1,000), Ezh2 (5246, Cell Signaling Technology, 1:1,000) and Ring1b (5694, Cell Signaling Technology, 1:1,000).
Cellular immunofluorescence microscopy
Cells were fixed in 4% paraformaldehyde for 15 min at room temperature and washed with PBS 3 times. The fixed cells were permeabilized with 0.5% Triton X-100/PBS for 30 min, washed with PBS and blocked with 3% BSA/PBS for 30 min. The Cells were incubated with primary antibodies in a humidified chamber overnight. The primary antibodies used in the study include: anti-Oct4 (ab19857, Abcam), anti-Sox2 (ab5603, EMD Millipore), anti-Nanog (ab80892, Abcam) and anti-Flag (F1804, clone M2, Sigma). After washing 3 times with PBS, cells were incubated with Rhodamine-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories). The cells were washed three times with PBS, stained with 4′,6-diamidino-2-phenylindole dihydrochloride and mounted on glass slides in fluorescent mounting medium (DAKO). The slides were viewed and photos were taken on an Axio Observer Z1 microscope (Carl Zeiss).
Protein expression and purification
Wild-type and CxxC-ZF-deleted Kdm2b cDNAs were cloned into pFASTBAC as described previously30. Each baculovirus expressing recombinant proteins was generated and amplified following the manufacturer’s protocol (Invitrogen). To purify the recombinant proteins, infected insect SF9 cells were collected and resuspended in F lysis buffer (20 mM Tris–HCl, at pH 7.9, 500 mM NaCl, 4 mM MgCl2, 0.4 mM EDTA, 2 mM dithiothreitol, 20% glycerol and 0.1% NP40) with proteinase inhibitors. Cells were homogenized with pestle A. The supernatant was recovered by centrifuging and was adjusted to 300 mM NaCl by adding dilution buffer (20 mM Tris–HCl, at pH 7.9), and subsequently incubated with M2 agarose (Sigma) for 4 h at 4 °C. After washing with F washing buffer (20 mM Tris–HCl at pH7.9, 150 mM NaCl, 2 mM MgCl2, 0.2 mM EDTA, 1 mM dithiothreitol, 15% glycerol and 0.01% NP40), the bound proteins were eluted with Flag peptide (0.2 mg ml−1).
EMSAs
A 146-base-pair (bp) DNA fragment containing 14 CpG dinucleotides was PCR amplified from the plasmid pGEM3z-601 (gift from R. Klose, University of Oxford, UK). The methylated DNA probe was generated by PCR amplification with 5-methylcytosine-containing dNTP mix (Zymo Research). The DNA fragments were end labelled with [γ-32P]ATP (Perkin Elmer) and T4 polynucleotide kinase (New England Biolab). Labelled probes were purified by Illustra MicroSpin G-25 columns (GE Healthcare Life Sciences). EMSA reactions were carried out by mixing the proteins in binding buffer (4% Ficoll-400, 20 mM HEPES at pH 7.9, 150 mM KCl, 1 mM EDTA, 0.5 mM dithiothreitol and 25 ng μl−1 poly-dAdT competitor DNA) for 10 min at room temperature. The radiolabelled probe was then added to the reaction and incubated for 20 min before being resolved in 0.8% agarose gel in 0.5× TBE.
Immunoprecipitation and co-immunoprecipitation
Nuclear extracts from the Kdm2b Flag-tagged mESCs were treated with 15U DNase I (Roche) at 4 °C for 1 h and incubated with M2 agarose beads (A2220, Sigma), Ring1b antibody (D139-3, clone 3-3, MBL International Cooperation) or control mouse IgG in BC150 binding buffer (50 mM Tris–HCl at pH 7.9, KCl 150 mM and 10% glycerol) for 4 h or overnight. For Ring1b and control IgG immunoprecipitation, protein-G-conjugated agarose beads (EMD Millipore) were added and incubated for another 2 h. The beads were washed 3 times with BC150 washing buffer (50 mM Tris–HCl at pH 7.9, KCl 150 mM and 10% glycerol). The bound proteins were dissolved in gel loading buffer.
Microarray and data analysis
Total RNA was extracted from the control and Kdm2b knockdown mESCs. The RNA processing and hybridization to the MoGene-1_0 gene expression array chip (Affymetrix) were carried out at the Functional Genomics Core Facility at UNC-CH. Hybridization for each sample was performed in two biological replicates. Microarray data were analysed using GeneSpring Software Version 11.5.1 (Agilent Technologies).
RT–qPCR and ChIP–qPCR assays
RNA was extracted and purified from cells using Qiashredder (Qiagen) and RNeasy (Qiagen) spin columns. Total RNA (1 μg) was subjected to reverse transcription using random primers (Promega) and Superscript II reverse transcriptase (Invitrogen). cDNA levels were assayed by real-time PCR using FAST SYBR Green PCR master mix (Applied Biosystems) and analysed on the ViiA 7 Real-Time PCR System with ViiA 7 Software v1.2. The expression of individual genes was normalized to the level of Gapdh. ChIP assays using Flag antibody (F7425, Sigma), anti-H3K36me2 (ab9049, Abcam), Ring1b antibody (5694, Cell Signaling Technology) and Ezh2 (5246, Cell Signaling Technology) were carried out as reported previously30. ChIPed DNA was analysed by qPCR and data are presented as the percentage of input using Applied Biosystems ViiA 7 Software v1.2. Primers for qPCR and ChIP assays are listed in Supplementary Table S4.
ChIP-seq sample preparation
For Kdm2b ChIP, Kdm2b Flag-tag knock-in mESCs were fixed with 2 mM ethylene glycol bis(succinimidylsuccinate) (Thermo Scientific) for 1 h, followed by 10 min in 1% formaldehyde and 5 min in 0.125 M glycine to sequence the reaction. Cells were lysed in 1% SDS, 10 mM EDTA and 50 mM Tris–HCl (pH 8.0) and the DNA was fragmented to approximately 200–400 bp by sonification (Branson Sonifier 450). Immunoprecipitation was performed with 5 μg rabbit polyclonal anti-Flag (F7425, Sigma), 5 μl rabbit polyclonal Ring1b antibody (5694, Cell Signaling Technology) and 5 μl rabbit polyclonal Ezh2 antibody (5246, Cell Signaling Technology) overnight at 4 °C. Antibody-bound protein was isolated by protein G plus/protein A agarose beads (EMD Millipore), washed, eluted and reverse cross-linked. DNA was extracted by phenol/chloroform and precipitated.
ChIP DNA preparation for HiSeq2000 sequencing
The ChIP DNA library for HiSeq2000 sequencing was constructed using the NEBNext DNA library Prep Master Mix Set for Illumina (New England BioLabs) according to the manufacturer’s instruction. Briefly, the ChIP DNA was blunt ended and a dA tail was added. The DNA with dA overhangs was ligated with home-made adaptors with individual index sequences. Adaptor-ligated DNA was amplified by PCR for 18 cycles, followed by size selection using agarose gel electrophoresis. The DNA was purified using the Qiaquick gel extraction kit (Qiagen) and quantified both with an Agilent Bioanalyzer and an Invitrogen Qubit. The DNA was diluted to 10 nM before sequencing. Sequencing on a HiSeq2000 instrument was carried out by the High Throughput Sequencing facility at UNC-CH.
ChIP-seq data analysis
ChIP-seq data analysis was performed essentially as described previously55. All sequencing reads (42 bp in length) were mapped to NCBI build 36 (mm8) of the mouse genome using the software Bowtie56. Mapped reads were subjected to the MACS program57 and bound regions (peaks) were determined using sequencing reads from IgG control experiments as negative controls. When multiple reads mapped to the same genomic position, a maximum of two reads were retained. The statistical cutoff used for peak calling was P <10−8 and >5-fold enrichment over the IgG control. ChIP-seq data sets of H3K4me3 and H3K27me3 were obtained from a previous publication54 and the DIP-seq data set of 5mC was obtained from a previous publication and reanalysed with Bowtie and MACS using identical parameters. The lengths of the CGIs were obtained from the UCSC Table Browser58. ChIP-seq read counts for each ChIP-seq experiment were binned into 50-bp windows along the genome and visualized in the UCSC genome browser59. To assign ChIP-seq enriched regions to genes, a complete set of Refseq genes was downloaded from the UCSC Table Browser58. For all data sets, genes with enriched regions within 5 kb of their TSSs were called bound. The ChIP-seq density profiles were normalized to the density per 10 million total reads with a resolution of 100 bp. Heat maps were generated using Java Treeview (http://jtreeview.sourceforge.net/).
Supplementary Material
Acknowledgments
We thank R. Klose for the plasmid pGEM3z-601. This project was supported by NIH GM068804 and U01DK089565. Y.Z. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Note: Supplementary Information is available in the online version of the paper
AUTHOR CONTRIBUTIONS
J.H. and Y.Z. designed all of the experiments and wrote the manuscript. J.H. performed most of the experiments. L.S. and H.W. performed the microarray and ChIP-seq data analysis. M.W. helped in characterizing the Kdm2b complex. O.T. helped in analysing Kdm2b expression in various cells.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at www.nature.com/reprints
Accession Numbers. The complete data sets of the microarray and ChIP-seq analyses are available from the NCBI GEO repository (GSE41316).
References
- 1.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 2.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci USA. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blair K, Wray J, Smith A. The liberation of embryonic stem cells. PLoS Genet. 2011;7:e1002019. doi: 10.1371/journal.pgen.1002019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wray J, Kalkan T, Smith AG. The ground state of pluripotency. Biochem Soc Trans. 2010;38:1027–1032. doi: 10.1042/BST0381027. [DOI] [PubMed] [Google Scholar]
- 5.Boyer LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 6.Jorgensen HF, et al. Stem cells primed for action: polycomb repressive complexes restrain the expression of lineage-specific regulators in embryonic stem cells. Cell Cycle. 2006;5:1411–1414. doi: 10.4161/cc.5.13.2927. [DOI] [PubMed] [Google Scholar]
- 7.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Gen Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bilodeau S, Kagey MH, Frampton GM, Rahl PB, Young RA. SetDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes Dev. 2009;23:2484–2489. doi: 10.1101/gad.1837309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chamberlain SJ, Yee D, Magnuson T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells. 2008;26:1496–1505. doi: 10.1634/stemcells.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morin-Kensicki EM, Faust C, LaMantia C, Magnuson T. Cell and tissue requirements for the gene eed during mouse gastrulation and organogenesis. Genesis. 2001;31:142–146. doi: 10.1002/gene.10017. [DOI] [PubMed] [Google Scholar]
- 11.Leeb M, et al. Polycomb complexes act redundantly to repress genomic repeats and genes. Genes Dev. 2010;24:265–276. doi: 10.1101/gad.544410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Carroll D, et al. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol. 2001;21:4330–4336. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J, Mager J, Schnedier E, Magnuson T. The mouse PcG gene eed is required for Hox gene repression and extraembryonic development. Mamm Genome. 2002;13:493–503. doi: 10.1007/s00335-002-2182-7. [DOI] [PubMed] [Google Scholar]
- 15.Voncken JW, et al. Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proc Natl Acad Sci USA. 2003;100:2468–2473. doi: 10.1073/pnas.0434312100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L, et al. Hierarchical recruitment of polycomb group silencing complexes. Mol Cell. 2004;14:637–646. doi: 10.1016/j.molcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 17.Peng JC, et al. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139:1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen X, et al. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell. 2009;139:1303–1314. doi: 10.1016/j.cell.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanhere A, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell. 2010;38:675–688. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsai MC, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 23.Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev. 2004;14:155–164. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 24.Yu M, et al. Direct recruitment of polycomb repressive complex 1 to chromatin by core binding transcription factors. Mol Cell. 2012;45:330–343. doi: 10.1016/j.molcel.2011.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tavares L, et al. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell. 2012;148:664–678. doi: 10.1016/j.cell.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schoeftner S, et al. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J. 2006;25:3110–3122. doi: 10.1038/sj.emboj.7601187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ku M, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mendenhall EM, et al. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet. 2010;6:e1001244. doi: 10.1371/journal.pgen.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He J, Kallin EM, Tsukada Y, Zhang Y. The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b) Nat Struct Mol Biol. 2008;15:1169–1175. doi: 10.1038/nsmb.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki T, Minehata K, Akagi K, Jenkins NA, Copeland NG. Tumour suppressor gene identification using retroviral insertional mutagenesis in Blm-deficient mice. EMBO J. 2006;25:3422–3431. doi: 10.1038/sj.emboj.7601215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blackledge NP, et al. CpG islands recruit a histone H3 lysine 36 demethylase. Mol Cell. 2010;38:179–190. doi: 10.1016/j.molcel.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tzatsos A, Pfau R, Kampranis SC, Tsichlis PN. Ndy1/KDM2B immortalizes mouse embryonic fibroblasts by repressing the Ink4a/Arf locus. Proc Natl Acad Sci USA. 2009;106:2641–2646. doi: 10.1073/pnas.0813139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He J, Nguyen AT, Zhang Y. KDM2b/JHDM1b, an H3K36me2-specific demethylase, is required for initiation and maintenance of acute myeloid leukaemia. Blood. 2011;117:3869–3880. doi: 10.1182/blood-2010-10-312736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang G, He J, Zhang Y. Kdm2b promotes induced pluripotent stem cell generation by facilitating gene activation early in reprogramming. Nat Cell Biol. 2012;14:457–466. doi: 10.1038/ncb2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang T, et al. The histone demethylases Jhdm1a/1b enhance somatic cell reprogramming in a vitamin-C-dependent manner. Cell Stem Cell. 2011;9:575–587. doi: 10.1016/j.stem.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 37.Agherbi H, et al. Polycomb mediated epigenetic silencing and replication timing at the INK4a/ARF locus during senescence. PLoS One. 2009;4:e5622. doi: 10.1371/journal.pone.0005622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 39.Bracken AP, et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007;21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fukuda T, Tokunaga A, Sakamoto R, Yoshida N. Fbxl10/Kdm2b deficiency accelerates neural progenitor cell death and leads to exencephaly. Mol Cell Neurosci. 2011;46:614–624. doi: 10.1016/j.mcn.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 41.Marson A, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cierpicki T, et al. Structure of the MLL CXXC domain-DNA complex and its functional role in MLL-AF9 leukaemia. Nat Struct Mol Biol. 2010;17:62–68. doi: 10.1038/nsmb.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu C, Bian C, Lam R, Dong A, Min J. The structural basis for selective binding of non-methylated CpG islands by the CFP1 CXXC domain. Nat Commun. 2011;2:227. doi: 10.1038/ncomms1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomson JP, et al. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature. 2010;464:1082–1086. doi: 10.1038/nature08924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ficz G, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 46.Gearhart MD, Corcoran CM, Wamstad JA, Bardwell VJ. Polycomb group and SCF ubiquitin ligases are found in a novel BCOR complex that is recruited to BCL6 targets. Mol Cell Biol. 2006;26:6880–6889. doi: 10.1128/MCB.00630-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanchez C, et al. Proteomics analysis of Ring1B/Rnf2 interactors identifies a novel complex with the Fbxl10/Jhdm1B histone demethylase and the Bcl6 interacting corepressor. Mol Cell Proteomics. 2007;6:820–834. doi: 10.1074/mcp.M600275-MCP200. [DOI] [PubMed] [Google Scholar]
- 48.Gao Z, et al. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell. 2012;45:344–356. doi: 10.1016/j.molcel.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morey L, et al. Nonoverlapping functions of the Polycomb group Cbx family of proteins in embryonic stem cells. Cell Stem Cell. 2012;10:47–62. doi: 10.1016/j.stem.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 50.Farcas AM, et al. KDM2B links the Polycomb Repressive Complex 1 (PRC1) to recognition of CpG islands. eLife. 2012;1:e00205. doi: 10.7554/eLife.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fouse SD, et al. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell. 2008;2:160–169. doi: 10.1016/j.stem.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arnold K, et al. Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell. 2011;9:317–329. doi: 10.1016/j.stem.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ikushima H, et al. Glioma-initiating cells retain their tumourigenicity through integration of the Sox axis and Oct4 protein. J Biol Chem. 2011;286:41434–41441. doi: 10.1074/jbc.M111.300863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu H, et al. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011;473:389–393. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Karolchik D, et al. The UCSC Table Browser data retrieval tool. Nucl Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.