Abstract
Laribacter hongkongensis is a gram-negative, facultative anaerobic, motile, S-shaped, asaccharolytic, urease-positive bacillus in the Neisseriaceae family of β-proteobacteria. To date, all patients with L. hongkongensis infection have survived, including the two patients with L. hongkongensis bacteremia and patients with L. hongkongensis gastroenteritis. In this study, we describe the clinical, microbiological and molecular characterization of the first fatal case associated with L. hongkongensis bacteremia in a patient with colonic carcinoma that metastasized to the liver. The identity of the isolate was confirmed via phenotypic tests and 16S rRNA gene sequencing. Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI–TOF MS), using the Bruker database extended with L. hongkongensis reference strains, also identified the isolate as L. hongkongensis, with a top match score of 2.473. Multilocus sequence typing revealed a new sequence type (ST), and phylogenetic analysis and eBURST demonstrated unambiguously that the ST of the isolate was clustered with two other STs found exclusively in human patients, consistent with the theory that some clones of L. hongkongensis could be more virulent than others. Underlying liver diseases and ascites potentially represent distinct risk factors for invasive L. hongkongensis infection. More widespread use of MALDI–TOF MS for identification and improvements of selective media should facilitate the identification of more cases of L. hongkongensis infection.
Keywords: bacteraemia, infections
INTRODUCTION
Laribacter hongkongensis is a gram-negative, facultative anaerobic, motile, S-shaped, asaccharolytic, urease-positive bacillus in the Neisseriaceae family of β-proteobacteria.1,2 This species was first isolated from the blood and thoracic empyema of an alcoholic liver cirrhosis patient in Hong Kong.3 This species was also recently recovered from a blood culture from a Korean patient with liver cirrhosis caused by Wilson's disease.4 These cases suggest that chronic liver disease is a risk factor for invasive L. hongkongensis infections. In addition to invasive bacteremic infections, L. hongkongensis is also associated with community-acquired gastroenteritis and traveler's diarrhea.2 L. hongkongensis is likely to be globally distributed, as travel histories from patients suggest that this bacterium is present on at least four continents: Asia, Europe, Africa and Central America.2,5,6,7 L. hongkongensis has been found in up to 60% of intestines from commonly consumed freshwater fish of the carp family.2,8 This species has also been isolated from drinking water reservoirs, Chinese tiger frogs and little egrets.9,10,11 Pulsed-field gel electrophoresis and multilocus sequence typing (MLST) demonstrated that the fish and patient isolates fall into separate clusters, suggesting that some clones could be more virulent or adapted to humans.8,12 The complete genome of L. hongkongensis was recently sequenced, and this bacterium's mechanism of acid resistance was characterized.1,13
To date, all patients with L. hongkongensis infection have survived, including the two patients with L. hongkongensis bacteremia and the patients with L. hongkongensis gastroenteritis. In this article, we report the first fatal case associated with L. hongkongensis bacteremia. In addition to phenotypic tests and 16S rRNA gene sequencing, the isolate was also characterized using matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI–TOF MS) and MLST. The antibiotic resistance mechanisms of this isolate were also determined via sequencing of the ampC and tetA genes.
MATERIALS AND METHODS
Patient and microbiological methods
All clinical data were collected prospectively, as described in our previous publication.14 The BacT/ALERT 3D blood culture system (Biomerieux, Marcy L'Etoile, Frace) was used. Phenotypic identification was performed using standard conventional biochemical methods.12 All tests were performed in triplicate with freshly prepared media on separate occasions. Antimicrobial susceptibility was tested using the Kirby Bauer disk diffusion method, and the results were interpreted according to the Clinical and Laboratory Standards Institute interpretive criteria for Enterobacteriaceae.
16S rRNA gene sequencing
16S rRNA gene sequencing was performed as described previously.7,10 To extract bacterial DNA, 80 µL of 0.05 M NaOH were added to 20 µL of bacterial cells suspended in distilled water, and the mixture was incubated at 60 °C for 45 min, followed by the addition of 6 µL of Tris-HCl (pH 7.0), achieving a final pH of 8.0. The resultant mixture was diluted ×100, and 5 µL of the diluted extract was used for polymerase chain reaction (PCR).
For PCR amplification, DNase I-treated distilled water and PCR master mix, which contains deoxynucleoside triphosphates, PCR buffer and Taq polymerase, were used in all PCR reactions by adding 1 U of DNase I (Pharmacia, Uppsala, Sweden) to 40 µL of distilled water or PCR master mix, incubating the mixture at 25 °C for 15 min and subsequently incubating the mixture at 95 °C for 10 min to inactivate the DNase I. The bacterial DNA extract and the control were amplified using 0.5 µM primers (LPW57 (5′-AGT TTG ATC CTG GCT CAG-3′) and LPW58 (5′-AGG CCC GGG AAC GTA TTC AC-3′)) (Sigma-Proligo, Singapore). The PCR mixture (50 µL) contained bacterial DNA, PCR buffer (10 mM Tris-HCl pH 8.3, 50 mM KCl, 2 mM MgCl2 and 0.01% gelatin), 200 µM of each deoxynucleoside triphosphate and 1.0 U of Taq polymerase (Boehringer Mannheim, Mannheim, Germany). The mixtures were amplified in an automated thermal cycler (Perkin-Elmer Cetus, Gouda, The Netherlands) via 40 cycles of 94 °C for 1 min, 55 °C for 1 min and 72 °C for 2 min and a final extension at 72 °C for 10 min. DNase I-treated distilled water was used as the negative control. A total of 10 µL of each amplified product was electrophoresed in a 1.0% (w/v) agarose gel, with the Lambda DNA AvaII digest molecular size marker (Boehringer Mannheim) electrophoresed in parallel. Electrophoresis was performed in Tris-borate-EDTA buffer at 100 V for 1.5 h. The gel was stained with 0.5 µg/mL of ethidium bromide for 15 min, rinsed and photographed under ultraviolet light illumination.
The PCR product was gel-purified using the QIAquick PCR purification kit (Qiagen, Hilden, Germany). Both strands of the PCR product were sequenced twice with an ABI 377 automated sequencer, according to the manufacturers' instructions (Perkin-Elmer, Foster City, CA, USA), using the PCR primers LPW57 and LPW58.
Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS)
MALDI-TOF MS was performed as described in our previous publication, with 21 L. hongkongensis strains added as reference strains.15 The bacterial isolate was grown on sheep blood agar at 35°C for 18–24 h and analyzed using the direct transfer method. The sample was processed in a Bruker Daltonik MALDI–TOF MS spectrometer with 1 µL of Bruker α-cyano matrix solution. The spectrum was obtained with an accelerating voltage of 20 kV in linear mode and analyzed within an m/z range 2000–20 000 Da. The spectrum was analyzed using MALDI Biotyper 3.0 and Reference Library v3.1.2.0 software (Bruker Daltonik, Bremen, Germany).
ampC and tetA gene sequencing
The ampC genes of the isolate were amplified and sequenced using primers LPW663 (5′-CGG ATT ACC CCA TTT TCC CG-3′) and LPW664 (5′-CAT GTC TGC CCG GAA TGT GC-3′), and the tetA genes of the isolate were amplified and sequenced using primers LPW1015 (5′-ATG TCC ACC AAC TTA TCA-3′) and LPW1697 (5′-TCA GCG ATC GGC TCG TTG-3′). The conditions used were those that were described in our previous publications.14,16
Multi-locus sequence typing (MLST)
MLST was performed according to the protocol described in our previous publication.12 Seven housekeeping genes (i.e., rho, acnB, ftsH, trpE, ilvC, thiC and eno) were amplified and sequenced using previously described primers.12 The results were analyzed with MLST data from other L. hongkongensis isolates from humans and fish that were described in our previous publication.12 The Sequence Type Analysis and Recombinational Tests version 2 program was used for Unweighted Pair Group Method with Arithmetic Mean dendrogram construction, using the nucleotide sequences of the seven gene loci. In addition, the eBURST program was used for lineage analysis.17,18
RESULTS
Patient
A 74-year-old Chinese man was admitted to the hospital due to nonspecific chest discomfort and dizziness. The patient had hypertension, diabetes mellitus and chronic renal failure and a history of ischemic heart disease with percutaneous intervention to the left anterior descending artery eight years prior to admission and right middle cerebral artery infarct with a recurrent cerebrovascular accident one year prior to admission. On admission, the patient's temperature was 37.9 °C. The patient also exhibited pallor and ascites. The patient developed right upper quadrant abdominal pain and septic shock, with a blood pressure of 80/40 mmHg and a heart rate of 130 beats/min, two days after admission. The total white cell count was 16.3×109/L, with a neutrophil count of 15.2×109/L, a lymphocyte count of 0.2×109/L and a monocyte count of 1.0×109/L. The hemoglobin level was 6.7 g/dL, and the platelet count 328×109/L. The serum urea level was 29.3 mmol/L, the creatinine level was 447 µmol/L, the albumin level was 16 g/L, the globulin level was 39 g/L, the total bilirubin level was 27 µmol/L, the alkaline phosphatase level was 165 IU/L, the alanine aminotransferase level was 8 IU/L and the aspartate aminotransferase level was 30 IU/L. Blood culture was performed, and empirical intravenous meropenem at a dose of 500 mg daily was initiated. On day 2 post-incubation, the aerobic blood culture bottle turned positive for a gram-negative bacillus. Ultrasound examination of the abdomen revealed gross ascites, two isoechoic masses of 4.2 cm×2.5 cm×2.5 cm and 8.0 cm×3.4 cm×6.1 cm in the peritoneal cavity anterior to the right hepatic lobe, and multiple vague hypoechoic masses of up to 3.7 cm in the liver. Serum tests for hepatitis B surface antigen and anti-hepatitis C virus antibody were negative. The serum α-fetoprotein level was <1 µg/L, and the carcinoembryonic antigen level was 1841 µg/L. A probable diagnosis of metastatic carcinoma of the colon was made, but the patient refused further investigation. The patient succumbed on day 10 after admission.
Phenotypic characteristics
The blood culture isolate was a gram-negative, S-shaped and motile bacillus. The isolate grew on horse blood agar as non-hemolytic, gray colonies with a 1 mm diameter after 24 h of incubation at 37 °C in ambient air. The isolate also grew on MacConkey agar and in an anaerobic environment. The bacillus produced catalase, cytochrome oxidase, urease and arginine dihydrolase and reduced nitrate. However, the isolate failed to ferment, oxidize or assimilate any sugar tested. The isolate was susceptible to gentamicin, amikacin, levofloxacin, cefuroxime, amoxicillin-clavulanate and imipenem but resistant to ampicillin, cefalothin, cefotaxime, ceftazidime, ceftriaxone, ceftibuten and tetracycline.
16S rRNA gene sequencing
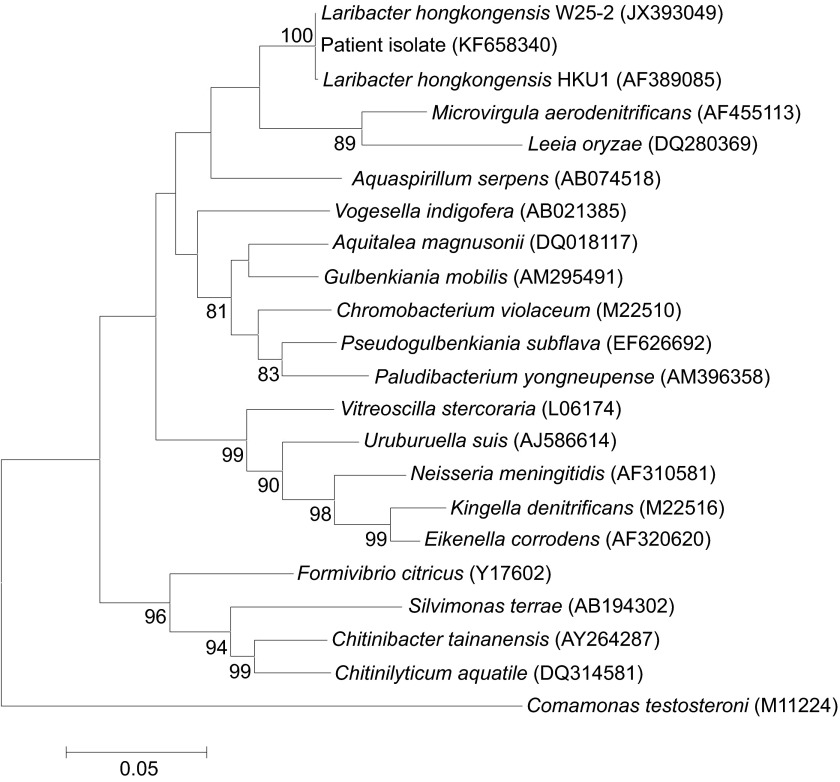
PCR amplification of the 16S rRNA gene of the isolate revealed a band with a size of approximately 1300 bp. Sequencing and comparative analysis revealed no differences between the 16S rRNA gene sequence of the blood culture isolate and that of L. hongkongensis (GenBank accession NO JX393049), and a single base differed between the 16S rRNA gene sequence of the blood culture isolate and that of L. hongkongensis HKU1T (GenBank accession NO AF389085). These findings indicated that the isolate was a strain of L. hongkongensis (Figure 1).
Figure 1.
Phylogenetic tree showing the relationship of the patient isolate to other related members of the β-subclass of Proteobacteria, inferred from 16S rRNA gene sequences. The tree was constructed using the neighbor-joining method and rooted using Comamonas testosteroni (M11224). The bootstrap values calculated from 1000 trees are shown when they are ≥70%. The scale bar indicates the estimated number of substitutions per 20 bases. Names and accession numbers are presented as cited in the GenBank database.
Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS)
Using the Bruker database extended with L. hongkongensis reference strains, the isolate was identified as L. hongkongensis, with a top match score of 2.473.
ampC and tetA gene sequencing
PCR amplification of the ampC and tetA genes of the isolate revealed bands of approximately 1350 bp and 1230 bp, respectively. Sequencing and comparative analysis demonstrated that the ampC gene of the isolate possessed 100% nucleotide and amino-acid identities to the ampC gene of L. hongkongensis strain HLHK-6 and that the tetA gene of the isolate possessed >99% nucleotide and amino-acid identities to the tetA genes of L. hongkongensis strains HLHK5 and HLHK22, as well as those of other bacteria, such as Salmonella enterica subsp. Enterica. These results indicated that the mechanisms of β-lactam and tetracycline resistance in the isolate were identical to those that we previously reported (Figure 2).14,16
Figure 2.
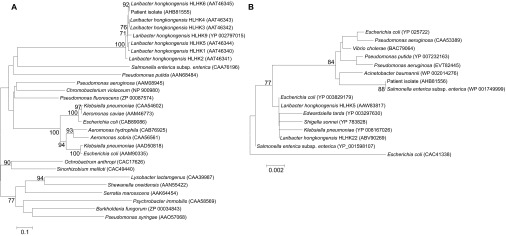
Phylogenetic tree showing the relationships of the amino acid sequences of the (A) AmpC and (B) TetA proteins from the patient isolate with other related species. The tree was constructed using the neighbor-joining method with Jukes–Cantor correction. The bootstrap values calculated from 1000 trees are shown when they are ≥70%. The scale bars indicate the estimated number of substitutions per 10 and 500 amino acids. Names and accession numbers are presented as cited in the GenBank database.
Multi-locus sequence typing (MLST)
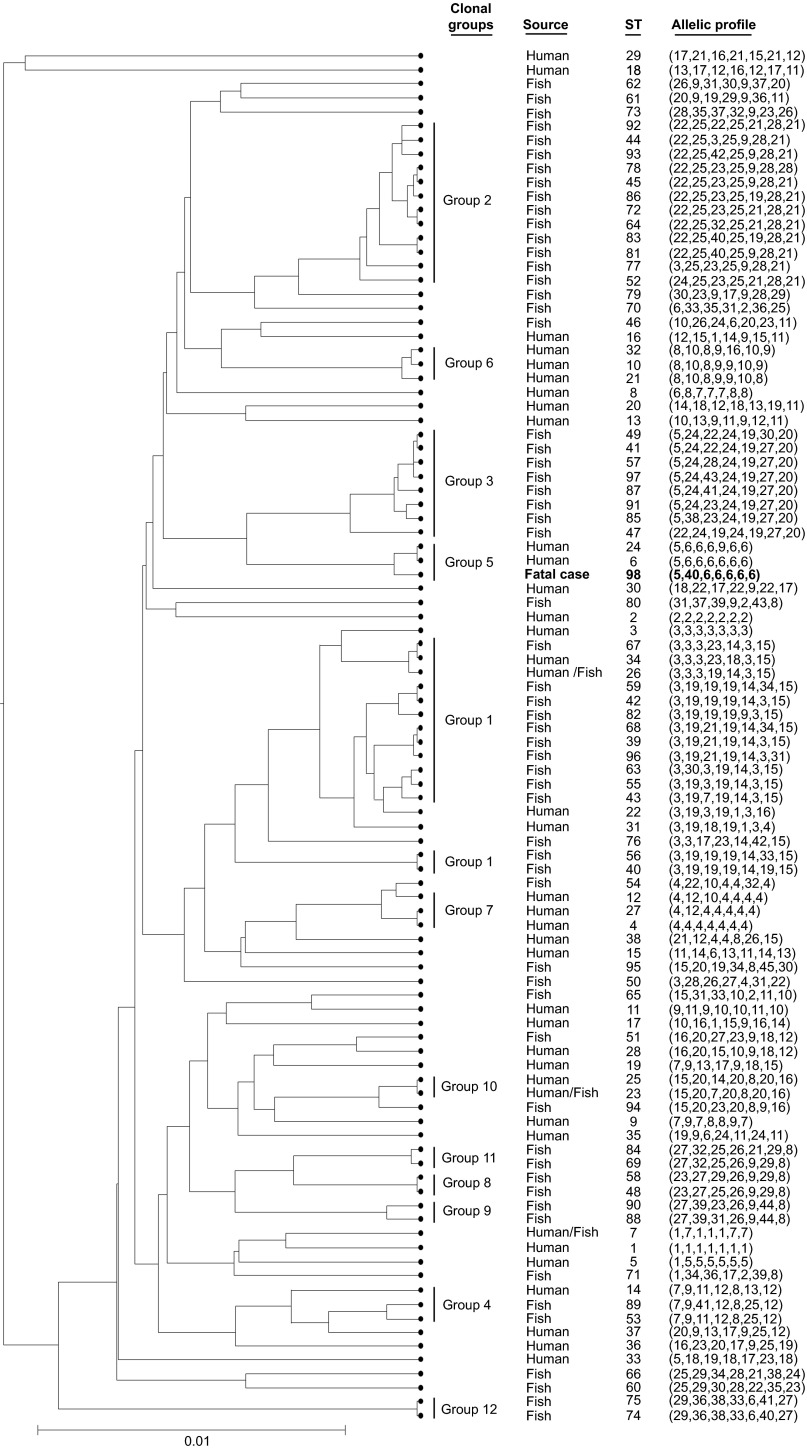
Sequence analysis demonstrated that the sequence of the isolate constituted a new sequence type (ST): ST-98 (5,40,6,6,6,6,6). Phylogenetic analysis demonstrated that the isolate was clustered with ST-6 (5,6,6,6,6,6,6) and ST-24 (5,6,6,6,9,6,6), forming clonal group 5 (Figure 3). eBURST also grouped ST-6, ST-24 and ST-98 into a single lineage, consistent with results of the phylogenetic analysis (Figure 4).
Figure 3.
UPGMA-generated dendrogram showing the relationships of the strain isolated from the fatal case with other L. hongkongensis strains isolated from humans and fish.12 The ST of the isolate from the fatal case is displayed in bold. The genetic relatedness among the STs was generated using START2 software (http://pubmlst.org/software/analysis/start2/).18 The clonal group displayed was assigned according to the eBURST clonal clusters to which the STs belong. START2, Sequence Type Analysis and Recombinational Tests version 2; UPGMA, Unweighted Pair Group Method with Arithmetic Mean.
Figure 4.
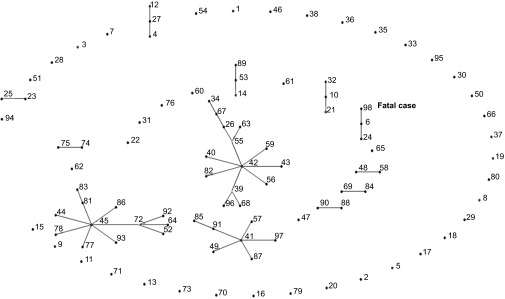
BURST analysis of the strain isolated from the fatal case in comparison to other L. hongkongensis strains isolated from humans and fish.12 The relationships between STs and clonal complexes are displayed. Each number represents an ST, and each line connects STs that are identical for at least six of the seven MLST loci. Boxed numbers represent STs that are found only in humans. The isolate from the fatal case belongs to ST 98 and is displayed in bold. The clonal cluster was generated using eBURST software version 3 (http://eburst.mlst.net/).
Nucleotide sequence accession number
The 16S rRNA, ampC and tetA gene sequences of the L. hongkongensis strain isolated in this study were deposited in the GenBank sequence database under accession numbers KF658340, KF658341 and KF658342, respectively.
DISCUSSION
In this study, we report the first case of fatal infection associated with L. hongkongensis bacteremia. To date, the most serious infection associated with L. hongkongensis was the first case of thoracic empyema and bacteremia.3 In this case, the patient required hospitalization for more than one month, with prolonged intravenous antibiotic treatment and chest drainage.3 With respect to the cases of community-acquired gastroenteritis and traveler's diarrhea associated with L. hongkongensis, approximately 20% and 80% of patients developed bloody and watery diarrhea, respectively, and systemic upset and vomiting occurred in approximately 30% of patients.2 In the most severe cases, patients can have diarrhea up to 30 times per day or the diarrhea can linger for up to three months.2 MLST, phylogenetic analysis and eBURST demonstrated unambiguously that the ST of the isolate in the present study was clustered with two other STs that were found exclusively in human patients. This result is consistent with the findings of our previous studies, which demonstrated clustering of isolates from humans and fish into different groups, suggesting that some clones of L. hongkongensis could be more virulent than others.8,12
Underlying liver diseases and ascites potentially represent distinct risk factors for invasive L. hongkongensis infection. Both patients with reported L. hongkongensis bacteremia had underlying liver cirrhosis and ascites due to alcoholism and Wilson's disease.3,4 The patient in the present study also had liver metastasis and ascites. Similar to the other reported cases of invasive and non-invasive L. hongkongensis infection, the gastrointestinal tract was likely the portal of entry in the present patient. Although a complete and functional urease cassette is present in the genome of L. hongkongensis, we recently found that the urease gene is of limited importance for the survival of L. hongkongensis in the acidic environment of the stomach.13 In contrast, the bacterium's arc gene cassettes, which each encode three enzymes and a membrane bound arginine-ornithine antiporter of the arginine deiminase pathway, are far more important than urease for acid resistance in L. hongkongensis, possibly due to the duplication of the arc gene cassette in the L. hongkongensis genome.13 We speculate that after surviving the acidic environment of the stomach and reaching the intestine, intestinal mucosal edema and local immunosuppression secondary to portal venous congestion vasculopathy caused by liver disease predisposed the patients to L. hongkongensis invasion through the gastrointestinal mucosa, resulting in patients with L. hongkongensis bacteremia. In conclusion, both microbial factors, as demonstrated by molecular typing, and patient factors, as demonstrated by the patients' underlying disease, are likely to make important contributions to the development of L. hongkongensis bacteremia.
More widespread use of MALDI–TOF MS for identification and improvements in selective media should help identify more cases of L. hongkongensis infection. In all previous cases of human L. hongkongensis infection and in reports of the detection of this species in animal specimens, the strains were identified using a combination of phenotypic tests and 16S rRNA gene sequencing.2,7,8,9,10,11,19,20,21 In recent years, MALDI–TOF MS has emerged as a revolutionary technique for rapid bacterial identification at a low cost. Using an expanded Bruker database with L. hongkongensis reference strains, we recently found that all 240 L. hongkongensis isolates could be correctly identified using MALDI–TOF MS, demonstrating the importance of expanding the MALDI–TOF MS database with bacteria endemic to different localities.15 This finding is also consistent with the correct identification of the present isolate by MALDI–TOF MS. With respect to the selective medium, it was recently observed that the L. hongkongensis strain recovered from the blood culture of the Korean patient was cefoperazone-susceptible.4 Further, a recent study from Guangzhou reported that 45.2% of the L. hongkongensis strains recovered from fish and frogs were susceptible to cefoperazone.21 These findings imply that a significant number of L. hongkongensis cases may have been missed in studies that used cefoperazone MacConkey agar as the selective medium and that the incidence of L. hongkongensis infection could have been underestimated.2,20 With bacterial identification becoming easier and more rapid via MALDI–TOF MS, the use of a less selective medium, such as MacConkey agar, in combination with picking more colonies for rapid identification via MALDI–TOF MS, could represent a strategy for diagnosing more cases of L. hongkongensis infection of the gastrointestinal tract.
Acknowledgments
We are grateful to Ms Eunice Lam for her generous donation to emerging infectious disease and microbial genetics research.
References
- Woo PC, Lau SK, Tse H, et al. The complete genome and proteome of Laribacter hongkongensis reveal potential mechanisms for adaptations to different temperatures and habitats. PLoS Genet. 2009;5:e1000416. doi: 10.1371/journal.pgen.1000416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo PC, Lau SK, Teng JL, et al. Association of Laribacter hongkongensis in community-acquired gastroenteritis with travel and eating fish: a multicentre case–control study. Lancet. 2004;363:1941–1947. doi: 10.1016/S0140-6736(04)16407-6. [DOI] [PubMed] [Google Scholar]
- Yuen KY, Woo PC, Teng JL, Leung KW, Wong MK, Lau SK. Laribacter hongkongensis gen. nov., sp. nov., a novel Gram-negative bacterium isolated from a cirrhotic patient with bacteremia and empyema. J Clin Microbiol. 2001;39:4227–4232. doi: 10.1128/JCM.39.12.4227-4232.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DS, Wi YM, Choi JY, Peck KR, Song JH, Ko KS. Bacteremia caused by Laribacter hongkongensis misidentified as Acinetobacter lwoffii: report of the first case in Korea. J Korean Med Sci. 2011;26:679–681. doi: 10.3346/jkms.2011.26.5.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verboeket SO, van den Berk GE, Arends JE, van Dam AP, Peringa J, Jansen RR. Hookworm with hypereosinophilia: atypical presentation of a typical disease. J Travel Med. 2013;20:265–267. doi: 10.1111/jtm.12042. [DOI] [PubMed] [Google Scholar]
- Ni XP, Ren SH, Sun JR, et al. Laribacter hongkongensis isolated from a patient with community-acquired gastroenteritis in Hangzhou City. J Clin Microbiol. 2007;45:255–256. doi: 10.1128/JCM.01400-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo PC, Kuhnert P, Burnens AP, et al. Laribacter hongkongensis: a potential cause of infectious diarrhea. Diagn Microbiol Infect Dis. 2003;47:551–556. doi: 10.1016/s0732-8893(03)00161-5. [DOI] [PubMed] [Google Scholar]
- Teng JL, Woo PC, Ma SS, et al. Ecoepidemiology of Laribacter hongkongensis, a novel bacterium associated with gastroenteritis. J Clin Microbiol. 2005;43:919–922. doi: 10.1128/JCM.43.2.919-922.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni X, Sun J, Kong Q, et al. Isolation of Laribacter hongkongensis from Little Egrets (Egretta garzetta) in Hangzhou, China. Lett Appl Microbiol. 2011;52:465–467. doi: 10.1111/j.1472-765X.2011.03024.x. [DOI] [PubMed] [Google Scholar]
- Lau SK, Woo PC, Fan RY, et al. Isolation of Laribacter hongkongensis, a novel bacterium associated with gastroenteritis, from drinking water reservoirs in Hong Kong. J Appl Microbiol. 2007;103:507–515. doi: 10.1111/j.1365-2672.2006.03263.x. [DOI] [PubMed] [Google Scholar]
- Lau SK, Lee LC, Fan RY, et al. Isolation of Laribacter hongkongensis, a novel bacterium associated with gastroenteritis, from Chinese tiger frog. Int J Food Microbiol. 2009;129:78–82. doi: 10.1016/j.ijfoodmicro.2008.10.021. [DOI] [PubMed] [Google Scholar]
- Woo PC, Teng JL, Tsang AK, et al. Development of a multi-locus sequence typing scheme for Laribacter hongkongensis, a novel bacterium associated with freshwater fish-borne gastroenteritis and traveler's diarrhea. BMC Microbiol. 2009;9:21. doi: 10.1186/1471-2180-9-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Teng JL, Watt RM, Kan B, Lau SK, Woo PC. Arginine deiminase pathway is far more important than urease for acid resistance and intracellular survival in Laribacter hongkongensis: a possible result of arc gene cassette duplication. BMC Microbiol. 2014;14:42. doi: 10.1186/1471-2180-14-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau SK, Wong GK, Li MW, Woo PC, Yuen KY. Distribution and molecular characterization of tetracycline resistance in Laribacter hongkongensis. J Antimicrob Chemother. 2008;61:488–497. doi: 10.1093/jac/dkm539. [DOI] [PubMed] [Google Scholar]
- Tang BS, Lau SK, Teng JL, et al. Matrix-assisted laser desorption ionisation–time of flight mass spectrometry for rapid identification of Laribacter hongkongensis. J Clin Pathol. 2013;66:1081–1083. doi: 10.1136/jclinpath-2013-201651. [DOI] [PubMed] [Google Scholar]
- Lau SK, Ho PL, Li MW, et al. Cloning and characterization of a chromosomal class C beta-lactamase and its regulatory gene in Laribacter hongkongensis. Antimicrob Agents Chemother. 2005;49:1957–1964. doi: 10.1128/AAC.49.5.1957-1964.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol. 2004;186:1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolley KA, Feil EJ, Chan MS, Maiden MC. Sequence type analysis and recombinational tests (START) Bioinformatics. 2001;17:1230–1231. doi: 10.1093/bioinformatics/17.12.1230. [DOI] [PubMed] [Google Scholar]
- Lau SK, Woo PC, Fan RY, Lee RC, Teng JL, Yuen KY. Seasonal and tissue distribution of Laribacter hongkongensis, a novel bacterium associated with gastroenteritis, in retail freshwater fish in Hong Kong. Int J Food Microbiol. 2007;113:62–66. doi: 10.1016/j.ijfoodmicro.2006.07.017. [DOI] [PubMed] [Google Scholar]
- Lau SK, Woo PC, Hui WT, et al. Use of cefoperazone MacConkey agar for selective isolation of Laribacter hongkongensis. J Clin Microbiol. 2003;41:4839–4841. doi: 10.1128/JCM.41.10.4839-4841.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng JL, Hu J, Lin JY, et al. The prevalence, antimicrobial resistance and PFGE profiles of Laribacter hongkongensis in retail freshwater fish and edible frogs of southern China. Food Microbiol. 2012;32:118–123. doi: 10.1016/j.fm.2012.04.018. [DOI] [PubMed] [Google Scholar]