Significance
Hemophilia A, a genetic bleeding disorder, is often caused by chromosomal inversions that involve a portion of the blood coagulation factor VIII (F8) gene that encodes one of the key enzymes in blood clotting. In this study, we developed enzymes known as transcription activator-like effector nucleases (TALENs) that cleave chromosomal DNA in a targeted manner to invert the 140-kbp chromosomal segment that spans the portion of the F8 gene in human induced pluripotent stem cells (iPSCs) to create a hemophilia A model cell line. In addition, we reverted the inverted segment back to its normal orientation using the same enzymes. This strategy provides an iPSC-based novel therapeutic option for the treatment of hemophilia A.
Keywords: genome editing, CRISPR, Cas9, ZFN
Abstract
Hemophilia A, one of the most common genetic bleeding disorders, is caused by various mutations in the blood coagulation factor VIII (F8) gene. Among the genotypes that result in hemophilia A, two different types of chromosomal inversions that involve a portion of the F8 gene are most frequent, accounting for almost half of all severe hemophilia A cases. In this study, we used a transcription activator-like effector nuclease (TALEN) pair to invert a 140-kbp chromosomal segment that spans the portion of the F8 gene in human induced pluripotent stem cells (iPSCs) to create a hemophilia A model cell line. In addition, we reverted the inverted segment back to its normal orientation in the hemophilia model iPSCs using the same TALEN pair. Importantly, we detected the F8 mRNA in cells derived from the reverted iPSCs lines, but not in those derived from the clones with the inverted segment. Thus, we showed that TALENs can be used both for creating disease models associated with chromosomal rearrangements in iPSCs and for correcting genetic defects caused by chromosomal inversions. This strategy provides an iPSC-based novel therapeutic option for the treatment of hemophilia A and other genetic diseases caused by chromosomal inversions.
Hemophilia A is one of the most common genetic bleeding disorders, with an incidence of 1 in 5,000 males worldwide (1). This disorder is caused by various genetic mutations, which include large deletions, insertions, inversions, and point mutations, in the X-linked coagulation factor VIII (F8) gene (Haemophilia A Mutation, Structure, Test and Resource Site; http://hadb.org.uk); a total of 1,492 different mutations are known to cause hemophilia A. Clinical symptoms vary widely according to the genotypes (2). Hemophilia A can be characterized as severe (<1% activity), moderate (1–5% activity), or mild (5–30% activity), depending on the relative amount of F8 activity in the patient’s plasma (1). Approximately 50% of severe hemophilia A cases are caused by two different types of chromosomal inversions that involve a part of the F8 gene (3–5).
Currently, there is no cure for hemophilia A. Recombinant F8 protein has been used for the treatment of this condition, but is limited by the formation of F8-inactivating antibodies, high cost, and the requirement for frequent injections. Gene therapy is a promising option for the cure of hemophilia. Remarkably, Nathwani et al. used an adeno-associated virus vector (AAV) to deliver the F9 cDNA, which encodes blood coagulation factor IX, to six patients with hemophilia B, a less common form of X-linked bleeding disorder (6). Unfortunately, however, this vector cannot be used to deliver the full-length F8 cDNA to patients with hemophilia A because AAV cannot accommodate the large size of the F8 cDNA (∼8 kbp). In contrast, the F9 cDNA is much smaller (∼1.4 kbp). Besides, gene therapy is ideally used to correct genetic defects rather than to deliver a functional gene that is not under endogenous regulatory control.
Patient-derived induced pluripotent stem cells (iPSCs) provide another promising option for the cure of hemophilia. Patient-derived iPSCs per se, however, cannot be used in cell therapy because they contain the original genetic defect. Importantly, the defective gene can be corrected in iPSCs by using programmable nucleases, which include zinc finger nucleases (ZFNs) (7–10), transcription activator-like effector nucleases (TALENs) (11–13), and clusters of regularly interspaced palindromic repeats (CRISPR)/Cas-derived RNA-guided endonucleases (RGENs; or engineered nucleases) (14–21). These programmable nucleases cleave chromosomal DNA in a targeted manner, producing DNA double-strand breaks (DSBs), whose repair via endogenous mechanisms, known as homologous recombination (HR) or nonhomologous end-joining (NHEJ), gives rise to targeted mutagenesis and chromosomal rearrangements such as deletions (22, 23), duplications, and inversions (24). Gene-corrected iPSCs are then differentiated into appropriate somatic cells before delivery to patients to ensure the expression of the corrected gene and to prevent teratoma formation in patients.
In this study, we show that TALENs can be used to invert the 140-kbp chromosomal segment in human iPSCs to create hemophilia A model cell lines that recapitulate one of the most frequent genotypes of hemophilia A and to flip-flop the inverted region back to the wild-type state. Importantly, the F8 mRNA is expressed in cells differentiated from reverted—i.e., genome-corrected—iPSCs but not in cells differentiated from the hemophilia model iPSCs. To the best of our knowledge, this report is the first demonstration that engineered nucleases can be used to rearrange large genomic segments in iPSCs and to isolate clones harboring such genomic rearrangements, providing a proof-of-principle for correcting genetic defects caused by genome rearrangements in iPSCs.
Results
Generation and Characterization of Human iPSCs.
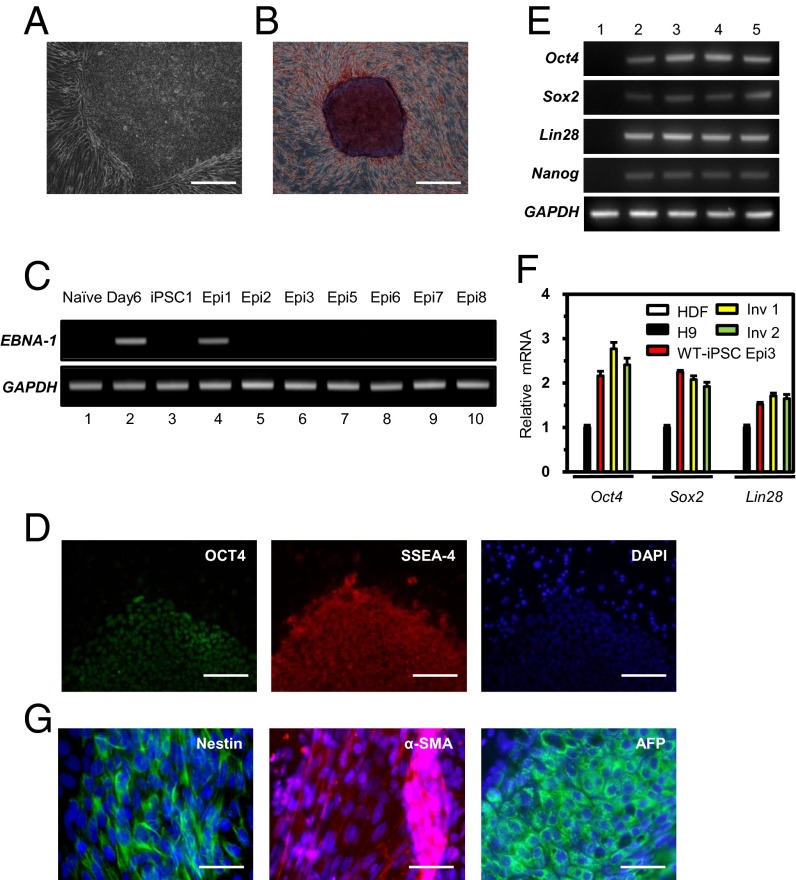
We derived wild-type iPSCs from human dermal fibroblasts (HDFs) using episomal vectors that encode the four Yamanaka factors, which we introduced into cells by electroporation. Embryonic stem cell (ESC)-like colonies appeared 10 d after replating of transfected cells onto a feeder cell layer. We selected a total of eight colonies (termed Epi1–Epi8) exhibiting alkaline phosphatase activities (Fig. 1 A and B). To confirm the absence of episomal vectors in these clones after seven or eight passages, we used PCR with specific primers for the EBNA-1 sequence, which is encoded in the vectors. Only one clone (Epi1) contained the EBNA-1 sequence; this clone was excluded from further analyses (Fig. 1C). Next, we checked the karyotypes of two iPSC lines (Epi3 and Epi8). As shown in Fig. S1A, they had a normal karyotype. We also confirmed that these iPSC lines were derived from parental HDFs using DNA fingerprinting analysis (Table S1). After these initial characterizations, we chose the Epi3 line for further experiments. This iPSC line expressed the typical ESC marker proteins such as OCT4, NANOG, SSEA-4, and TRA-1-60 (Fig. 1D and Fig. S1B). RT-PCR and quantitative PCR (qPCR) analyses showed that pluripotent marker genes were expressed at higher levels in this iPSC line than in the human ESC line H9 (Fig. 1 E and F).
Fig. 1.
Generation of iPSC clones from HDFs using episomal reprogramming vectors. (A) Morphology of the expanded human iPSCs (clone Epi3). (Scale bar, 200 μm.) (B) Alkaline phosphatase staining of iPSCs (clone Epi3). (Scale bar, 500 μm.) (C) Detection of an episomal vector sequence (EBNA-1) that remained in established iPSC lines (Epi1–Epi8). The GAPDH gene was used as a quality control for isolated total DNA. Total DNA isolated from the cells before (naïve) and after (day 6) electroporation was used as negative and positive controls for episomal vector DNA. A retrovirus-derived wild-type iPSC line (iPSC1) was also analyzed as a negative control. (D) The expression of OCT4 and SSEA-4, which are human ESC-specific markers, was detected by immunocytochemistry. DAPI signals indicate the total cell presence in the image. (Scale bars, 100 μm.) (E) RT-PCR analysis to determine the transcriptional levels of OCT4, SOX2, LIN28, NANOG, and GAPDH using gene-specific primers (listed in Table S3). mRNA levels were measured in HDFs, human ES line (H9), a wild-type iPSC line (WT-iPSCEpi3), and inversion clones (Inv 1 and Inv 2) derived from the WT-iPSCEpi3 line (1, HDFs; 2, H9; 3, WT-iPSC; 4, Inv 1; 5, Inv 2). (F) Quantification of OCT4, SOX2, and LIN28 mRNAs in the indicated cell lines as determined by qPCR and normalized to GAPDH expression. (G) Expression of marker proteins representing ectoderm (Nestin), mesoderm (α-smooth muscle actin; α-SMA), and endoderm (α-fetoprotein; AFP). (Scale bars, 50 μm.)
Next, we determined the differentiation potential of the Epi3 iPSC line. Embryonic bodies were derived and attached to gelatin-coated culture plates for spontaneous differentiation into three germ layers in vitro. As expected, marker proteins for ectoderm (Nestin and Pax6), mesoderm [α-smooth muscle actin (α-SMA) and Brachyury], and endoderm [α-fetoprotein (AFP) and hepatocyte nuclear factor 3-β (HNF3β)] lineages were expressed in the differentiated cells (Fig. 1G and Fig. S1C). These data indicate that the Epi3 line derived from adult HDFs is pluripotent.
Targeted Inversion of the F8 Locus in iPSCs Using a TALEN Pair.
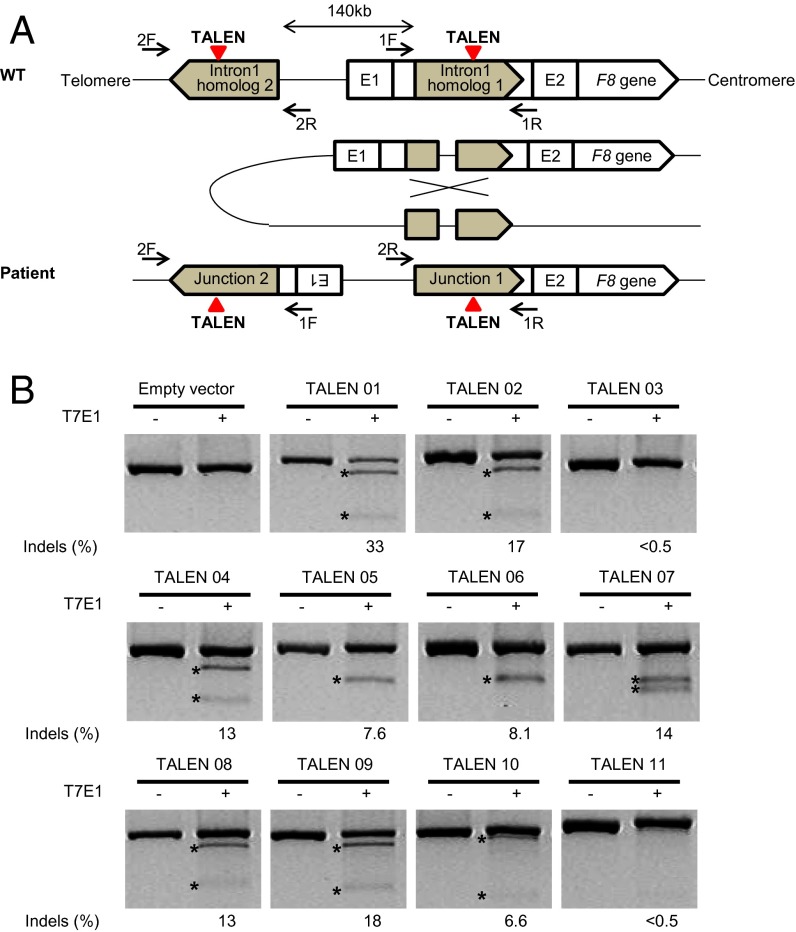
Structural variations (SVs) such as inversions are associated with genetic diseases including hemophilia A (25). Almost half of all severe hemophilia A cases are caused by two different types of inversions that disrupt the integrity of the X-linked F8 gene. These inversions result from nonallelic HR (NAHR) that involves sequences present in intron 1 (1–4% of severe hemophilia A cases) or intron 22 (up to 50% of severe hemophilia A cases) and their corresponding homologous sequences located far upstream of the F8 gene (referred to as the intron 1 or 22 inversion, respectively) (3, 4). In this study, we focused on the intron 1 inversion and constructed 11 pairs of TALENs that target the intron 1 homolog (Fig. 2A). The genome-editing activities of these TALENs were tested in HEK 293T cells by using T7 endonuclease I (T7E1) assays (10) (Fig. 2B). We chose the most active TALEN pair (termed TALEN 01) that induced mutations with a frequency of 33% at the target site. Importantly, this TALEN induced the 140-kb inversion that involves the intron 1 homolog in HEK 293T cells at a frequency of 1.9% (Fig. S2). Next, we tested whether this TALEN had off-target effects at highly homologous sites. No off-target mutations were detected at these sites by using T7E1 assays (Fig. S3 and Table S2).
Fig. 2.
TALEN-mediated inversion of the F8 gene in HEK 293T cells. (A) Proposed mechanism of a chromosomal inversion found in patients with severe hemophilia A. Inversions of 140-kbp chromosomal segments spanning the F8 gene are associated with two homologous regions oriented in opposite directions: homolog 1 located in intron1 of the F8 gene and homolog 2 located in the 140-kbp upstream region. Colored triangles show TALEN target sites, and arrows indicate the primers designed to detect 140-kbp inversions. (B) T7E1 assay results of the 11 TALEN pairs we designed. The predicted positions of DNA bands cleaved by T7E1 are indicated by asterisks.
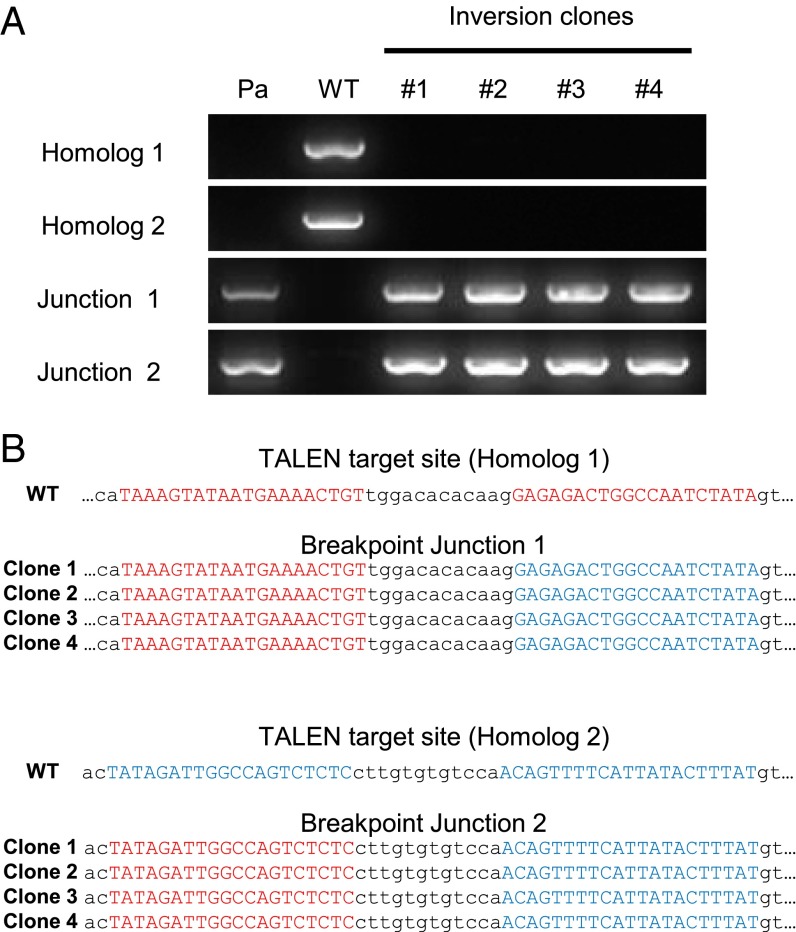
We then used the same TALEN pair to induce the 140-kb inversion in iPSCs and to create a hemophilia model cell line. Wild-type iPSCs were electroporated with the TALEN plasmids and cultured for 10 d to form colonies. Genomic DNA samples isolated from each colony were subjected to PCR by using specific primer sets that detect the inversion event. Six colonies of 432 (1.4%, comparable to that in HEK 293 cells) showed positive PCR bands for the two inversion breakpoint junctions. Four colonies were then further cultured to derive single cell clones. These clones produced PCR bands that are diagnostic of the 140-kb inversion but, importantly, did not produce PCR bands that correspond to the wild-type genotype (Fig. 3A). Next, we cloned these PCR products and determined their DNA sequences to confirm the inversion genotype. No indels were found at the TALEN target sites (Fig. 3B). This result suggests that a single DSB that was induced by the TALEN in either the intron 1 homolog 1 or homolog 2 triggered DNA inversion via error-free NAHR. However, we cannot rule out the possibility that the TALEN produced two concurrent DSBs—one in the intron homolog 1 and the other in homolog 2—and that these DSBs were joined seamlessly by NHEJ without leaving secondary mutations.
Fig. 3.
TALEN-mediated inversion of the F8 locus in iPSCs. (A) PCR analysis of genomic DNA from four inversion clones. Genomic DNA samples isolated from hemophilia A patient cells (Pa) or wild-type iPSCs (WT) served as positive controls for the inversion or normal genotypes, respectively. (B) DNA sequences of breakpoint junctions in inversion clones. TALEN binding sites are shown in red (homolog 1) or blue (homolog 2).
Targeted Reversion of the Inverted Segment in the iPSC System.
In our previous report, we induced the targeted chromosomal inversion that involves the intron 1 homolog in the HEK 293 cell line using a ZFN pair and isolated heterozygous clones that harbor the inversion (24). However, HEK 293 cells do not express the F8 gene and cannot be used in cell therapy. Furthermore, HEK 293 cells carry three copies of the X chromosome. These limitations hampered our efforts to revert the inverted region back to the normal orientation to restore expression of the F8 gene, a demonstration required for therapeutic applications.
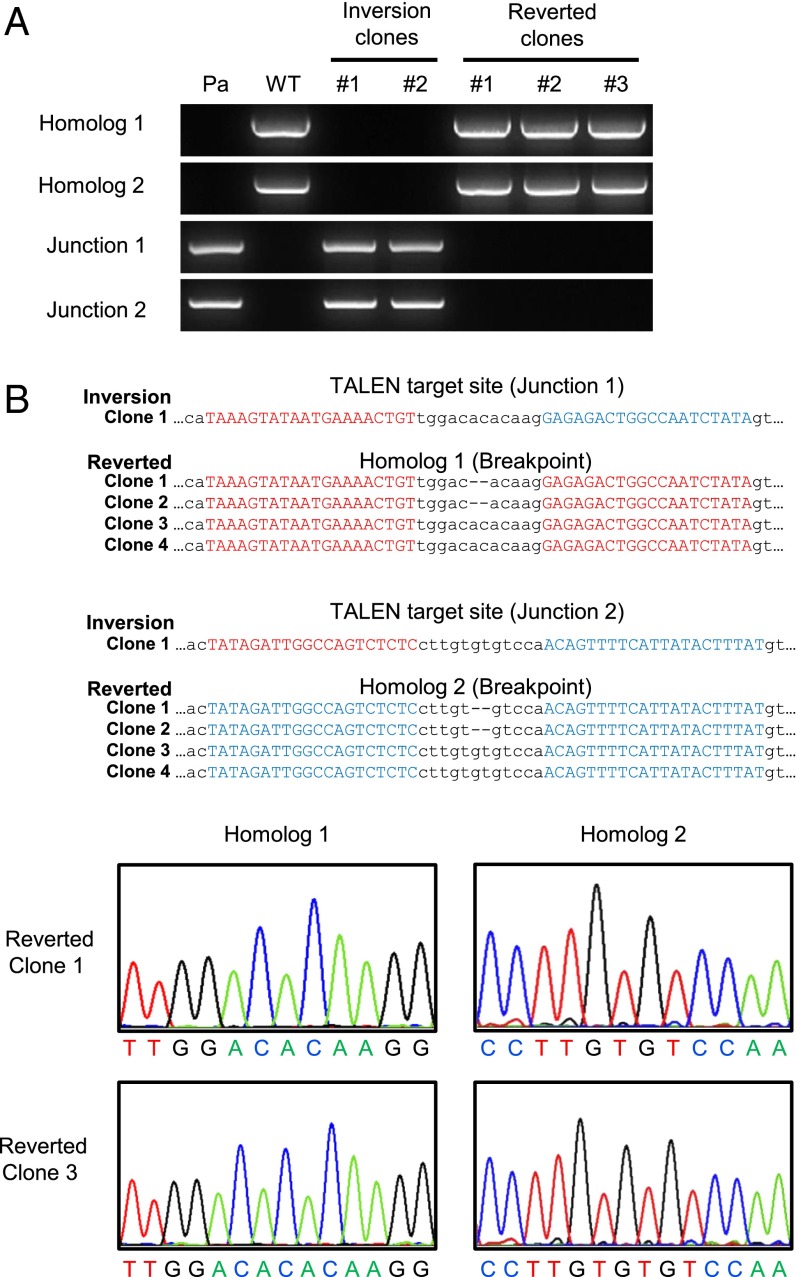
In this study, we investigated whether the inverted 140-kbp segment in the hemophilia model iPSC line could be corrected by reversion using the same TALEN pair. (Note that the TALEN site remains intact in the model cell line.) The TALEN plasmids were transfected into two iPSC clones containing the inversion (referred to here as “inversion clones”), and then genomic DNA samples isolated from several colonies were subjected to PCR to identify reverted cells. We obtained two reverted clones from each of the iPSC clones after screening a total of 300 colonies. Thus, the reversion frequency was 1.3% (4 of 300), on par with the inversion frequency. PCR analysis revealed that the genotype of these reverted clones was consistent with a reversion to wild type: No inversion-specific PCR bands were detected in the samples from these clones (Fig. 4A). We then cloned and sequenced these PCR products containing homolog 1 or 2. Two clones had no additional mutations, but the other two clones had 2-bp deletions at the two TALEN sites in both homologs 1 and 2 (Fig. 4B). These results show that the inversion genotype found in severe hemophilia A can be corrected by using the same TALEN pair that was used to generate the disease model.
Fig. 4.
Reversions of the F8 gene inversion. (A) PCR analysis is of genomic DNA from three reverted clones. Genomic DNAs isolated from hemophilia A patient cells (Pa) or wild-type iPSCs (WT) served as positive controls for inversion or normal genotypes, respectively. (B) DNA sequences of breakpoint junctions in reverted clones. TALEN binding sites are shown in red (junction 1) or blue (junction 2). Dashes indicate deleted bases. The chromatograms show the sequences (of homolog 1 and 2, respectively) between two TALEN binding sites in reverted clones (clones 1 and 3).
In addition, we investigated whether both the inversion clones and reverted clones remained pluripotent by checking their expression of human ES marker genes and their ability to undergo differentiation into the three primary germ layers. These clones expressed stem cell marker genes at levels comparable with those in wild-type iPSCs (Fig. 1F and Fig. S4) and differentiated into three germ layers in vitro (Fig. S5). These results show that TALEN-mediated genome engineering does not negatively affect iPSC pluripotency.
F8 Gene Expression in Cells Differentiated from Reverted iPSCs.
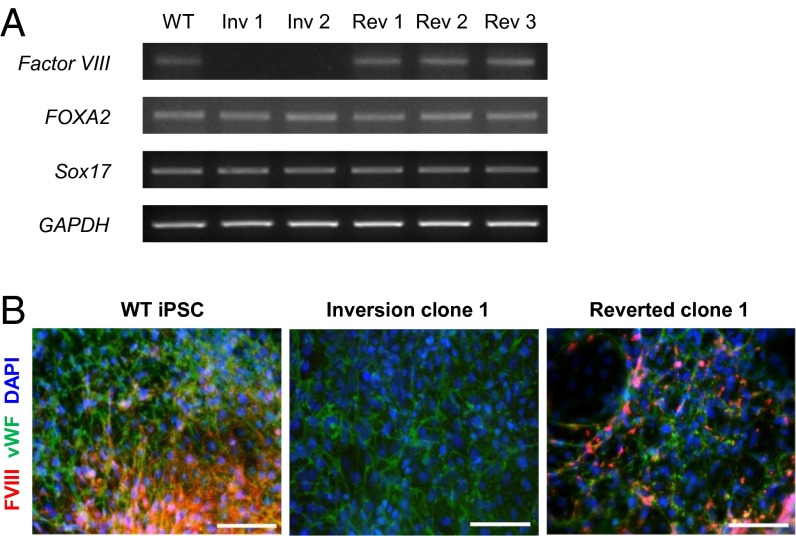
The F8 gene is expressed in hepatocytes and endothelial cells (26–29), which are derived from endoderm and mesoderm, respectively. First, we examined whether the F8 gene could be expressed in endodermal cells derived from the wild-type and reverted iPSC clones. We differentiated iPSCs into endoderm and performed an RT-PCR analysis to detect the F8 mRNA. As expected, the F8 mRNA was detected in cells differentiated from the wild-type and reverted iPSC clones (Fig. 5A). By contrast, no F8 mRNA was detected in cells derived from the iPSCs with the inversion, although these cells could differentiate into endoderm as efficiently as wild-type and reverted iPSCs. Next, we examined the expression of the F8 protein in endothelial cells, which are the main source of production of the F8 protein (28). We differentiated iPSCs into endothelial cells and performed immunostaining to detect the F8 protein. As expected, the cells differentiated from wild-type and reverted iPSC clones expressed the F8 protein (Fig. 5B). However, the F8 protein was not detected in the cells differentiated from the inversion clone, although this iPSC clone differentiated successfully into endothelial cells as shown by the expression of von Willebrand factor, a mature endothelial cell marker protein. These results prove that the integrity of the F8 gene is restored in reverted iPSCs, which supports expression of the F8 gene in endodermal cells and mesoderm-derived endothelial cells.
Fig. 5.
Characterization of inverted and reverted clones. (A) F8 gene expression in cells derived from inverted and reverted clones. RT-PCR was used to detect expression of F8 and endoderm marker genes (FOXA2 and Sox17) in cells derived from wild-type iPSCs (WT), inversion clones (Inv 1 and 2), and reverted clones (Rev 1, 2, and 3). GAPDH served as a loading control. (B) Expression of the F8 protein in endothelial cells differentiated from inverted and reverted clones. The differentiated cells were fixed and stained with the indicated antibodies. DAPI signals indicate the total cell presence in the image. FVIII, F8 protein; vWF, von Willebrand factor (a mature endothelial marker protein). (Scale bars, 100 μm.)
Discussion
Over the past decade, next-generation sequencing technologies have revealed numerous SVs or copy number variations in individual human genomes (30, 31). SVs include deletions, duplications, and inversions that involve chromosomal segments whose sizes range from hundreds to millions of base pairs. These variations are associated with diverse phenotypes such as disease susceptibility. To study the biological consequences of individual SVs of interest in cultured cells such as iPSCs or model organisms, one must be able to construct genome-modified clones that harbor such variations and compare them with isogenic wild-type controls.
Three different molecular mechanisms have been postulated to explain the etiology of these variations: NAHR, NHEJ, and DNA replication errors (32). Among these, NAHR and NHEJ are triggered by DNA DSBs, which are produced randomly in the genome by various environmental, chemical, and biological stresses. DSBs can also be produced by programmable nucleases in a targeted manner. Indeed, we and others have shown that programmable nucleases can be used to create SVs in human cancer cell lines and animals (22, 24, 33, 34). In this study, we extended this approach further by inverting the 140-kbp chromosomal segment that includes the upstream sequence in the F8 gene in human iPSCs to create model cell lines that recapitulate one of the most frequent genotypes of hemophilia A and, then, reverting the segment back to the normal state to demonstrate a proof-of-principle for gene/cell therapy.
Three different programmable nucleases are now available to create SVs: ZFNs, TALENs, and Cas9 RGENs (35). ZFNs and TALENs share the same nuclease domain—which is derived from FokI, a type IIS restriction endonuclease—but differ in their DNA-binding domains: ZFNs contain zinc finger proteins, and TALENs contain TAL effector arrays derived from Xanthomonas, a plant pathogen. Unlike these FokI-based nucleases, RGENs consist of Cas9 protein and guide RNA, and their DNA cleavage specificities are governed by Watson–Crick base pairing of guide RNA with target DNA sequences. We have developed all of these nucleases for the last several years (10, 12, 17, 36–39) and found that functional ZFNs were the most difficult to make and were often associated with cytotoxicity (10). Still, we were able to use a ZFN pair to invert the 140-kbp segment that contains the promoter and upstream sequence in the F8 gene in HEK 293 cells (24). However, the efficiency of this inversion, even in the highly transfectable HEK 293 cell line, was very low (0.1%). We isolated 3 inversion clones among 3,000 single cells using this ZFN. To improve the frequency of inversion, we tested 11 TALENs in this study that are designed to target the F8 intron homolog 1 and chose the best-performing TALEN. This TALEN inverted the 140-kbp chromosomal segment at a frequency of 1.9%. This high efficiency allowed us to isolate iPSC clones with the inversion genotype and those with the restored genotype. The use of surrogate reporters (40–42) may facilitate the isolation of clones that harbor chromosomal inversions further. Importantly, we showed that F8 was expressed in cells derived from reverted iPSCs, but not in cells derived from inverted iPSCs.
We do not know whether the chromosomal flip-flop was mediated by NAHR or NHEJ. Because we used a TALEN pair to target the homolog sequence, it is equally possible that two concurrent DSBs were produced, which were then repaired by NHEJ, or that a single DSB triggered DNA repair via NAHR. DSB repair via NHEJ often gives rise to indels at the breakpoint junction. We did find two-base deletions at the TALEN target site in some reverted clones. These deletions can be produced during inversion or after inversion. Thus, these deletions do not rule out the possibility that chromosomal inversions were mediated by NAHR. Importantly, these small deletions provide strong evidence that reverted clones were generated by TALEN-mediated chromosomal flip-flop from the inversion clone rather than derived from rare contaminating wild-type cells. To the best of our knowledge, this is the first report that chromosomal segments can be inverted and then reverted in cells or organisms.
In summary, we demonstrated that TALENs can be used to induce targeted inversions in human iPSCs that recapitulate one of the most frequent genotypes responsible for severe hemophilia A. Furthermore, we restored the inverted 140-kbp DNA segment back to the normal orientation using the same TALEN, which resulted in expression of the F8 gene in cells differentiated from genome-corrected iPSCs. This approach can be used for autologous stem cell therapy to treat hemophilia A patients harboring chromosomal inversions. Genomic inversions are also associated with cancer (43) and genetic diseases such as Hunter syndrome (44). Programmable nucleases can be used to correct these genetic defects in patient-derived cells. In addition, we expect that engineered nucleases will be widely used to create SVs in iPSCs to study the resulting phenotypes.
Materials and Methods
Plasmids Encoding TALENs.
TALEN plasmids in this study were synthesized by using TAL effector array plasmids constructed for one-step Golden-Gate assembly as described (12). Each TALEN plasmid encodes the N-terminal 135 amino acids of AvrBs3, an array of RVD modules, one of the four RVD half-repeats, and the Sharkey FokI domain (45). TALEN sites were designed to target the intron 1 homolog of the F8 gene; potential off-target sites were identified as described (12).
Isolation of Genomic DNA from Hemophilia A Patient.
Seoul National University Institutional Review Board approval was obtained for the analysis of blood cells of a hemophilia A patient. The blood sample was provided by Korea Hemophilia Foundation Clinic, and genomic DNA was isolated as described (24).
Measuring the Frequencies of Targeted Inversions.
The frequencies of targeted inversions were estimated by digital PCR analysis as described (23). The genomic DNA samples isolated from cell transfected with TALEN plasmids were serially diluted, and the diluted samples were subjected to nested PCR by using appropriate primers (Table S3). The fraction of positive bands at each dilution point was counted, and the results were analyzed by using the Extreme Limiting Dilution Analysis program (46).
Cell Cultures.
HEK 293T/17 (ATCC; CRL-11268) and adult HDFs (Invitrogen; C-004-5C) were cultured in DMEM supplemented with FBS (10% vol/vol) and antibiotics (1%). Human ESC (hESC) lines (H9) obtained from WiCell, retrovirus-derived wild-type iPSCs (iPSC1), and iPSCs generated in this study were maintained in hESC medium composed of DMEM/F12 medium supplemented with 20% (vol/vol) knockout serum replacement (Invitrogen), 4.5 g/L l-glutamine, 1% nonessential amino acids, 0.1 mM 2-mercaptoethanol, and 4 ng/mL basic FGF (PeproTech) as described (47, 48).
Validation of TALENs Targeting the F8 Locus in HEK 293T Cells.
To validate the genome-editing activities of the TALENs designed for this study, each TALEN pair was transfected into HEK 293T cells, and their activities were measured by using the T7E1 assay (10). To measure the frequency of targeted inversions induced by TALENs targeting the F8 locus, HEK 293T/17 cells were seeded at 80% confluency before transfection and transfected by using Lipofectamine 2000 (Invitrogen) with TALEN-encoding plasmids. Genomic DNA samples were isolated and subjected to PCR analysis to confirm chromosomal inversion as described (23).
Generation of iPSCs and in Vitro Differentiation into Three Germ Layers.
Episomal vectors encoding defined reprogramming factors were used as reported (49). In brief, HDFs grown in DMEM supplemented with 10% FBS were electroporated by using a microporator system (Neon; Invitrogen) with episomal vector mixtures (total 3 μg) according to the manufacturer’s instructions. After being pulsed three times with a voltage of 1,650 for 10 ms, the cells were grown further in DMEM (containing 10% FBS). Seven days after transfection, cells were transferred onto a feeder layer. iPSC colonies that looked similar to hESCs were picked up mechanically and further cultured for characterization.
In vitro differentiation of the iPSCs into three germ layers was induced as described (50, 51). Embryoid bodies (EBs), formed by partially dissociating iPSCs using collagenase type IV (Invitrogen), were transferred to ultralow attachment plates (Corning) and cultured in DMEM/F12 (1:1) medium supplemented with 20% knockout serum (Invitrogen), 4.5 g/L l-glutamine, 1% nonessential amino acids, 0.1 mM 2-mercaptoethanol, and 5% FBS. After a week of cultivation under these conditions, EBs were attached onto Matrigel-coated culture dishes and further cultured for 10 d. Spontaneous differentiation of EBs into cells representing the three germ layer lineages was detected by immunostaining with appropriate antibodies.
Differentiation of iPSCs.
To induce differentiation of iPSCs into the endoderm lineage, we used a described method (52). In brief, iPSC colonies were cultured in mTeSR-1 hESC growth medium (StemCell Technology) for feeder-free culture (53). Undifferentiated iPSCs were incubated to obtain definitive endoderm cells in RPMI/B27 (RPMI-1640 from Sigma; B27 supplement from Invitrogen) medium supplemented with 100 ng/mL Activin A (PeproTech) and 5 μM phosphatidylinositol 3–kinase inhibitor (LY-294002; Sigma) for 5 d. Cells that had differentiated into endoderm were harvested for isolation of total RNAs, which were used as template for cDNA synthesis.
To induce differentiation of iPSCs into the endothelial cells, we used a described method (54) with slight modifications. In brief, EBs were cultured in hESC medium supplemented with 20 ng/mL bone morphogenic protein 4 (R&D Systems) and 10 ng/mL Activin A (PeproTech). On day 3 of EB formation, the EBs were attached onto Matrigel-coated dishes and induced to differentiate into endothelial cells for up to 10 d in medium supplemented with 100 ng/mL VEGF (PeproTech) and 50 ng/mL basic FGF (R&D Systems).
TALEN Transfections for Inducing Inversion and Reversion in iPSCs.
Cultured iPSCs were harvested by treating with collagenase type IV. After washing with PBS, the cells were further treated with Accutase (Invitrogen) to create single-cell suspensions as described (55). These single cells were mixed with 10 μg of TALEN-encoding plasmids (5 μg of each plasmid) and pulsed with a voltage of 850 for 30 ms. Cells were then seeded onto feeder cells and allowed to grow for 10 d. To detect genomic inversion or reversion events, cells from individual colonies were lysed in 20 μL of lysis buffer [1× Ex-taq buffer (pH 8.0) containing proteinase K] at 56 °C for 3 h. After inactivation of proteinase K, 2 μL of genomic DNA solution was subjected to PCR by using Ex-taq DNA polymerase (Takara) and specific primers. PCR products were analyzed by agarose gel electrophoresis. Specific primer sequences are shown in Table S3.
Isolation of Clonal Populations of Cells, PCR Analysis, and DNA Sequencing of Breakpoints.
To isolate clonal populations of inverted (or reverted) cells, each colony that had been identified by PCR as containing the desired genomic event was dissociated into single cells by using collagenase and Accutase as described above and replated. After three rounds of passaging, several clones (six clones for inversion, four clones for reversion) were chosen for sequencing and further experiments. For sequence determination, amplified PCR products were electrophoresed, eluted from the agarose gel by using a Gel Extraction kit (SolGent), and cloned into the pGEM-T vector (Promega). Cloned PCR products were sequenced by using T7 primers.
RNA Isolation, RT-PCR, and qPCR.
Total RNAs were purified from cells by using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. cDNAs were synthesized from total RNAs (1 μg) by using the DiaStar cDNA synthesis kit (SolGent). To confirm the expression of Factor VIII, FOXA2, Sox17, and GAPDH, PCR was performed with Ex-Taq (Takara) by using the synthesized cDNAs as template. For qPCR, SYBR Premix Ex-Taq (Takara) was used according to the manufacturer’s instructions. Specific primer sequences used for RT-PCR or qPCR are shown in Table S3.
Alkaline Phosphatase Staining and Immunostaining.
Alkaline phosphatase activity was measured with the leukocyte alkaline phosphatase staining kit (Sigma) according to the manufacturer’s instructions. For the immunostaining of pluripotent stem cell markers, cells were fixed in 4% paraformaldehyde solution and permeabilized with 0.2% Triton X-100. After washing with PBS, the cells were incubated with a PBS solution containing 5% normal goat serum and 2% BSA. The cells were then incubated with primary antibodies for 2 h at room temperature, washed with PBS, and incubated with fluorescence-conjugated secondary antibodies (Alexa Fluor 488 or 594; Invitrogen) for 1 h at room temperature. The cells were mounted with an antifade mounting medium containing DAPI (Vector Laboratories) for nuclei visualization. The images were captured and analyzed by using an Olympus IX71 microscope or FSX system.
DNA Fingerprinting and Karyotype Analysis.
To confirm the dermal fibroblast origin of iPSC lines, PCR-based short tandem repeat (STR) analysis was carried out at the Gene-Analysis Institute of Human Pass Inc. In brief, STR loci were amplified from genomic DNA samples isolated from iPSC lines and their parental cells by using the AmpFISTR PCR system (Applied Biosystems). The amplified products were analyzed by using an ABI PRISM 3130XL genetic analyzer and Genemapper (Version 3.2; Applied Biosystems). For karyotype analysis, chromosomes were stained with Giemsa for G-banding analysis and analyzed by the Chromosome Image Processing System at GenDix.
Statistical Analysis.
Data are presented as means ± SEs. Student t test was used for statistical analysis. P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
J.-S.K. was supported by National Research Foundation of Korea Grant 2013000718. D.-W.K. was supported by National Research Foundation of Korea Bio and Medical Technology Development Program Grants 2012M3A9B4028631 and 2012M3A9C7050126 and Korean Ministry of Health and Welfare Grant A120254.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1323941111/-/DCSupplemental.
References
- 1.Graw J, et al. Haemophilia A: From mutation analysis to new therapies. Nat Rev Genet. 2005;6(6):488–501. doi: 10.1038/nrg1617. [DOI] [PubMed] [Google Scholar]
- 2.Bolton-Maggs PH, Pasi KJ. Haemophilias A and B. Lancet. 2003;361(9371):1801–1809. doi: 10.1016/S0140-6736(03)13405-8. [DOI] [PubMed] [Google Scholar]
- 3.Lakich D, Kazazian HH, Jr, Antonarakis SE, Gitschier J. Inversions disrupting the factor VIII gene are a common cause of severe haemophilia A. Nat Genet. 1993;5(3):236–241. doi: 10.1038/ng1193-236. [DOI] [PubMed] [Google Scholar]
- 4.Naylor J, Brinke A, Hassock S, Green PM, Giannelli F. Characteristic mRNA abnormality found in half the patients with severe haemophilia A is due to large DNA inversions. Hum Mol Genet. 1993;2(11):1773–1778. doi: 10.1093/hmg/2.11.1773. [DOI] [PubMed] [Google Scholar]
- 5.Bagnall RD, Waseem N, Green PM, Giannelli F. Recurrent inversion breaking intron 1 of the factor VIII gene is a frequent cause of severe hemophilia A. Blood. 2002;99(1):168–174. doi: 10.1182/blood.v99.1.168. [DOI] [PubMed] [Google Scholar]
- 6.Nathwani AC, et al. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N Engl J Med. 2011;365(25):2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300(5620):763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- 8.Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300(5620):764. doi: 10.1126/science.1079512. [DOI] [PubMed] [Google Scholar]
- 9.Urnov FD, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435(7042):646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 10.Kim HJ, Lee HJ, Kim H, Cho SW, Kim JS. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 2009;19(7):1279–1288. doi: 10.1101/gr.089417.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller JC, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29(2):143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 12.Kim Y, et al. A library of TAL effector nucleases spanning the human genome. Nat Biotechnol. 2013;31(3):251–258. doi: 10.1038/nbt.2517. [DOI] [PubMed] [Google Scholar]
- 13.Kim YK, et al. TALEN-based knockout library for human microRNAs. Nat Struct Mol Biol. 2013;20(12):1458–1464. doi: 10.1038/nsmb.2701. [DOI] [PubMed] [Google Scholar]
- 14.Cho SW, Lee J, Carroll D, Kim JS, Lee J. Heritable gene knockout in Caenorhabditis elegans by direct injection of Cas9-sgRNA ribonucleoproteins. Genetics. 2013;195(3):1177–1180. doi: 10.1534/genetics.113.155853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sung YH, et al. Highly efficient gene knockout in mice and zebrafish with RNA-guided endonucleases. Genome Res. 2014;24(1):125–131. doi: 10.1101/gr.163394.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho SW, et al. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24(1):132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31(3):230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 18.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hwang WY, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31(3):227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31(3):233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee HJ, Kim E, Kim JS. Targeted chromosomal deletions in human cells using zinc finger nucleases. Genome Res. 2010;20(1):81–89. doi: 10.1101/gr.099747.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim S, Lee HJ, Kim E, Kim JS. Analysis of targeted chromosomal deletions induced by zinc finger nucleases. Cold Spring Harb Protoc. 2010 doi: 10.1101/pdb.prot5477. [DOI] [PubMed] [Google Scholar]
- 24.Lee HJ, Kweon J, Kim E, Kim S, Kim JS. Targeted chromosomal duplications and inversions in the human genome using zinc finger nucleases. Genome Res. 2012;22(3):539–548. doi: 10.1101/gr.129635.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet. 2006;7(2):85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 26.Zelechowska MG, van Mourik JA, Brodniewicz-Proba T. Ultrastructural localization of factor VIII procoagulant antigen in human liver hepatocytes. Nature. 1985;317(6039):729–730. doi: 10.1038/317729a0. [DOI] [PubMed] [Google Scholar]
- 27.Hollestelle MJ, et al. Tissue distribution of factor VIII gene expression in vivo—a closer look. Thromb Haemost. 2001;86(3):855–861. [PubMed] [Google Scholar]
- 28.Shahani T, et al. Activation of human endothelial cells from specific vascular beds induces the release of a FVIII storage pool. Blood. 2010;115(23):4902–4909. doi: 10.1182/blood-2009-07-232546. [DOI] [PubMed] [Google Scholar]
- 29.Terraube V, O’Donnell JS, Jenkins PV. Factor VIII and von Willebrand factor interaction: Biological, clinical and therapeutic importance. Haemophilia. 2010;16(1):3–13. doi: 10.1111/j.1365-2516.2009.02005.x. [DOI] [PubMed] [Google Scholar]
- 30.Iafrate AJ, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36(9):949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 31.Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444(7118):444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annu Rev Med. 2010;61:437–455. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- 33.Carlson DF, et al. Efficient TALEN-mediated gene knockout in livestock. Proc Natl Acad Sci USA. 2012;109(43):17382–17387. doi: 10.1073/pnas.1211446109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gupta A, et al. Targeted chromosomal deletions and inversions in zebrafish. Genome Res. 2013;23(6):1008–1017. doi: 10.1101/gr.154070.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim H, Kim JS. A guide to genome engineering with programmable nucleases. Nat Rev Genet. 2014;15(5):321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- 36.Kim S, Lee MJ, Kim H, Kang M, Kim JS. Preassembled zinc-finger arrays for rapid construction of ZFNs. Nat Methods. 2011;8(1):7. doi: 10.1038/nmeth0111-7a. [DOI] [PubMed] [Google Scholar]
- 37.Kim E, et al. Precision genome engineering with programmable DNA-nicking enzymes. Genome Res. 2012;22(7):1327–1333. doi: 10.1101/gr.138792.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim Y, Kweon J, Kim JS. TALENs and ZFNs are associated with different mutation signatures. Nat Methods. 2013;10(3):185. doi: 10.1038/nmeth.2364. [DOI] [PubMed] [Google Scholar]
- 39.Sung YH, et al. Knockout mice created by TALEN-mediated gene targeting. Nat Biotechnol. 2013;31(1):23–24. doi: 10.1038/nbt.2477. [DOI] [PubMed] [Google Scholar]
- 40.Kim H, et al. Surrogate reporters for enrichment of cells with nuclease-induced mutations. Nat Methods. 2011;8(11):941–943. doi: 10.1038/nmeth.1733. [DOI] [PubMed] [Google Scholar]
- 41.Kim H, et al. Magnetic separation and antibiotics selection enable enrichment of cells with ZFN/TALEN-induced mutations. PLoS ONE. 2013;8(2):e56476. doi: 10.1371/journal.pone.0056476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramakrishna S, et al. Surrogate reporter-based enrichment of cells containing RNA-guided Cas9 nuclease-induced mutations. Nat Commun. 2014;5:3378. doi: 10.1038/ncomms4378. [DOI] [PubMed] [Google Scholar]
- 43.Nikiforova MN, et al. Proximity of chromosomal loci that participate in radiation-induced rearrangements in human cells. Science. 2000;290(5489):138–141. doi: 10.1126/science.290.5489.138. [DOI] [PubMed] [Google Scholar]
- 44.Bondeson ML, et al. Inversion of the IDS gene resulting from recombination with IDS-related sequences is a common cause of the Hunter syndrome. Hum Mol Genet. 1995;4(4):615–621. doi: 10.1093/hmg/4.4.615. [DOI] [PubMed] [Google Scholar]
- 45.Guo J, Gaj T, Barbas CF., 3rd Directed evolution of an enhanced and highly efficient FokI cleavage domain for zinc finger nucleases. J Mol Biol. 2010;400(1):96–107. doi: 10.1016/j.jmb.2010.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu Y, Smyth GK. ELDA: Extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347(1-2):70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 47.Kim DS, et al. Robust enhancement of neural differentiation from human ES and iPS cells regardless of their innate difference in differentiation propensity. Stem Cell Rev. 2010;6(2):270–281. doi: 10.1007/s12015-010-9138-1. [DOI] [PubMed] [Google Scholar]
- 48.Jang J, et al. Disease-specific induced pluripotent stem cells: A platform for human disease modeling and drug discovery. Exp Mol Med. 2012;44(3):202–213. doi: 10.3858/emm.2012.44.3.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okita K, et al. A more efficient method to generate integration-free human iPS cells. Nat Methods. 2011;8(5):409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- 50.Sugii S, et al. Human and mouse adipose-derived cells support feeder-independent induction of pluripotent stem cells. Proc Natl Acad Sci USA. 2010;107(8):3558–3563. doi: 10.1073/pnas.0910172106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jang J, et al. Induced pluripotent stem cell models from X-linked adrenoleukodystrophy patients. Ann Neurol. 2011;70(3):402–409. doi: 10.1002/ana.22486. [DOI] [PubMed] [Google Scholar]
- 52.Si-Tayeb K, et al. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology. 2010;51(1):297–305. doi: 10.1002/hep.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jia F, et al. A nonviral minicircle vector for deriving human iPS cells. Nat Methods. 2010;7(3):197–199. doi: 10.1038/nmeth.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoo CH, et al. Endothelial progenitor cells from human dental pulp-derived iPS cells as a therapeutic target for ischemic vascular diseases. Biomaterials. 2013;34(33):8149–8160. doi: 10.1016/j.biomaterials.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 55.Desbordes SC, et al. High-throughput screening assay for the identification of compounds regulating self-renewal and differentiation in human embryonic stem cells. Cell Stem Cell. 2008;2(6):602–612. doi: 10.1016/j.stem.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.