Significance
Reversible protein phosphorylation is a major regulatory mechanism by which bacteria sense and respond to changes in their environment. In Mycobacterium tuberculosis (Mtb), however, protein phosphorylation on Tyr residues had not been described and was thought to be absent. We show that Mtb phosphorylates diverse proteins on Tyr, suggesting a broad functional role. We identify the Ser/Thr kinases as the kinases responsible for phosphorylation on Tyr and show that Tyr phosphorylation regulates Ser/Thr protein kinase activity. Together, our study provides the basis for understanding how this new Mtb posttranslational modification affects physiology and pathogenesis.
Abstract
Reversible protein phosphorylation determines growth and adaptive decisions in Mycobacterium tuberculosis (Mtb). At least 11 two-component systems and 11 Ser/Thr protein kinases (STPKs) mediate phosphorylation on Asp, His, Ser, and Thr. In contrast, protein phosphorylation on Tyr has not been described previously in Mtb. Here, using a combination of phospho-enrichment and highly sensitive mass spectrometry, we show extensive protein Tyr phosphorylation of diverse Mtb proteins, including STPKs. Several STPKs function as dual-specificity kinases that phosphorylate Tyr in cis and in trans, suggesting that dual-specificity kinases have a major role in bacterial phospho-signaling. Mutation of a phosphotyrosine site of the essential STPK PknB reduces its activity in vitro and in live Mtb, indicating that Tyr phosphorylation has a functional role in bacterial growth. These data identify a previously unrecognized phosphorylation system in a human pathogen that claims ∼1.4 million lives every year.
Reversible phosphorylation is the main signaling mechanism underlying the dynamic adaptive responses necessary for Mycobacterium tuberculosis (Mtb) survival in the host. Although phosphorylation on Ser, Thr, and Tyr was long considered exclusive to the eukaryotic lineage, it now is recognized as being ancient and ubiquitous (1). In the major human pathogen Mtb, phosphorylation on Ser and Thr residues is essential for growth and adaptation, and the essential growth regulator protein kinase PknB is under development as a therapeutic target (2, 3). In contrast, the prevalence and functional consequences of protein phosphorylation on Tyr in prokaryotes are less defined. The best-understood role of bacterial Tyr phosphorylation is the regulation of capsule exopolysaccharide production and transport in several species, directly linking Tyr phosphorylation to pathogenesis (4–7).
In Mtb, the phospho-signaling landscape is not well defined, and to date there has been no conclusive molecular evidence for protein Tyr phosphorylation. One study detected a small number of bands in Mtb lysate by Western blotting with a phospho-Tyr (pTyr)-specific antibody (4G10), but these proteins were not identified, and phosphorylation on Tyr was not confirmed by other methods (8). Another study showed in vitro Tyr phosphorylation of two Mtb proteins (9). Several phosphoproteomic studies in Mtb identified hundreds of pSer/pThr sites but did not detect pTyr sites (10, 11). The prokaryotic Phosphorylation Site Database (12) lists 167 pTyr sites from a number of bacterial species but none from Mtb. Bacterial Tyr phosphorylation is thought to be catalyzed largely by a distinct class of Tyr kinases, the BY kinases, that share no similarity with eukaryotic kinases (13). Mtb is not predicted to encode functional BY kinases but produces two canonical protein Tyr phosphatases of the low molecular weight and classical types (14). The Tyr phosphatases, however, are thought to be secreted and to act on host substrates (15). The absence of functional BY kinases and the presumed secretion of the phosphatases into the host supported the notion that Mtb does not support intrinsic protein Tyr phosphorylation.
Phosphorylation on Tyr is the least abundant, most labile, and technically least tractable of Ser/Thr/Tyr phosphorylation events. In eukaryotes, pTyr contributes 0.5–2% of phosphorylation sites (16, 17). In prokaryotes, the abundance of Tyr phosphorylation differs widely among species. Most studies reported 2.7–25.8% of Ser/Thr/Tyr phosphorylation events occurring on Tyr (18), including 8.6% in Escherichia coli (19). However, a recent study in E. coli identified >500 pTyr sites, a number far exceeding that of Ser/Thr phosphosites, suggesting that Tyr phosphorylation is much more prevalent than previously believed (20). The same study suggests a much smaller role for the E. coli BY kinases Etk and Wzc than previously thought.
The presence of Tyr phosphorylation in many bacterial phyla and its presumed absence in others led us to reconsider the possibility of Tyr phosphorylation in Mtb. Here, we show that Mtb does indeed phosphorylate protein on Tyr. We found that several Ser/Thr protein kinases (STPKs) phosphorylate protein on Tyr in cis and in trans and that Tyr phosphorylation regulates STPK activity. Using a gel-free, high-accuracy mass spectrometry (MS) approach, we identified several Tyr-phosphorylated proteins in Mtb, indicating potentially broad regulation of Mtb physiology by this posttranslational modification.
Results
Mtb STPKs Are Dual-Specificity Kinases.
Protein phosphorylation is a major regulatory mechanism in Mtb. Protein Tyr phosphorylation is widespread among prokaryotes from Archaea to Firmicutes and Proteobacteria, but there was no previous evidence for Tyr phosphorylation in Mtb. In contrast to the prevailing notion, we hypothesized that Mtb may regulate cellular processes by Tyr phosphorylation. To explore this idea, we first investigated whether Mtb encodes BY kinases, a distinct family of Tyr kinases with no sequence or structural similarity to eukaryotic Tyr kinases. To identify potential BY kinases in Mtb, we searched the Mtb proteome using the BYKdb prediction program (21). BYKdb did not predict any functional Mtb BY kinases but did identify two proteins with similarity to BY kinases, Rv1708 and Rv3918c. However, neither Rv1708 nor Rv3918c contains the BY kinase signature Walker A′ motif. In a chemical biology screen for ATPases (22), we previously identified Rv1708 as a functional ATPase, a prerequisite for kinase function. To verify the prediction that Rv1708 is nonfunctional even though it has ATPase activity, we expressed Rv1708 recombinantly and tested for kinase activity. Rv1708 did not have any detectable kinase activity (Fig. S1A). In an in vitro study, Rv2232 previously has been shown to phosphorylate protein on Tyr (9). Rv2232 is predicted to be a haloacid dehydrogenase, a hydrolase family with no similarity to known kinases. Consistent with the structural prediction for Rv2232, we did not detect kinase activity by measuring 32P incorporation (Fig. S1A). Also, we did not detect any Tyr-phosphorylated residues of Rv2232 by MS after incubation with ATP and a range of cofactors.
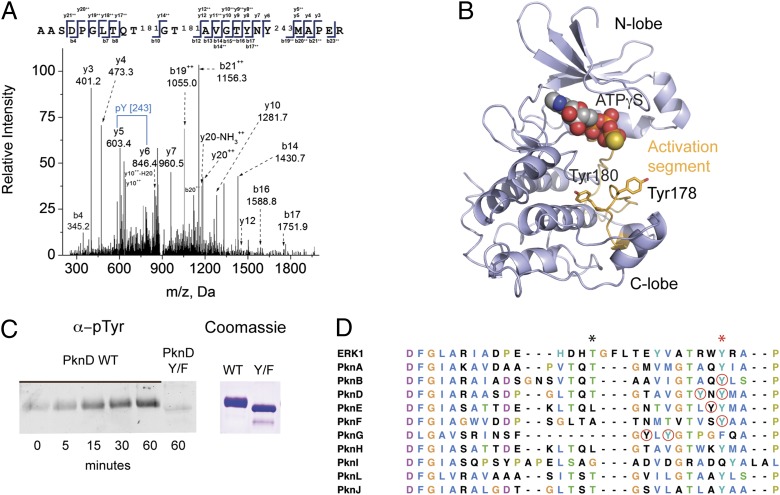
Although our findings suggested that Mtb does not produce BY kinases, Tyr phosphorylation might be carried out by noncanonical mechanisms, as would be consistent with a recent study of Tyr phosphorylation in E. coli indicating that, despite their biochemical capacity, BY kinases actually play a limited role in Tyr phosphorylation in vivo (20). In yeast, Tyr phosphorylation is carried out exclusively by dual-specificity kinases that have both Ser/Thr and Tyr kinase activity. Although rare, dual-specificity kinases also have been described in bacteria (23, 24), raising the possibility that some Mtb STPKs also could have dual specificity. To test this possibility, we measured in vitro Tyr phosphorylation of the kinase domain (KD) of the Mtb transcriptional regulator kinase PknD. Surprisingly, in addition to several previously described pSer and pThr sites, we observed MS spectra for phosphorylation on the activation-segment residues Tyr178 and Tyr180 (Fig. 1 A and B) as well as on residues outside the activation segment. Because E. coli also phosphorylates protein on Tyr, we sought to rule out the possibility that PknD phosphorylation resulted from E. coli phosphorylation during recombinant expression by testing for PknD Tyr autophosphorylation activity. PknD showed clear autophosphorylation on Tyr by Western blotting when recombinant PknD was incubated with ATP and manganese (Fig. 1C), confirming that PknD autophosphorylates on Tyr and can act as a dual-specificity kinase. Mutation of Tyr178 and Tyr180 to Phe abrogated autophosphorylation, showing that these sites indeed are phosphorylated (Fig. 1C).
Fig. 1.
STPKs are dual-specificity kinases. (A) MS/MS spectrum showing phosphorylation of recombinant PknD KD on Tyr180. (B) A prediction of the PknD structure (PHYRE) shows the predicted position of pTyr178 and -180 on the activation segment (orange) relative to the ATP-binding site, N lobe, and C lobe. (C) Autophosphorylation of recombinant PknD KD on Tyr. Western blotting with anti-pTyr antibody shows an increase in PknD Tyr phosphorylation after incubation with ATP and Mn2+ and loss of Tyr phosphorylation in the Tyr(178,180)Phe double mutant, showing intrinsic Tyr kinase activity. (D) Sequence alignment of Mtb STPK activation segments. Red circles indicate pTyr sites identified in this study. The black asterisk marks the position of phospho-amino acids that regulate activity in some Mtb STPKs, and a red asterisk marks conserved Tyr.
Activation-segment phosphorylation is a central regulator of kinase activity. To gauge the possibility for Tyr phosphorylation on other Mtb STPK activation segments, we aligned the activation segments of the 11 Mtb STPKs (Fig. 1D). All except one activation segment contained Tyr residues, including the Tyr residue toward the C-terminal end that is conserved in some STPK families and is present in all Mtb STPKs except PknG. Several other Tyr residues are in the vicinity of pThr sites known to control STPK activation. To test for Tyr phosphorylation in the STPK-activation segment sequences, we phosphorylated recombinant PknB, PknD, PknE, PknF, and PknH KDs and full-length PknG in vitro and investigated Tyr phosphorylation by MS/MS. All tested STPKs were phosphorylated on Tyr (Fig. 1D and Table S1), suggesting that most, if not all, Mtb STPKs have dual specificity. PknB, -D, -E, -F, and -G were phosphorylated on Tyr in the activation segment, suggesting that several STPKs also may be regulated by Tyr phosphorylation. To rule out the possibility that Tyr phosphorylation of these STPKs during recombinant expression results from E. coli Tyr kinases rather than intrinsic Tyr kinase activity, we compared Tyr phosphorylation on kinase-dead PknB, -F, and -H with Tyr phosphorylation on WT kinase; all Tyr phosphorylation on the kinase-dead mutants would be the result of E. coli phosphorylation. We detected no or one (PknH) pTyr on the dead kinases but numerous pTyr on the WT kinases, indicating that the pTyr detected in recombinant STPKs indeed results from STPK Tyr kinase activity (Fig. S1B).
The STPKs Phosphorylate Tyr in trans.
To test whether the STPKs also phosphorylate Tyr in trans, we mapped the Tyr phosphosites on structural models predicted by the PHYRE Protein Fold Recognition Server. Three pTyr sites of PknD mapped to the back side of the N lobe, and one site mapped to the C lobe. Most of these pTyr residues are oriented in the opposite direction or are >10 Å away from the predicted position of the ATP γ-phosphate, precluding intramolecular autophosphorylation of these sites. PknH was Tyr-phosphorylated at residues 16 and 31 in the N lobe, which also are distant and oriented away from the active site. We detected similar phosphorylation sites that are not accessible for intramolecular autophosphorylation in PknB and PknE. These data suggest that the STPKs can phosphorylate substrates in trans.
Mtb Phosphorylates Proteins on Tyr.
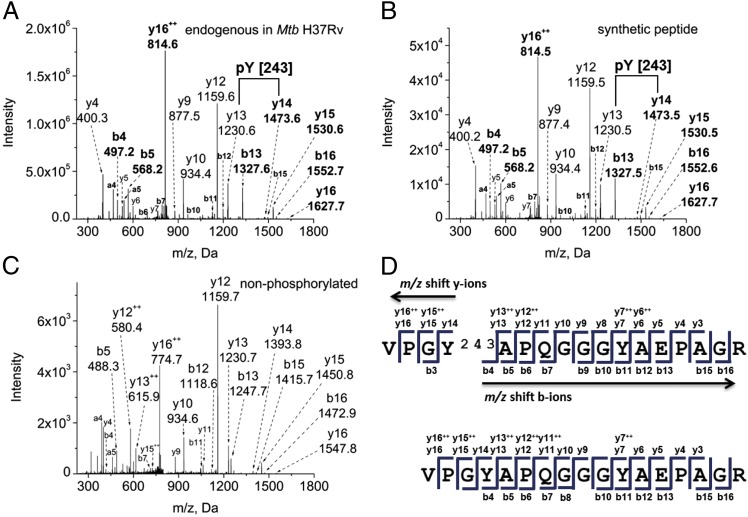
Given our finding that the Mtb STPKs can phosphorylate on Tyr in trans, we next investigated whether Mtb supports broader in vivo protein phosphorylation on Tyr. By using a targeted, gel-free, MS-based proteomics approach in combination with affinity-based phosphopeptide isolation, we probed the Mtb phosphoproteome specifically for phosphorylation on Tyr. Mtb cultures were grown to late log phase, lysed, proteolytically digested, and subsequently enriched for phosphopeptides by tandem immobilized metal-affinity chromatography (IMAC) using Fe(III) and TiO2 procedures to reduce sample complexity. We performed high-mass accuracy nano liquid chromatography MS (LC-MS) analysis with collision-induced dissociation (LC-MS/MS) of the isolated analytes to study the phosphoproteome comprehensively. Data were analyzed with the proteomics software tool suite Trans-Proteomic-Pipeline (TPP) (25). We detected 30 unambiguous Tyr phosphorylation sites on 17 proteins in Mtb lysates (Table S2). A representative fragmentation spectrum of the Tyr-phosphorylated peptide VPGpYAPQGGGYAEPAGR derived from the protein FhaA is shown in Fig. 2A. Nearly the entire b and y ion series in the spectrum could be annotated. The two most intense fragments, y12+ and y16++, result from internal Pro fragmentation in the peptide sequence, a well-known collision-induced fragmentation behavior of this amino acid (26). Less common but not unusual is a doubly charged fragment derived from a doubly charged precursor such as the y16++ ion, which may be caused by a mobile proton. The probability of phosphorylation on Tyr388 was determined to be 1.000 by the PTMProphet algorithm of the TPP (25). To further verify Tyr phosphorylation at position 388 and other positions identified in Mtb lysate, we chemically synthesized the FhaA phosphopeptide and a subset of the phosphopeptides listed in Table S2 and subjected the synthetic peptides to identical LC-MS/MS analysis. The spectrum obtained from the synthetic Tyr-phosphorylated peptide VPGpYAPQGGGYAEPAGR (Fig. 2B) is identical to the fragmentation pattern of the endogenous peptide observed in Mtb cell lysate and provides conclusive evidence of the pTyr assignment. The MS/MS spectrum of the nonphosphorylated peptide is shown for comparison in Fig. 2C. The observed b and y fragment ions for the phosphorylated and nonphosphorylated form of this peptide are annotated in the peptide sequence. The mass shift caused by phosphorylation is shown by the different y ion masses upstream and the different b ion masses downstream of the pTyr residue, as highlighted by arrows in Fig. 2D. Spectra of additional synthetic pTyr peptides also were nearly identical to the spectra obtained from the endogenous peptides in Mtb lysate, confirming Tyr phosphorylation of these sites (Fig. S2). In addition, we fragmented peptides from Mtb lysate by higher-energy collisional dissociation in a Q Exactive mass spectrometer. The low molecular weight pTyr-specific immonium reporter ion of m/z 216.0426 (27) was observed, as shown in a representative spectrum in Fig. S3, further confirming the presence of pTyr in these samples.
Fig. 2.
FhaA is phosphorylated on Tyr. Precursor ion masses were measured in the Orbitrap Elite mass analyzer, and MS/MS spectra were acquired in the LTQ mass spectrometer. (A) Endogenous peptide identified in the phosphopeptide-enriched fraction of total Mtb cell lysate (precursor ion mass accuracy −0.69 ppm). (B) Synthetic peptide confirming the fragmentation pattern of the endogenous peptide (mass accuracy 1.04 ppm). (C) Nonphosphorylated peptide VPGYAPQGGGYAEPAGR, shown for comparison (mass accuracy 0.61 ppm). (D) Sequence annotation of identified fragment ions of the phosphorylated and nonphosphorylated peptides. The mass shift caused by phosphorylation is shown by the different y ion masses observed upstream and the different b ion masses observed downstream of the pTyr residue, as highlighted by arrows in the phosphorylated peptide sequence and shown in bold in the spectra.
In five biological replicates, we detected 30% of phosphorylation sites multiple times, indicating nonstochastic, specific phosphorylation events (Table S2). The overlap between different biological replicates was similar to that reported in analyses of eukaryotic phosphoproteomes (28). The Mtb proteins phosphorylated on Tyr span all functional categories except for the PE/PPE family, and essentials are enriched by approximately twofold compared with all genes (Table S2). The most highly phosphorylated protein by both number of different sites and total abundance is FhaA. FhaA is a protein containing a forkhead-associated (FHA) domain that is also a substrate of several Ser/Thr kinases (29). Of note, FhaA is a 57-kDa protein and thus likely is the major protein detected by anti-pTyr Western blot in a previous study (8). All FhaA phosphorylation sites mapped to the unstructured intermediate domain of the protein that is Pro-, Gly-, and Tyr-rich (30). Based on structure predictions using PHYRE, other Tyr phosphorylation sites are predicted to be in well-structured regions, precluding a general preference for Mtb Tyr phosphorylation of disordered regions. Other Tyr-phosphorylated proteins with known or predicted functions include the chaperone GroEL2, the proteasomal subunit PrcB, and the probable aminopeptidase PepB, raising the possibility that protein folding and degradation are regulated by Tyr phosphorylation. Consistent with our in vitro data, we also detected phosphorylation of Tyr178 of PknD, showing that STPK activation-segment phosphorylation occurs in live Mtb.
To determine the ratio of pTyr to pSer and pThr in Mtb, we compared the number of Tyr phosphorylation sites with the number of pSer and pThr sites detected in the same samples. In agreement with previous studies of bacterial Ser/Thr/Tyr phosphorylation, we found that 5.8% of Mtb proteins are phosphorylated, with a strong bias toward pThr (pSer:pThr:pTyr ratio of 34%:62%:4%). At 4%, the percentage of pTyr in Mtb is within the range found in other bacterial phosphoproteomes (18, 31).
Protein Tyr Phosphorylation in Published Mtb Proteomic Data.
In light of our identification of Tyr phosphorylation in vitro and in vivo, it is puzzling that numerous previous studies did not detect this modification. To test if phosphorylated Tyr peptides also may be contained, albeit overlooked, in previous data, we analyzed four datasets available through the public MS data repositories PeptideAtlas (www.peptideatlas.org) and Peptidome (ftp://ftp.ncbi.nih.gov/pub/peptidome/studies/PSEnnn/PSE133). We considered two recently published datasets on Mtb strain H37Rv described by Kelkar et al. (10) (123 LC-MS/MS runs, 1,385,970 spectra) and Schubert et al. (32) (24 LC-MS/MS runs, 236,475 spectra), one Mycobacteria bovis bacillus Calmette–Guérin set by Schubert et al. (32) (24 LC-MS/MS runs, 213,183 spectra), and one set on Mtb strain A7494 and A12998 (250 LC-MS/MS runs, 1,784,650 spectra, Peptidome PSE133). Each study used different types of sample fractionation including OffGel electrophoresis, strong cation exchange chromatography, C18 chromatography, multidimensional protein identification technology, or in-gel fractionation of total cell lysate and culture filtrate. Because of the different scope of each study and the assumption that Mtb does not support phosphorylation on Tyr, none of these data, to the best of our knowledge, were analyzed extensively with regard to Tyr phosphorylation. The search of the more than 3.6 million spectra from these four datasets resulted in the identification of 35 Tyr phosphorylation sites from 34 proteins from a broad range of functional categories (Table S3). Two peptides from FhaA (R.GGYPPETGGYPPQPGpYPRPR.H and R.VPGpYAPQGGGYAEPAGR.D) were identified both in the public dataset and in our experiments, further confirming assignment of this pTyr site. Of note, the former peptide was identified in each of the four public datasets as well as in our study. Rv0440 also was identified through different pTyr peptides in our experiments and the public data. All other phosphorylation sites were different in our and published datasets, likely because of the large differences in sample type, sample preparation, and fractionation techniques used, MS approaches and instrumentation, and data analysis. These data, which demonstrate the utility of reanalysis of public datasets for addressing new biological questions, further expand our view of the Tyr phosphoproteome and bring the total number of Mtb Tyr-phosphorylated sites to 63 in 49 different proteins.
To test if phosphorylation on Tyr is conserved in other mycobacteria, we also measured pTyr in the soil-dwelling Mycobacterium smegmatis. Indeed, we identified four proteins (Table S4), including MSMEG_5785, the Mtb ortholog of Rv2145c, which also was identified in the public dataset as a Tyr-phosphorylated protein. These data show that Tyr phosphorylation is a conserved regulatory mechanism in mycobacteria. Because M. smegmatis is a soil-dwelling, nonpathogenic species, pTyr does not seem to be linked exclusively to pathogenesis but likely supports basic bacterial functions.
Phosphosite Mutation Affects PknB Activity.
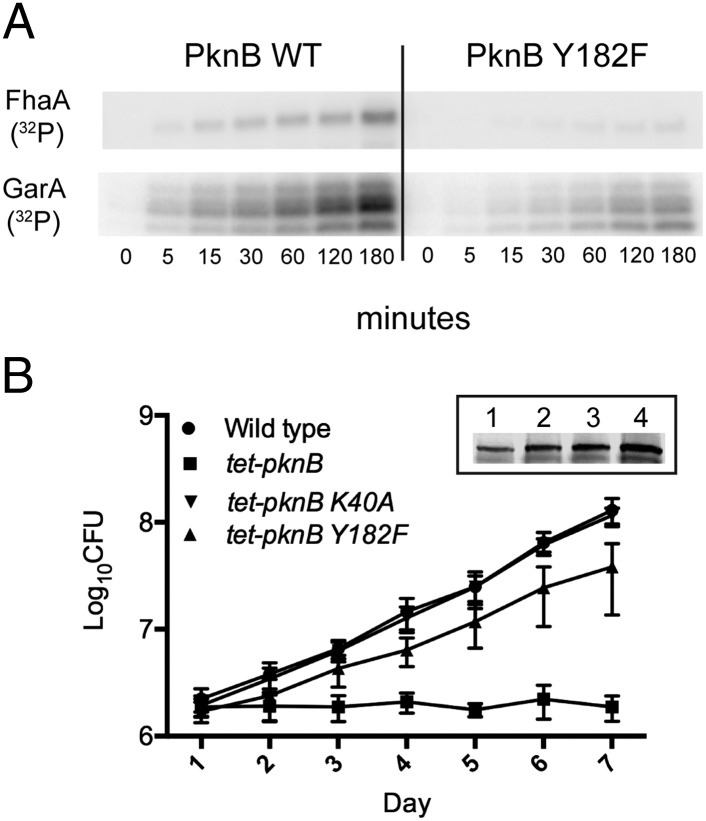
Kinases are widely regulated by phosphorylation in their activation segments. To test the effect of Tyr phosphorylation of the activation segment on Mtb kinase activity, we mutated the PknB KD pTyr site 182 to Phe and generated a Thr173Ala mutation, which is known to regulate PknB activity (33). All proteins were purified by metal-affinity and size-exclusion chromatography and eluted at a similar molecular weight (Fig. S4A). We tested for autophosphorylating activity by measuring 32P incorporation after incubation of PknB with [γ-32P] ATP. Unlike the PknB Thr173Ala mutation, the autophosphorylating activity of the Tyr182Phe mutation did not differ from that of WT (Fig. S4B), suggesting that activation-segment Tyr phosphorylation is not required for this STPK’s autophosphorylating activity. However, transphosphorylation of FhaA and GarA, two known PknB substrates that are phosphorylated on Ser and Thr residues (29, 34), was reduced in the Tyr182Phe mutant (Fig. 3A). PknB is essential for Mtb growth, and we recently showed that growth is highly sensitive to altered PknB activity (35). To test whether the reduced activity of the Tyr182Phe mutant in vitro affects live Mtb, we measured the growth of Mtb strains overexpressing WT PknB, the kinase-dead Lys40Ala mutant, and the Tyr182Phe mutant (Fig. 3B, Inset). As previously shown, overexpression of WT PknB led to complete growth arrest, whereas the kinase-dead mutant did not affect Mtb growth. Overexpression of Tyr182Phe led to a >20-fold reduction of toxicity compared with overexpression of WT PknB on day 7 (Fig. 3B). These data show that PknB Tyr182Phe function is impaired in live Mtb, suggesting that Tyr phosphorylation of the activation segment regulates some essential functions of PknB relating to growth.
Fig. 3.
Phosphosite deletion affects STPK activity. (A) Mutation of Tyr182 to Phe in PknB reduces PknB activity in vitro. Autoradiogram shows loss of phosphorylation of the PknB substrates FhaA and GarA upon Tyr mutation. (B) Overexpression of PknB is toxic to Mtb, but overexpression of the Tyr182Phe mutant leads to >20-fold decreased toxicity compared with overexpression of WT PknB. (Inset) Western blot of PknB expression in WT Mtb (lane 1), tet-pknB overexpressor (lane 2), K40A mutant (lane 3), and Y182F mutant (lane 4). Equal protein loading was controlled by Bradford assay.
Collectively, we present multiple lines of conclusive evidence for Tyr phosphorylation in Mtb, providing a roadmap for defining new Tyr phosphorylation-dependent regulatory mechanisms in this major human pathogen.
Discussion
Protein Tyr phosphorylation in prokaryotes has long been elusive, and phosphorylation on Tyr was thought to be absent in Mtb. Until now, both phosphoproteins and functional Tyr kinases were considered absent, and the orphan Tyr phosphatases were explained plausibly as virulence factors targeting host proteins. We now show conclusive evidence for protein Tyr phosphorylation in Mtb. Along with the recent identification of >500 pTyr sites in E. coli (20), these findings suggest a more pervasive presence of Tyr phosphorylation in prokaryotes and reveal a new layer of Mtb phospho-signaling.
Until recently, most pTyr activity in bacteria was ascribed to BY kinases. However, we found no evidence of functional BY kinases in Mtb but instead identified Tyr-phosphorylation activity of a number of STPKs, providing a set of candidate kinases that may mediate selective Tyr phosphorylation in vivo. Individual bacterial dual-specificity kinases have been described previously in Chlamydophila and Bacillus, but our data suggest that dual-specificity kinases may play a prominent role in bacterial phospho-signaling. The BY kinases, on the other hand, may have a more peripheral role in Tyr phosphorylation than previously thought. This idea is consistent with the role of the two known E. coli BY kinases, Etk and Wzc, in E. coli Tyr phosphorylation: Deletion of both kinases had a limited effect on overall Tyr phosphorylation, suggesting the presence of additional, noncanonical Tyr kinases in E. coli (20).
Our data suggest that Tyr phosphorylation of the activation segment may regulate STPK activity in a way that is reminiscent of other dual-specificity kinases. Mutation of the PknB Tyr site 182 led to loss of function in vitro and in live Mtb, similar to the loss of function previously described for deletion of the Thr173 phosphosite. However, we cannot rule out the possibility that the mutagenic removal of a hydroxyl group from Tyr182 has other, phospho-independent effects on activity or substrate binding. Because PknB and PknD activation-segment Tyr mutations did not affect autophosphorylation, these Tyr phosphosites might function in other ways, for example by recruiting specific substrates. Our finding of intrinsic Tyr phosphorylation in Mtb also puts the Tyr phosphatases PtpA and PtpB in a new light. Although dephosphorylation of host substrates by PtpA has been substantiated, it now appears likely that PtpA has an additional role in intrinsic dephosphorylation. PtpA deletion does not affect in vitro growth in rich medium (36) but causes attenuation of Mtb in activated human macrophages (37), indicating that PtpA has an intracellular role in processes that affect host–pathogen interactions. With Tyr phosphorylation regulating the cell wall in other bacteria and with cell wall enzymes phosphorylated on Tyr in Mtb, the effect of PtpA on the survival in the host also may relate to control of cell wall biosynthesis.
We identified Tyr phosphorylation sites on proteins with a wide range of functions. Consistent with a role of Tyr phosphorylation in capsule biosynthesis in other bacteria, two essential Mtb enzymes generating cell wall-associated polysaccharides also are phosphorylated on Tyr: GlfT2, which catalyzes the polymerization of arabinogalactan with peptidoglycan to form an essential component of the mycobacterial cell wall (38), and the 1,4-alpha-glucan branching enzyme GlgB (39) that is involved in the biosynthesis of glycogen and capsular α-d-glucan. These data suggest that the regulation of cell wall-associated carbohydrates by Tyr phosphorylation might be conserved in Mtb. The most highly Tyr-phosphorylated protein, FhaA, is an FHA-domain–containing protein that also is a substrate of several Ser/Thr kinases. A recent study showed a role for FhaA in assembling a signaling complex with PknB and Rv3910 that affects cell wall synthesis (40). Interestingly, FhaA contains a long intermediate domain in addition to the C-terminal FHA domain and an N-terminal domain of unknown function. The intermediate region has a highly repetitive, Tyr-rich sequence that is unstructured. All Tyr phosphorylation sites mapped to this intermediate domain. The FhaA intermediate domain could link FHA-mediated functions to other pathways by serving as a switchboard through combinatorial Tyr phosphorylation. In addition, FhaA and several other Tyr-phosphorylated proteins also are phosphorylated on Ser and Thr, showing that both modifications can decorate the same protein simultaneously.
The detection of Tyr phosphorylation in Mtb appears to be more challenging than in other organisms. We found that pTyr-specific antibodies cannot readily detect Tyr phosphorylated proteins in cell lysate, and their reactivity even with recombinant Tyr-phosphorylated protein is limited. However, our finding of Tyr phosphorylation sites in existing MS datasets using vastly different fractionation methods shows that the current absence of evidence for Tyr phosphorylation was not caused primarily by technical difficulties.
The next challenge for understanding the role of Tyr phosphorylation in Mtb is the identification of specific functions of Tyr phosphorylation events. More than 20 years after the discovery of Tyr phosphorylation in bacteria, this study redefines phospho-signaling in Mtb and should encourage research into the function of this previously unidentified arm of signal transduction.
Experimental Procedures
Cloning, Expression, and Purification of Recombinant Mtb Proteins.
The genes for STPK kinase domains, full-length PknG, Rv1708, and Rv2232 were amplified from genomic Mtb H37Rv DNA and cloned into the pET28b expression vector in-frame with an N-terminal six-His tag and a SUMO tag for protein stability (FhaA). The kinase domains contain the following residues: PknB amino acids 1–307; PknD amino acids 1–378; PknE amino acids 1–289; PknF amino acids 1–301; and PknH amino acids 1–399. Site-directed mutagenesis was carried out using the QuikChange protocol (Stratagene). Proteins were expressed and purified as described previously (22).
Western Blotting.
Kinase reactions were carried out with 200–500 ng kinase, 150 mM NaCl, 20 mM Tris (pH 7.5), 5% (vol/vol) glycerol, 1 mM MnCl2 (1 mM each of MnCl2 and MgCl2 for Rv1708 and Rv2232, respectively), 500 μM Tris (2-carboxyethyl) phosphine hydrochloride, and 1 mM ATP. The reactions were incubated for the indicated time at 37 °C and stopped by adding SDS/PAGE sample buffer to a final 2-Mercaptoethanol concentration of 1%. The samples were separated by SDS/PAGE, transferred to nitrocellulose membrane, and probed with P-Tyr-100 antibody (Cell Signaling Technology). The blots were probed with secondary antibody goat anti-mouse IRDye 800cw (Licor) and scanned using the Odyssey digital imagining system.
32P Incorporation Kinase Assay.
Kinase reactions were carried out as above, using the indicated amounts of kinase, 5 µg GarA or FhaA substrate, 50 µM ATP, and 5 µCi [γ-32P] ATP per reaction. The reactions were incubated for the indicated time at room temperature and stopped by adding SDS/PAGE sample buffer. The samples were separated by SDS/PAGE, the gels were dried, and the 32P incorporation was visualized by autoradiography.
Mtb Strains, Culture, and Lysate Preparation.
Mtb strain H37Rv (ATCC) was grown in liquid 7H9 medium with 10% OADC in rolling cultures. The overexpressing strains tet-pknB, tet-pknB Lsy40Ala, and tet-pknB Tyr182Phe were generated as previously described (35). For cfu assays, cultures at OD600 0.05 were induced with 20ng/mL ATc and were plated daily for 7 d. For LC-MS/MS analysis, cultures were harvested by centrifugation for 5 min at 4000 × g and were washed once in PBS. Cells were pelleted and resuspended in 50 mM NH4HCO3/50% (vol/vol) 2,2,2-Trifluoroethanol. Cells were lysed by bead beating and inactivated by heat killing. M. smegmatis strain MC2 155 was grown and lysate was prepared as described above for Mtb.
Tryptic Digestion and Phosphopeptide Enrichment.
Proteins were reduced with 5 mM dithiothreitol (DTT) for 30 min at 55 °C, alkylated with 14 mM iodoacetamide (IAM) for 30 min at room temperature in darkness, followed by quenching of unreacted IAM with 5 mM DTT. The sample was diluted with 125 mM NH4HCO3 and digested with trypsin at an enzyme:substrate ratio of 1:50 at 37 °C overnight. The digest was dried under centrifugal evaporation (Savant), resolubilized in 500 µL 1% trifluoroacetic acid, and peptides were desalted with tC18 SepPak cartridges (Waters). To enrich for phosphopeptides, we performed IMAC using PHOS-Select Iron Affinity Gel (Sigma-Aldrich) following a similar protocol (41) and then the Titansphere Phos-TiO kit (GL Sciences Inc.). Before MS analysis, peptides were desalted with a tC18 SepPak cartridge. Alternatively, Tyr-phosphorylated proteins were purified by immunoprecipitation with a 1:1 mixture of the anti-pTyr antibodies PY99 agarose and 4G10 Sepharose before tryptic digestion. After three washes in lysis buffer, protein was eluted with 0.2% trifluoroacetic acid, and samples were processed further as described above. Recombinant proteins were reduced with 1 mM DDT, alkylated with 10 mM IAM, and quenched with 5 mM DDT. Samples were diluted 1:1 with 125 mM NH4HCO3 and digested with trypsin (1:50) overnight.
LC-MS/MS Analysis.
Peptides were analyzed on either an LTQ-Velos Orbitrap or an LTQ-Velos Pro Orbitrap Elite (Thermo Fisher Scientific) mass spectrometer equipped with a nano LC system. Peptide separation was performed on C18 (ReproSil-Pur C18-AQ, 120 Å, 3 μm; Dr. Maisch GmbH, Germany) capillary columns packed in house using 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B) with a gradient from 3–25% (vol/vol) B in 90 min at a flow rate of 0.3 µL/min. Survey full-scan MS spectra were acquired in the mass range m/z 400–2,000 in the Orbitrap analyzer at a resolution of 60,000. The 10 most intense ions determined in the survey scan were fragmented by collision-induced dissociation in the LTQ. Dynamic exclusion was enabled. In addition, a portion of phospho-enriched Mtb lysate was analyzed on a Q Exactive (Thermo Fisher Scientific) by higher-energy collisional dissociation and nano LC conditions as described above.
Data Analysis.
Instrument-native data files were converted to mzML or mzXML files using the ProteoWizard msconvert program (42, 43). MS/MS spectra were associated with peptide sequences using SEQUEST (version UW2012.01.2) and a database comprising 3,996 protein entries plus common contaminants and a sequence-shuffled decoy counterpart. Peptides were allowed to be semitryptic with up to two internal cleavage sites. The search parameters included a fixed modification of +57.021464 to account for carbamidomethylated cysteines and differential modifications of +15.9949 for oxidized methionines and +79.966331 for phosphorylated Ser, Thr, and Tyr. The search results were processed with the TPP (v4.6 rev1 and 4.6.2) including PeptideProphet (44) and iProphet (45). Public MS data were analyzed using the TPP with open source SEQUEST called Comet (46) and X!Tandem (47).
Extended experimental procedures are provided in SI Experimental Procedures.
Supplementary Material
Acknowledgments
We thank Zhi Sun and David Shteynberg for expert assistance and Seemay Chou for support and discussions. This work has been funded in part with federal funds from National Science Foundation Major Research Instrumentation Grant 0923536, by funds from the American Recovery and Reinvestment Act through Grant R01 HG005805 from the National Human Genome Research Institute, Grant S10 RR027584 from the National Institute of General Medical Sciences, Grant 2P50 GM076547 from the Center for Systems Biology, and Grant OPP1039684 from the Global Health Initiative of the Bill & Melinda Gates Foundation. C.G. was supported by the Paul G. Allen Family Foundation Grant 8999 and by National Institute of Allergy and Infections Diseases Grant 10802993. C.O. is the recipient of an American Society of Microbiology Robert D. Watkins Graduate Research Fellowship and a Bank of America Endowed Minority Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Raw mass spectrometry data have been deposited in the PeptideAtlas (www.peptideatlas.org/PASS/PASS00489).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1323894111/-/DCSupplemental.
References
- 1.Pérez J, Castañeda-García A, Jenke-Kodama H, Müller R, Muñoz-Dorado J. Eukaryotic-like protein kinases in the prokaryotes and the myxobacterial kinome. Proc Natl Acad Sci USA. 2008;105(41):15950–15955. doi: 10.1073/pnas.0806851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fernandez P, et al. The Ser/Thr protein kinase PknB is essential for sustaining mycobacterial growth. J Bacteriol. 2006;188(22):7778–7784. doi: 10.1128/JB.00963-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wehenkel A, et al. Mycobacterial Ser/Thr protein kinases and phosphatases: Physiological roles and therapeutic potential. Biochim Biophys Acta. 2008;1784(1):193–202. doi: 10.1016/j.bbapap.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 4.Bender MH, Cartee RT, Yother J. Positive correlation between tyrosine phosphorylation of CpsD and capsular polysaccharide production in Streptococcus pneumoniae. J Bacteriol. 2003;185(20):6057–6066. doi: 10.1128/JB.185.20.6057-6066.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ilan O, et al. Protein tyrosine kinases in bacterial pathogens are associated with virulence and production of exopolysaccharide. EMBO J. 1999;18(12):3241–3248. doi: 10.1093/emboj/18.12.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vincent C, et al. Relationship between exopolysaccharide production and protein-tyrosine phosphorylation in gram-negative bacteria. J Mol Biol. 2000;304(3):311–321. doi: 10.1006/jmbi.2000.4217. [DOI] [PubMed] [Google Scholar]
- 7.Wugeditsch T, et al. Phosphorylation of Wzc, a tyrosine autokinase, is essential for assembly of group 1 capsular polysaccharides in Escherichia coli. J Biol Chem. 2001;276(4):2361–2371. doi: 10.1074/jbc.M009092200. [DOI] [PubMed] [Google Scholar]
- 8.Chow K, Ng D, Stokes R, Johnson P. Protein tyrosine phosphorylation in Mycobacterium tuberculosis. FEMS Microbiol Lett. 1994;124(2):203–207. doi: 10.1111/j.1574-6968.1994.tb07285.x. [DOI] [PubMed] [Google Scholar]
- 9.Bach H, Wong D, Av-Gay Y. Mycobacterium tuberculosis PtkA is a novel protein tyrosine kinase whose substrate is PtpA. Biochem J. 2009;420(2):155–160. doi: 10.1042/BJ20090478. [DOI] [PubMed] [Google Scholar]
- 10.Kelkar DS, et al. Proteogenomic analysis of Mycobacterium tuberculosis by high resolution mass spectrometry. Mol Cell Proteomics. 2011;10(12):M111.011627. doi: 10.1074/mcp.M111.011627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prisic S, et al. Extensive phosphorylation with overlapping specificity by Mycobacterium tuberculosis serine/threonine protein kinases. Proc Natl Acad Sci USA. 2010;107(16):7521–7526. doi: 10.1073/pnas.0913482107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gnad F, Gunawardena J, Mann M. PHOSIDA 2011: The posttranslational modification database. Nucleic Acids Res. 2011;39(Database issue):D253–D260. doi: 10.1093/nar/gkq1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee DC, Jia Z. Emerging structural insights into bacterial tyrosine kinases. Trends Biochem Sci. 2009;34(7):351–357. doi: 10.1016/j.tibs.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 14.Koul A, et al. Cloning and characterization of secretory tyrosine phosphatases of Mycobacterium tuberculosis. J Bacteriol. 2000;182(19):5425–5432. doi: 10.1128/jb.182.19.5425-5432.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong D, Chao JD, Av-Gay Y. Mycobacterium tuberculosis-secreted phosphatases: From pathogenesis to targets for TB drug development. Trends Microbiol. 2013;21(2):100–109. doi: 10.1016/j.tim.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 16.Ballif BA, Villén J, Beausoleil SA, Schwartz D, Gygi SP. Phosphoproteomic analysis of the developing mouse brain. Mol Cell Proteomics. 2004;3(11):1093–1101. doi: 10.1074/mcp.M400085-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Olsen JV, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127(3):635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 18.Ge R, Shan W. Bacterial phosphoproteomic analysis reveals the correlation between protein phosphorylation and bacterial pathogenicity. Genomics Proteomics Bioinformatics. 2011;9(4-5):119–127. doi: 10.1016/S1672-0229(11)60015-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macek B, et al. Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Mol Cell Proteomics. 2008;7(2):299–307. doi: 10.1074/mcp.M700311-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Hansen AM, et al. The Escherichia coli phosphotyrosine proteome relates to core pathways and virulence. PLoS Pathog. 2013;9(6):e1003403. doi: 10.1371/journal.ppat.1003403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jadeau F, et al. BYKdb: The Bacterial protein tYrosine Kinase database. Nucleic Acids Res. 2012;40(Database issue):D321–D324. doi: 10.1093/nar/gkr915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ansong C, et al. Identification of widespread adenosine nucleotide binding in Mycobacterium tuberculosis. Chem Biol. 2013;20(1):123–133. doi: 10.1016/j.chembiol.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arora G, et al. Unveiling the novel dual specificity protein kinases in Bacillus anthracis: Identification of the first prokaryotic dual specificity tyrosine phosphorylation-regulated kinase (DYRK)-like kinase. J Biol Chem. 2012;287(32):26749–26763. doi: 10.1074/jbc.M112.351304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson DL, Mahony JB. Chlamydophila pneumoniae PknD exhibits dual amino acid specificity and phosphorylates Cpn0712, a putative type III secretion YscD homolog. J Bacteriol. 2007;189(21):7549–7555. doi: 10.1128/JB.00893-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deutsch EW, et al. A guided tour of the Trans-Proteomic Pipeline. Proteomics. 2010;10(6):1150–1159. doi: 10.1002/pmic.200900375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kapp EA, et al. Mining a tandem mass spectrometry database to determine the trends and global factors influencing peptide fragmentation. Anal Chem. 2003;75(22):6251–6264. doi: 10.1021/ac034616t. [DOI] [PubMed] [Google Scholar]
- 27.Olsen JV, et al. Higher-energy C-trap dissociation for peptide modification analysis. Nat Methods. 2007;4(9):709–712. doi: 10.1038/nmeth1060. [DOI] [PubMed] [Google Scholar]
- 28.Zhang G, Neubert TA. Comparison of three quantitative phosphoproteomic strategies to study receptor tyrosine kinase signaling. J Proteome Res. 2011;10(12):5454–5462. doi: 10.1021/pr200697x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grundner C, Gay LM, Alber T. Mycobacterium tuberculosis serine/threonine kinases PknB, PknD, PknE, and PknF phosphorylate multiple FHA domains. Protein Sci. 2005;14(7):1918–1921. doi: 10.1110/ps.051413405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roumestand C, et al. Structural insight into the Mycobacterium tuberculosis Rv0020c protein and its interaction with the PknB kinase. Structure. 2011;19(10):1525–1534. doi: 10.1016/j.str.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 31.Macek B, et al. The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol Cell Proteomics. 2007;6(4):697–707. doi: 10.1074/mcp.M600464-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Schubert OT, et al. The Mtb proteome library: A resource of assays to quantify the complete proteome of Mycobacterium tuberculosis. Cell Host Microbe. 2013;13(5):602–612. doi: 10.1016/j.chom.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Durán R, et al. Conserved autophosphorylation pattern in activation loops and juxtamembrane regions of Mycobacterium tuberculosis Ser/Thr protein kinases. Biochem Biophys Res Commun. 2005;333(3):858–867. doi: 10.1016/j.bbrc.2005.05.173. [DOI] [PubMed] [Google Scholar]
- 34.Villarino A, et al. Proteomic identification of M. tuberculosis protein kinase substrates: PknB recruits GarA, a FHA domain-containing protein, through activation loop-mediated interactions. J Mol Biol. 2005;350(5):953–963. doi: 10.1016/j.jmb.2005.05.049. [DOI] [PubMed] [Google Scholar]
- 35.Ortega C, et al. Mycobacterium tuberculosis Ser/Thr protein kinase B mediates an oxygen-dependent replication switch. PLoS Biol. 2014;12(1):e1001746. doi: 10.1371/journal.pbio.1001746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grundner C, Cox JS, Alber T. Protein tyrosine phosphatase PtpA is not required for Mycobacterium tuberculosis growth in mice. FEMS Microbiol Lett. 2008;287(2):181–184. doi: 10.1111/j.1574-6968.2008.01309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bach H, Papavinasasundaram KG, Wong D, Hmama Z, Av-Gay Y. Mycobacterium tuberculosis virulence is mediated by PtpA dephosphorylation of human vacuolar protein sorting 33B. Cell Host Microbe. 2008;3(5):316–322. doi: 10.1016/j.chom.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 38.Kremer L, et al. Galactan biosynthesis in Mycobacterium tuberculosis. Identification of a bifunctional UDP-galactofuranosyltransferase. J Biol Chem. 2001;276(28):26430–26440. doi: 10.1074/jbc.M102022200. [DOI] [PubMed] [Google Scholar]
- 39.Sambou T, et al. Capsular glucan and intracellular glycogen of Mycobacterium tuberculosis: Biosynthesis and impact on the persistence in mice. Mol Microbiol. 2008;70(3):762–774. doi: 10.1111/j.1365-2958.2008.06445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gee CL, et al. A phosphorylated pseudokinase complex controls cell wall synthesis in mycobacteria. Sci Signal. 2012;5(208):ra7. doi: 10.1126/scisignal.2002525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Villén J, Gygi SP. The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat Protoc. 2008;3(10):1630–1638. doi: 10.1038/nprot.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kessner D, Chambers M, Burke R, Agus D, Mallick P. ProteoWizard: Open source software for rapid proteomics tools development. Bioinformatics. 2008;24(21):2534–2536. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martens L, et al. mzML - a community standard for mass spectrometry data. Mol Cell Proteomics. 2011;10(1):R110.000133. doi: 10.1074/mcp.R110.000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74(20):5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 45.Shteynberg D, et al. iProphet: Multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates. Mol Cell Proteomics. 2011;10(12):M111.007690. doi: 10.1074/mcp.M111.007690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eng JK, Jahan TA, Hoopmann MR. Comet: An open-source MS/MS sequence database search tool. Proteomics. 2013;13(1):22–24. doi: 10.1002/pmic.201200439. [DOI] [PubMed] [Google Scholar]
- 47.Craig R, Beavis RC. TANDEM: Matching proteins with tandem mass spectra. Bioinformatics. 2004;20(9):1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.