Significance
This study demonstrates the widespread distribution of nonribosomal peptide synthetase and modular polyketide synthase biosynthetic pathways across the three domains of life, by cataloging a total of 3,339 gene clusters from 2,699 genomes. Our analysis suggests that noncanonical nonmodular biosynthetic enzymes are common in bacteria. Proteobacteria, Actinobacteria, Firmicutes, and Cyanobacteria in bacteria and Ascomycota in fungi contained higher number of these gene clusters and are likely to produce a wide variety of nonribosomal peptide and polyketide types of natural products. The data generated here provide a basis for the exploration of nonribosomal peptide and polyketide biosynthetic capacity and present a compelling wealth of new information for natural product discovery.
Keywords: biosynthetic gene cluster, data mining, distribution, bioactive compound
Abstract
Nonribosomal peptides and polyketides are a diverse group of natural products with complex chemical structures and enormous pharmaceutical potential. They are synthesized on modular nonribosomal peptide synthetase (NRPS) and polyketide synthase (PKS) enzyme complexes by a conserved thiotemplate mechanism. Here, we report the widespread occurrence of NRPS and PKS genetic machinery across the three domains of life with the discovery of 3,339 gene clusters from 991 organisms, by examining a total of 2,699 genomes. These gene clusters display extraordinarily diverse organizations, and a total of 1,147 hybrid NRPS/PKS clusters were found. Surprisingly, 10% of bacterial gene clusters lacked modular organization, and instead catalytic domains were mostly encoded as separate proteins. The finding of common occurrence of nonmodular NRPS differs substantially from the current classification. Sequence analysis indicates that the evolution of NRPS machineries was driven by a combination of common descent and horizontal gene transfer. We identified related siderophore NRPS gene clusters that encoded modular and nonmodular NRPS enzymes organized in a gradient. A higher frequency of the NRPS and PKS gene clusters was detected from bacteria compared with archaea or eukarya. They commonly occurred in the phyla of Proteobacteria, Actinobacteria, Firmicutes, and Cyanobacteria in bacteria and the phylum of Ascomycota in fungi. The majority of these NRPS and PKS gene clusters have unknown end products highlighting the power of genome mining in identifying novel genetic machinery for the biosynthesis of secondary metabolites.
Nonribosomal peptides and polyketides are two diverse families of natural products with a broad range of biological activities and pharmacological properties (1). They include toxins, siderophores, pigments, antibiotics, cytostatics, and immunosuppressants (2, 3). Nonribosomal peptide and polyketide natural products have remarkably diverse structures and can be linear or cyclic or have branched structures (4). They can be further reengineered to produce complex products with exotic chemical structures and biological activities (5).
Nonribosomal peptides and polyketides are synthesized on large nonribosomal peptide synthetase (NRPS) and polyketide synthase (PKS) enzyme complexes, respectively. PKSs are currently classified into three types that differ in their organization of catalytic domains (6). Type I PKSs are large multidomain enzymes using a modular strategy, with each module being comprised of catalytic domains responsible for recognition, activation, and condensation of acyl-CoA (7). The catalytic sites of type II and type III PKSs are organized into separate proteins (6, 8). NRPSs are usually defined as modular multidomain enzymes (7). However, a nonmodular NRPS enzyme, a stand-alone peptidyl carrier protein (BlmI) from the bleomycin gene cluster, has been reported (9). Nonmodular NRPS enzymes are found in well-known siderophore biosynthetic pathways: e.g., EntE (adenylation), VibH (condensation), and VibE (adenylation) in enterobactin and vibriobactin clusters, respectively (10). The condensation, adenylation, and acyl carrier domains for brucebactin biosynthesis in Brucella abortus strain 2308 are encoded as fully separated proteins (11); thus, these NRPSs could be considered to have type II architecture. The arrangement of modules within the NRPS and type I PKS enzymes often determines the number and order of the monomer constituents of the product (12), despite deviations in module iteration (13, 14) and skipping (15). A growing number of gene clusters encoding both NRPSs and type I PKSs have been identified for biosynthesis of complex natural products (16).
The bulk of natural products in clinical use today come from a handful of bacterial and fungal lineages (17–21). However, genomics studies imply that the ability to make these compounds is much more widespread (22–24). To gain insights into the occurrence and distribution of the ability to produce nonribosomal peptides and polyketides, we undertook a systematic genome-mining study. Here, we show the widespread occurrence of NRPS and PKS genetic machinery across the three domains of life with the discovery of 3,339 gene clusters from 991 organisms, by examining a total of 2,699 genomes. Our data mining further revealed that more than half of the NRPS and type I PKS enzymes have a nonmodular composition. A total of 314 gene clusters that are comprised mostly of these nonmodular enzymes were discovered in noncanonical organizations, which deviate from the present definition.
Results
Widespread Distribution of NRPSs and Type I PKSs.
Our survey demonstrated the widespread distribution of NRPS and type I PKS biosynthetic pathways in all three domains of life (Table 1 and Fig. 1). A total of 3,339 NRPS, PKS, and hybrid NRPS/PKS gene clusters, which amount to 102.35 Mb in size, were discovered by mining of 15.72 Gb of genomic sequences from 2,699 organisms (Table 1 and Datasets S1 and S2). The majority of the gene clusters (2,976, 89%) were detected in bacteria. They were less frequent in eukarya and rare in archaea.
Table 1.
Summary of NRPS/PKS gene clusters and genomes analyzed in this study
| Domain | Gene cluster | Genome | ||||
| No. | No. of proteins | No. of domains | Cumulative size, Mb | No. | Cumulative size, Gb | |
| Bacteria | 2,976 | 15,889 | 56,269 | 95.88 | 2,478 | 8.69 |
| Archaea | 3 | 6 | 27 | 0.03 | 160 | 0.38 |
| Eukarya | 360 | 699 | 3,304 | 6.44 | 61 | 6.65 |
| Total | 3,339 | 16,594 | 59,600 | 102.35 | 2,699 | 15.72 |
Fig. 1.
The widespread distribution of NRPSs and PKSs across the three domains of life. The phylogenetic analysis is based on 16S or 18S rRNA genes from selected organisms (Table S3) for representative phyla in bacteria and eukarya, and classes in archaea. The midpoint tree was constructed by PhyML 3.0 using the GTR substitution model and with 100 bootstrap replications for each branch. The lineages containing both NRPSs and PKSs, or hybrid NRPS-PKS enzymes are indicated in red, the ones containing only NRPSs are indicated in green, and those containing only PKSs are in blue. The numbers of examined genomes and discovered gene clusters for each phylum or class are next to the taxon name and separated by a slash. The biosynthetic pathways of NRPS and modular PKS not only were found densely distributed among bacterial phyla and fungi, but also were found in animals, plants, and protists in eukarya and archaean strains.
NRPS and PKS gene clusters show a nonuniform distribution in bacteria (Fig. 1 and Table S1). They are common in the phyla Proteobacteria, Actinobacteria, Firmicutes, and Cyanobacteria. However, the gene clusters are noticeably absent in some lineages with small genomes, such as the phyla of Tenericutes and Spirochaetes, which have many sequenced genomes of intracellular pathogens. In addition, we showed that Chlamydiae, Deinococcus-Thermus, Chlorobi, Verrucomicrobia, Nitrospirae, and Elusimicrobia could be potential producers of nonribosomal peptides and polyketides. There appeared a 2-Mb threshold for genomes with the gene clusters (Fig. S1). Genome size seems not to be the only requirement because some bacteria with large genomes (>8 Mb) still lack these gene clusters (Fig. S1). We found that the average numbers of clusters are highly correlated (P < 0.00001) with the average genome sizes among strains in the orders of Proteobacteria, Actinobacteria, Firmicutes, and Cyanobacteria (Fig. S2). NRPS and PKS gene clusters were found throughout the eukaryotic lineages. Fungi, known producers of nonribosomal peptides and polyketides, were a rich source of NRPSs and type I PKSs. A total of 307 gene clusters were located from 12 strains in the phylum Ascomycota (Table S2). However, no NRPS or PKS gene clusters were found in the other two Microsporidia and three Basidiomycota strains (Fig. 1). NRPS and PKS gene clusters were also distributed sporadically in many other lineages of protist, plant, and animal (Fig. 1). Both NRPSs and PKSs were found from phyla of Amoebozoa, Dinoflagellata, Apicomplexa, Nematoda, Arthropoda, and Mollusca. In addition, we also located NRPS gene clusters in phyla of alga Stramenopiles and Chlorophyta, as well as PKSs from slime-molds and metazoan phyla of Annelida and Chordata. Archaea appears to rarely contain these biosynthetic pathways in the 128 genomes analyzed in this study. Only three NRPS gene clusters were found in two strains, which belong to classes Methanobacteria and Methanomicrobia, respectively (Fig. 1). Meanwhile, modular PKSs were absent from these sequenced archaean genomes.
Overall, despite biases in the number of genomes available for each lineage, NRPS and type I PKS gene clusters were more frequent in the phyla of Proteobacteria, Firmicutes, Actinobacteria, and Cyanobacteria in bacteria (Table S1) and Ascomycota in fungi (Table S2). The NRPS and PKS gene clusters identified in this study can be accessed from an associated web database (http://npgc.biocenter.helsinki.fi).
Gene-Cluster Composition Reveals High Diversity of NRPS and PKS Biosynthetic Machineries and Their Close Relationship.
A total of 3,339 gene clusters were detected in this genome mining study. By examining the composition of the biosynthetic and tailoring enzymes of these gene clusters, we collectively obtained 59,600 domains (Table 1), among which the core biosynthetic domains accounted for the majority (Fig. S3).
These gene clusters were classified into NRPS, PKS, and hybrid types, according to the presence of core domains of NRPS, PKS, or both systems (Fig. 2). One third (1,147, 34.4%) of gene clusters belonged to the hybrid type and encoded 462 hybrid proteins that contain both NRPS and PKS core domains (Fig. 2 and Dataset S3). The hybrid clusters tend to be larger and possess more domains, compared with stand-alone NRPS and PKS gene clusters (Fig. S4). Therefore, they may produce more complex products than NRPS and PKS clusters on their own. Moreover, nearly all domain types are presented in hybrid gene clusters, especially in bacteria where domains from hybrid gene clusters are more frequently observed than in fungi (Fig. S3). For example, a total of 113 bacterial gene clusters were found with free-standing acyltransferases (AT), which iteratively load the monomers in trans for other modular PKSs lacking an AT domain (25). They are almost all the hybrid type, illustrated by the fact that their docking and dehydratase domains are entirely from hybrid clusters (Fig. S3). Among the 298 AT-less PKS module-containing enzymes found from these gene clusters, 117 are hybrid enzymes with NRPS modules. Interestingly, we also found three enzymes in which the normal PKS module and the AT-less module are fused together; one of them (SBI_00671) even has a middle NRPS module residing between normal and AT-less PKS modules.
Fig. 2.
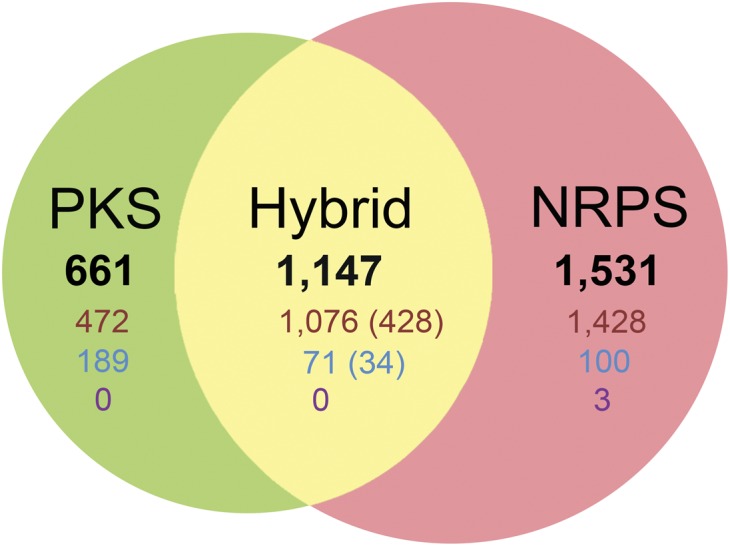
A Venn diagram of PKS, NRPS, and hybrid gene-cluster numbers. The gene-cluster numbers of bacteria, archaea, and eukarya are shown in red, purple, and blue, respectively. The values in parentheses represent the numbers of hybrid enzymes that contain both NRPS and PKS core domains.
The NRPS/PKS gene clusters displayed highly diverse organizational architectures, based on their domain and protein composition (Fig. S5). The number of biosynthetic enzymes and domains of these gene clusters was mostly fewer than 10 and 20, respectively. On the other hand, some hybrid clusters coded for over 100 domains (Fig. S4). Based on our data, the largest PKS (MULP_065, 17,019 aa) has nine modules constituting 47 domains whereas the largest NRPS (plu2670, 16,367 aa) was found with 15 modules consisting of 46 domains, and the longest hybrid enzyme (MXAN_3779, 14,274 aa) possesses 39 domains fused into 1 PKS and 11 NRPS modules. These enzymes are in a highly modular structure and may represent the upper limits of the NRPS and PKS modularity level.
Evolutionary Analysis of NRPS Biosynthetic Enzymes.
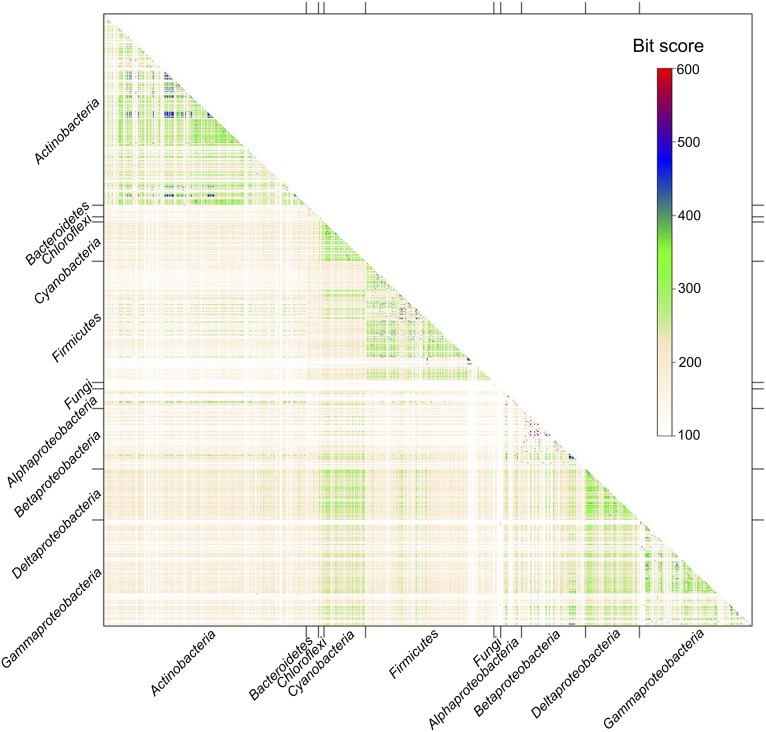
Phylogenetic analysis was conducted based on the sequences of the most abundant condensation domain type LCL, which catalyzes the formation of a peptide bond between two adjacent l-amino acids (Fig. S6). The condensation domains from archaea, bacteria, fungi, and other eukaryotic organisms did not form monophyletic clades and were instead mixed. A heat map showing similarities between l-amino acid condensation domains among the major lineages was constructed based on 4,087 LCL domain sequences (Fig. 3). Homologies could be evidently observed within the lineages of Actinobacteria, Cyanobacteria, Firmicutes, Deltaproteobacteria,and Gammaproteobacteria. The high levels of similarity (indicated by the regions in blue and red in Fig. 3) clearly demonstrate events of inner lineage duplication. Moreover, the data also suggest horizontal transfers among these lineages: for example, between Cyanobacteria and Proteobacteria.
Fig. 3.
A heat map showing sequence similarities between l-amino acid condensation domains among the major lineages. Sequence similarities are measured by the bit scores of the reciprocal bla2seq alignments and indicated in color as the scheme shows. Bit scores are shown in red if they equal 600 or more and in white if they equal 100 or less. The self-hits in the diagonal line were omitted for clarity.
Common Occurrence of Nonmodular Biosynthetic Enzymes in Bacteria.
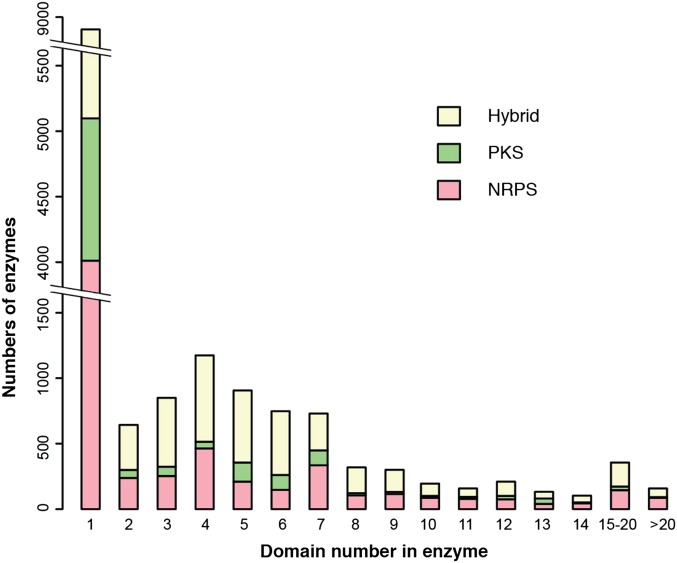
There are a total of 15,889 biosynthetic and tailoring enzymes identified from the 2,976 bacterial gene clusters (Table 1). Surprisingly, nonmodular enzymes showed an extraordinary abundance (8,906) compared with others with multiple domains (Fig. 4). Among these nonmodular enzymes, 4,012 are from NRPS gene clusters, 1,086 from PKS, and 3,808 from hybrid clusters. Nearly every domain type was found as a nonmodular enzyme (Fig. S7). Some modification domains of aminotransferase and polyketide cyclase were found almost exclusively as separate proteins. Once again, many of the nonmodular enzymes are also present in hybrid clusters, such as the AT proteins from trans-AT clusters (Fig. S7).
Fig. 4.
Distribution of bacterial NRPS/PKS enzymes ranked according to their domain numbers. A total of 15,889 enzymes were found in 2,976 gene clusters in bacteria. These enzymes were grouped according to the number of their domains. The nonmodular enzymes showed high levels of abundance (8,906) compared with other multidomain enzymes. Enzymes from NRPS gene clusters are indicated in red, PKS gene clusters in green, and hybrid gene clusters in yellow.
Most of the nonmodular enzymes are found together with multidomain enzymes in gene clusters. However, one fifth of them (1,803) occurred in close genomic vicinity with each other and form into gene clusters (Fig. 5). Our data mining located a total of 314 (10.6%) bacterial NRPS and PKS gene clusters that are comprised mostly of nonmodular enzymes (Dataset S4; also indicated by the blue circle in Fig. S5). These gene clusters are structurally similar to type II PKSs (26). The majority (260, 82.8%) of them are the NRPS type. There are also 37 hybrid clusters, in which NRPS and type I PKS nonmodular enzymes were present together (Fig. 5), in addition to 17 PKS clusters. Although these type II-like gene clusters were found in multiple lineages of bacteria (Dataset S4), many of the NRPS clusters are from proteobacteria and appear to be related to siderophore biosynthesis. For example, a total of 16 putative brucebactin gene clusters, in which the condensation, adenylation, and carrier domains are presented as separated proteins, were found from sequenced Brucella genomes.
Fig. 5.

Examples of gene clusters comprised of nonmodular enzymes. A total of 314 gene clusters formed mostly by nonmodular enzymes were found in bacteria. Examples of two hybrid gene clusters and a putative acinetobactin biosynthesis gene cluster are shown. The domains are indicated by abbreviations as adenylation (A), peptidyl carrier domain (PCP), condensation (C), acyltransferase (AT), acyl carrier or peptidyl carrier domain (PP), ketosynthase (KS), thioesterase (TE), epimerization (E), heterocyclization (H), ketoreductase (KR), enoylreductase (ER), aminotransferase (Amino), and 4′-phosphopantetheinyl transferase (ACPS). The ABC transport system proteins are indicated in blue, other tailoring enzymes in light green, and hypothetical proteins in gray.
Phylogenetic Analysis of the Siderophore Biosynthesis NRPS Machineries with Gradient-Domain Organizations.
Our analysis showed that the domains of NRPS and type I PKS display gradient organizations among gene clusters, from fully dissociated nonmodular enzymes to a highly modular mode in massive multidomain enzymes. For example, the iron-chelating siderophore acinetobactin biosynthesis gene clusters in Acinetobacter baumannii were found with many nonmodular enzymes and grouped as type II-like gene clusters in this study (Dataset S4). The mono- and didomain enzymes of these gene clusters can be mapped to the six-domain VibF in the vibriobactin biosynthesis gene cluster (27). Recently, the well-conserved tandem heterocyclization domains were also found in the siderophore serratiochelin gene clusters (28). These siderophores share one or more 2,3-dihydroxypheny-5-methyloxazoline-acyl groups, which is synthesized by the domains of modular VibF in Vibrio cholerae (29), and also likely by similar domains but organized as SchF1, -2, and -3 in Serratia (28) and BsaD, -A, and -B in A. baumannii (27) (Fig. 6). Although the biosynthesis of parabactin in Paracoccus denitrificans is still unknown, its 2-hydroxypheny-5-methyloxazoline-acyl group appears to be synthesized by the shown modular enzyme (Fig. 6). Gene clusters with similar domains were also detected from strains of Pseudomonas, Marinomonas, Halomonas, and Agrobacterium, which may also produce siderophores with similar structures (Fig. 6). The first condensation domain C1 of VibF was missing from some gene clusters, likely due to its dispensability in biosynthesis (29).
Fig. 6.
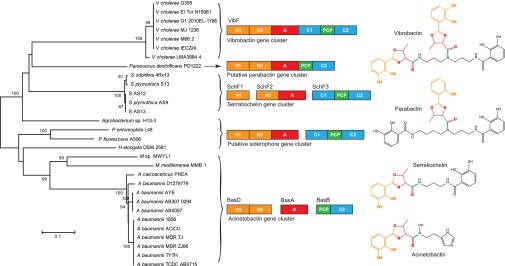
Phylogenetic analysis of condensation domains C2 from siderophore biosynthetic enzymes with gradient-domain organizations. The neighbor-joining tree was constructed by MEGA5.1 (46) using Poisson model and with 100 bootstrap replications for each branch. These siderophore biosynthetic pathways share an unusual pair of tandem heterocyclization domains and others with similar composition but in gradient organizations, which are congruent with the phylogeny of the C2 domains. These domains are responsible for the biosynthesis of the 2,3-dihydroxypheny-5-methyloxazoline-acyl group, which is synthesized by the double heterocyclization (H) domains from a 2,3-dihydroxybenzoate and an l-threonine that is activated by the adenylation (A) domain, and fused by the condensation (C) domain C2 with other substrates. The activated l-threonine and the derived group are both tethered by the peptidyl carrier (PCP) domain.
A neighbor-joining tree was constructed based on the C2 domains of VibF in V. cholerae and their homologs from other siderophore gene clusters (Fig. 6), which possess similar domains that, however, arranged in a gradient of organizations, from fully modular enzymes to nonmodular separate proteins (Fig. 6). The phylogeny of the C2 domains is congruent with the gradient-domain organizations of these siderophore biosynthetic pathways.
Discussion
Nonribosomal peptides and polyketides account for a significant portion of known natural products, which are the major source of drug candidates (30). Over 80% of these natural products were derived from fungi and actinomycetes (31). Recent studies showed the rich sources of nonribosomal peptides and polyketides in bacterial lineages of myxobacteria (18), pseudomonads (32), cyanobacteria (17), and streptomycetes (33). Genomic analysis further revealed the presence of NRPSs and PKSs in archaea (22), metazoa (23), and dinoflagellates (24). In this systematic study, we have demonstrated that NRPS and type I PKS biosynthetic pathways are widely distributed in bacteria and found sporadically in archaea and eukarya (Fig. 1 and Table 1). We have shown the high frequency of these gene clusters in bacteria and fungi and their spreading into specific archaean classes and many eukaryotic phyla. The accumulation of NRPS and PKS gene clusters in some bacteria phyla may partly reflect the biased distribution of sequenced genomes toward pathogens and environmentally important strains (19). Nevertheless, there is no doubt that this distribution will be inevitably expanded, because of the exponential increasing of genome data.
The archaean and eukaryotic NRPSs and PKSs appeared to be acquired from bacteria via horizontal gene transfer according to sequence and phylogenetic analysis in this study (Fig. S6) and others (23, 34). However, more evidence is needed to prove whether they are recently spread from bacteria or are remnants of ancient pathways after genome reduction. On the other hand, sequence analysis indicates that the evolution of nonribosomal machineries is consistent with the phylogenetic classifications based on ribosomal sequences (Fig. 3). In addition, the plasmid-borne gene clusters (Table S1) might be associated with horizontal gene transfer events in bacteria.
A previous study analyzing 141 strains implied that bacterial strains with PKSs usually have genomes larger than 2 Mb (26). Based on a much larger dataset, our analysis conclusively indicates that this threshold applies to both NRPSs and PKSs. We therefore suggest that bacterial strains with genomes fewer than 2 Mb may not be able to sustain these secondary metabolic pathways, perhaps due to the extra metabolic burdens. Myxococcales (proteobacteria), Actinomycetales (actinobacteria), Pleurocapsales, and Nostocales (cyanobacteria) are rich sources of NRPS and PKS gene clusters (Fig. S2); thus, new genomes from these orders would most likely provide novel natural products.
NRPSs and type I PKSs produce distinct groups of natural products with different enzyme systems, despite their common assembly line-like mechanism (7). Surprisingly, a high number of hybrid NRPS-PKS gene clusters were found in this study (Fig. 2), and all NRPS/PKS domain types are presented in hybrid gene clusters (Fig. S3). These analyses demonstrated their close relationship, which might be an outcome of long-term convergent interaction of the two systems and might yield a higher magnitude of structural diversity for the derived hybrid peptide-polyketide compounds that could be exploited as potential biological agents and drug leads (35, 36).
By definition, type I PKSs are modular enzyme systems with multidomain organization (7) whereas type II and III PKSs are nonmodular enzymes (6, 8). In contrast, NRPSs are defined only in a modular synthetic scheme (7) although there are occurrences of many NRPSs with a single domain (9, 10). In this study, we revealed that such nonmodular enzymes of NRPS and type I PKS are abundantly present in bacteria (Fig. 4), including both core and tailoring domains of the two systems (Fig. S7). Moreover, our analysis showed that 18% (260 out of 1,428) of NRPS gene clusters in bacteria were found in a type II-like organization (Dataset S4). Unlike NRPS-like enzymes (37, 38), these type II-like NRPS clusters have a complete core domain set, such as the identified putative brucebactin gene clusters that contain one fully dissociated NRPS module. Previously, reclassification of PKSs had been proposed (39), due to the discoveries of transition states between type I and II PKSs, the trans-AT PKS systems (25), and that between type II and III PKSs shown to be independent of the acyl carrier protein (40). The type II-like PKS clusters found in this study also appeared as transition states between type I and type II PKSs. They are more similar to type II PKSs than the trans-AT PKS systems, which have only a discrete AT enzyme. Note that NRPS/PKS hybrids also occurred in these clusters (Fig. 5). Consequently, the structural diversity of these enzyme systems presented here suggests that reclassification of NRPSs and PKSs is needed.
In conclusion, we have demonstrated the power of genome mining in studying natural product biosynthesis by showing the widespread distribution of NRPS/PKS gene clusters and by the finding of previously unidentified pathways. Our analysis showed that these various domain architectures from nonmodular to modular organization are evolutionarily related, indicating their similar enzymology. These type II-like gene clusters further expand the spectrum of the known NRPS and PKS machineries, which could be used in the future for natural product research.
Materials and Methods
Datasets.
A total of 2,699 genomes across three domains of life (Dataset S1) were downloaded from the National Center for Biotechnology Information genome database (ftp://ftp.ncbi.nlm.nih.gov/genomes/). To obtain reliable information, only completed genomes of bacteria, archaea, fungi, and protist were studied. Repeatedly sequenced genomes were detected by examining the strain codes, and only the newest versions were analyzed. For higher eukaryotic organisms, partial genomes were used because there are no complete genomes available.
Gene-Cluster Finding.
Two stand-alone tools for secondary metabolite biosynthetic enzyme detection were used: 2metDB (41) and antiSMASH (42) version 2.0.2. FASTA format protein sequences were used as input of 2metDB whereas GenBank format files were queried by antiSMASH. Gene-cluster organizations were determined by antiSMASH with a 20-kb genomic span. Their results were combined, clusters found by antiSMASH were compared with the hits of 2metDB output, and the clusters predicted by both tools were counted. The gene clusters were determined as the NRPS type only if the adenylation or condensation domains were present, as the PKS type only if the ketosynthase or acyltransferase domains were found, and the hybrid type if they contained both the PKS and NRPS core domains. The type II-like gene clusters were defined as at least half of the biosynthetic enzymes are nonmodular enzymes and the ratio of detected domain number versus protein number was no more than 1.5. We created a web database (http://npgc.biocenter.helsinki.fi) to deposit all identified gene clusters, which can be queried by taxonomic groups, organism names, or user-defined criteria.
Tree Building.
The rRNA sequences of representative species from the three domains of life (Table S3) were aligned by MUSCLE (43) with default parameters, and nonconserved positions were removed by Gblocks (44) (0.91b) with stringent criteria. The midpoint tree was constructed by PhyML 3.0 (45) using the GTR substitution model and with 100 bootstrap replications for each branch. The condensation domain sequences were aligned by MUSCLE (43) with default parameters. The neighbor-joining tree was constructed by MEGA5.1 (46) using Poisson model and with 100 bootstrap replications for each branch.
Heat-Map Construction.
A homologous map was constructed according to the bit scores of the reciprocal bla2seq results of the LCL domain sequences, whose lengths were examined, and the ones larger or smaller than the average size over 10% were excluded.
Supplementary Material
Acknowledgments
We thank Dr. Jouni Jokela for helpful comments on the manuscript. This work was financially supported by Academy of Finland Grants 258827 and 118637 (to K.S.) and by the Viikki Doctoral Programme in Molecular Biosciences in years 2010–2012 (H.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1401734111/-/DCSupplemental.
References
- 1.Cane DE, Walsh CT, Khosla C. Harnessing the biosynthetic code: Combinations, permutations, and mutations. Science. 1998;282(5386):63–68. doi: 10.1126/science.282.5386.63. [DOI] [PubMed] [Google Scholar]
- 2.Finking R, Marahiel MA. Biosynthesis of nonribosomal peptides1. Annu Rev Microbiol. 2004;58:453–488. doi: 10.1146/annurev.micro.58.030603.123615. [DOI] [PubMed] [Google Scholar]
- 3.Weissman KJ, Leadlay PF. Combinatorial biosynthesis of reduced polyketides. Nat Rev Microbiol. 2005;3(12):925–936. doi: 10.1038/nrmicro1287. [DOI] [PubMed] [Google Scholar]
- 4.Kopp F, Marahiel MA. Where chemistry meets biology: the chemoenzymatic synthesis of nonribosomal peptides and polyketides. Curr Opin Biotechnol. 2007;18(6):513–520. doi: 10.1016/j.copbio.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Walsh CT. The chemical versatility of natural-product assembly lines. Acc Chem Res. 2008;41(1):4–10. doi: 10.1021/ar7000414. [DOI] [PubMed] [Google Scholar]
- 6.Shen B. Polyketide biosynthesis beyond the type I, II and III polyketide synthase paradigms. Curr Opin Chem Biol. 2003;7(2):285–295. doi: 10.1016/s1367-5931(03)00020-6. [DOI] [PubMed] [Google Scholar]
- 7.Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: Logic, machinery, and mechanisms. Chem Rev. 2006;106(8):3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 8.Yu D, Xu F, Zeng J, Zhan J. Type III polyketide synthases in natural product biosynthesis. IUBMB Life. 2012;64(4):285–295. doi: 10.1002/iub.1005. [DOI] [PubMed] [Google Scholar]
- 9.Du L, Shen B. Identification and characterization of a type II peptidyl carrier protein from the bleomycin producer Streptomyces verticillus ATCC 15003. Chem Biol. 1999;6(8):507–517. doi: 10.1016/S1074-5521(99)80083-0. [DOI] [PubMed] [Google Scholar]
- 10.Crosa JH, Walsh CT. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol Mol Biol Rev. 2002;66(2):223–249. doi: 10.1128/MMBR.66.2.223-249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bellaire BH, et al. Genetic organization and iron-responsive regulation of the Brucella abortus 2,3-dihydroxybenzoic acid biosynthesis operon, a cluster of genes required for wild-type virulence in pregnant cattle. Infect Immun. 2003;71(4):1794–1803. doi: 10.1128/IAI.71.4.1794-1803.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marahiel MA, Stachelhaus T, Mootz HD. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem Rev. 1997;97(7):2651–2674. doi: 10.1021/cr960029e. [DOI] [PubMed] [Google Scholar]
- 13.Mootz HD, Schwarzer D, Marahiel MA. Ways of assembling complex natural products on modular nonribosomal peptide synthetases. ChemBioChem. 2002;3(6):490–504. doi: 10.1002/1439-7633(20020603)3:6<490::AID-CBIC490>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 14.Fisch KM. Biosynthesis of natural products by microbial iterative hybrid PKS-NRPS. RSC Adv. 2013;3(40):18228–18247. [Google Scholar]
- 15.Wenzel SC, Müller R. Formation of novel secondary metabolites by bacterial multimodular assembly lines: deviations from textbook biosynthetic logic. Curr Opin Chem Biol. 2005;9(5):447–458. doi: 10.1016/j.cbpa.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 16.Du L, Shen B. Biosynthesis of hybrid peptide-polyketide natural products. Curr Opin Drug Discov Devel. 2001;4(2):215–228. [PubMed] [Google Scholar]
- 17.Welker M, von Döhren H. Cyanobacterial peptides - nature’s own combinatorial biosynthesis. FEMS Microbiol Rev. 2006;30(4):530–563. doi: 10.1111/j.1574-6976.2006.00022.x. [DOI] [PubMed] [Google Scholar]
- 18.Wenzel SC, Müller R. Myxobacterial natural product assembly lines: fascinating examples of curious biochemistry. Nat Prod Rep. 2007;24(6):1211–1224. doi: 10.1039/b706416k. [DOI] [PubMed] [Google Scholar]
- 19.Donadio S, Monciardini P, Sosio M. Polyketide synthases and nonribosomal peptide synthetases: the emerging view from bacterial genomics. Nat Prod Rep. 2007;24(5):1073–1109. doi: 10.1039/b514050c. [DOI] [PubMed] [Google Scholar]
- 20.Doroghazi JR, Metcalf WW. Comparative genomics of actinomycetes with a focus on natural product biosynthetic genes. BMC Genomics. 2013;14:611. doi: 10.1186/1471-2164-14-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bushley KE, et al. The genome of tolypocladium inflatum: evolution, organization, and expression of the cyclosporin biosynthetic gene cluster. PLoS Genet. 2013;9(6):e1003496. doi: 10.1371/journal.pgen.1003496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leahy SC, et al. The genome sequence of the rumen methanogen Methanobrevibacter ruminantium reveals new possibilities for controlling ruminant methane emissions. PLoS ONE. 2010;5(1):e8926. doi: 10.1371/journal.pone.0008926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gladyshev EA, Meselson M, Arkhipova IR. Massive horizontal gene transfer in bdelloid rotifers. Science. 2008;320(5880):1210–1213. doi: 10.1126/science.1156407. [DOI] [PubMed] [Google Scholar]
- 24.López-Legentil S, Song B, DeTure M, Baden DG. Characterization and localization of a hybrid non-ribosomal peptide synthetase and polyketide synthase gene from the toxic dinoflagellate Karenia brevis. Mar Biotechnol (NY) 2010;12(1):32–41. doi: 10.1007/s10126-009-9197-y. [DOI] [PubMed] [Google Scholar]
- 25.Cheng YQ, Tang GL, Shen B. Type I polyketide synthase requiring a discrete acyltransferase for polyketide biosynthesis. Proc Natl Acad Sci USA. 2003;100(6):3149–3154. doi: 10.1073/pnas.0537286100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jenke-Kodama H, Sandmann A, Müller R, Dittmann E. Evolutionary implications of bacterial polyketide synthases. Mol Biol Evol. 2005;22(10):2027–2039. doi: 10.1093/molbev/msi193. [DOI] [PubMed] [Google Scholar]
- 27.Mihara K, et al. Identification and transcriptional organization of a gene cluster involved in biosynthesis and transport of acinetobactin, a siderophore produced by Acinetobacter baumannii ATCC 19606T. Microbiology. 2004;150:2587–2597. doi: 10.1099/mic.0.27141-0. [DOI] [PubMed] [Google Scholar]
- 28.Seyedsayamdost MR, et al. Mixing and matching siderophore clusters: structure and biosynthesis of serratiochelins from Serratia sp. V4. J Am Chem Soc. 2012;134(33):13550–13553. doi: 10.1021/ja304941d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marshall CG, Hillson NJ, Walsh CT. Catalytic mapping of the vibriobactin biosynthetic enzyme VibF. Biochemistry. 2002;41(1):244–250. doi: 10.1021/bi011852u. [DOI] [PubMed] [Google Scholar]
- 30.Walsh CT. Polyketide and nonribosomal peptide antibiotics: modularity and versatility. Science. 2004;303(5665):1805–1810. doi: 10.1126/science.1094318. [DOI] [PubMed] [Google Scholar]
- 31.Donadio S, et al. Sources of polyketides and non-ribosomal peptides. In: Wohlleben W, Spellig T, Müller-Tiemann B, editors. Biocombinatorial Approaches for Drug Finding. Heidelberg: Springer; 2005. pp. 19–41. [Google Scholar]
- 32.Gross H, Loper JE. Genomics of secondary metabolite production by Pseudomonas spp. Nat Prod Rep. 2009;26(11):1408–1446. doi: 10.1039/b817075b. [DOI] [PubMed] [Google Scholar]
- 33.Weber T, Welzel K, Pelzer S, Vente A, Wohlleben W. Exploiting the genetic potential of polyketide producing streptomycetes. J Biotechnol. 2003;106(2-3):221–232. doi: 10.1016/j.jbiotec.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 34.Lawrence DP, Kroken S, Pryor BM, Arnold AE. Interkingdom gene transfer of a hybrid NPS/PKS from bacteria to filamentous Ascomycota. PLoS ONE. 2011;6(11):e28231. doi: 10.1371/journal.pone.0028231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cane DE, Walsh CT. The parallel and convergent universes of polyketide synthases and nonribosomal peptide synthetases. Chem Biol. 1999;6(12):R319–R325. doi: 10.1016/s1074-5521(00)80001-0. [DOI] [PubMed] [Google Scholar]
- 36.Du L, Sánchez C, Shen B. Hybrid peptide-polyketide natural products: biosynthesis and prospects toward engineering novel molecules. Metab Eng. 2001;3(1):78–95. doi: 10.1006/mben.2000.0171. [DOI] [PubMed] [Google Scholar]
- 37.Balskus EP, Walsh CT. The genetic and molecular basis for sunscreen biosynthesis in cyanobacteria. Science. 2010;329(5999):1653–1656. doi: 10.1126/science.1193637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Forseth RR, et al. Homologous NRPS-like gene clusters mediate redundant small-molecule biosynthesis in Aspergillus flavus. Angew Chem Int Ed Engl. 2013;52(5):1590–1594. doi: 10.1002/anie.201207456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Müller R. Don’t classify polyketide synthases. Chem Biol. 2004;11(1):4–6. doi: 10.1016/j.chembiol.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 40.Kwon HJ, Smith WC, Xiang L, Shen B. Cloning and heterologous expression of the macrotetrolide biosynthetic gene cluster revealed a novel polyketide synthase that lacks an acyl carrier protein. J Am Chem Soc. 2001;123(14):3385–3386. doi: 10.1021/ja0100827. [DOI] [PubMed] [Google Scholar]
- 41.Bachmann BO, Ravel J. Chapter 8. Methods for in silico prediction of microbial polyketide and nonribosomal peptide biosynthetic pathways from DNA sequence data. Methods Enzymol. 2009;458:181–217. doi: 10.1016/S0076-6879(09)04808-3. [DOI] [PubMed] [Google Scholar]
- 42.Medema MH, et al. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011;39(Web Server issue):W339–W346. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17(4):540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 45.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 46.Tamura K, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.