Significance
The primary cilium is an organelle emanating from the cell surface, and recent evidence suggests that it regulates various cellular signaling pathways and development and that aberrations in its assembly and function could result in tumorigenesis. Skeletal muscle differentiation proceeds through a temporally defined series of events to form multinucleated myofibers, and in rhabdomyosarcomas (RMS), myoblasts fail to differentiate. However, whether primary cilia are functionally linked to normal muscle differentiation or RMS is not known. In this study we show that in skeletal myoblasts, primary cilia are important for proliferation, hedgehog signaling, and differentiation. Furthermore, the deregulation of cilia and hedgehog in RMS could suggest the utility of therapeutically targeting ciliary components in these tumors.
Keywords: centrosome, ciliogenesis, myotube
Abstract
The primary cilium acts as a cellular antenna, transducing diverse signaling pathways, and recent evidence suggests that primary cilia are important in development and cancer. However, a role for cilia in normal muscle development and rhabdomyosarcoma (RMS) has not been explored. Here we implicate primary cilia in proliferation, hedgehog (Hh) signaling, and differentiation of skeletal muscle cells. Cilia and Hh signaling are highly dynamic during the differentiation of myoblasts. We show that cilia are assembled during the initial stages of myogenic differentiation but disappear as cells progress through myogenesis, concomitant with the destruction of proteins critical for cilia assembly and shortly after the Hh effector, Gli3, leaves the cilium. Importantly, we show that ablation of primary cilia strongly suppresses Hh signaling and myogenic differentiation while enhancing proliferation. Interestingly, our data further indicate that both cilia assembly and Hh signaling are deregulated in RMS, and cilia respond to Hh ligand in certain subsets of RMS cells but not others. Together, these findings provide evidence for an essential role for both primary cilia assembly and disassembly in the control of Hh signaling and early differentiation in muscle cells. We suggest that the temporally orchestrated destruction of centrosomal and ciliary proteins is a necessary antecedent for removal of the primary cilium and cessation of Hh signaling during myogenic differentiation. Additionally, our results further stratify RMS populations and highlight cilia assembly and disassembly as potential RMS drug targets.
Primary cilia are microtubule-based solitary and nonmotile organelles that protrude from the surface of most quiescent mammalian cells, and their assembly and disassembly are controlled by multiple pathways involving intraflagellar transport (IFT) proteins (1). The primary cilium serves as a signaling center during vertebrate development, and considerable evidence suggests that the cilium is specialized for hedgehog (Hh) responsiveness (2). Sonic hedgehog (Shh) is the most thoroughly investigated ligand for Hh signaling. Patched1 (Ptc1), the Shh receptor, is thought to prevent the accumulation of smoothened (Smo) at the cilium. Shh binding to Ptc1 relieves its inhibitory activity, and Smo accumulates at cilia and activates Hh signaling (3). In vertebrates, three transcription factors, Gli1, Gli2, and Gli3, are responsible for transduction of Hh signals (4). In addition to Hh, PDGF receptor-α, Wnt, and Notch signaling pathways depend on the assembly of a primary cilium (2, 5).
The links between cilia and differentiation are still largely unknown, although cilia are thought to be required for the differentiation of diverse cell types and tissues. For example, cilium-associated polycystins are required for proper growth and differentiation of kidney epithelial cells (6). During skin development, cilia-dependent Notch signaling directs epidermal differentiation (5). In addition, the primary cilium promotes the early phase of cardiomyocyte differentiation (7). Ciliogenesis also regulates adipogenic, osteogenic, and chondrogenic differentiation (8–10), as well as mesenchymal stem cell differentiation (11).
Skeletal muscle differentiation occurs through a temporally defined series of events commencing with expression of myogenic regulatory factors (MRFs), cell cycle exit, and fusion of myoblasts into multinucleated myofibers. Rhabdomyosarcomas (RMS) are thought to arise from skeletal muscle cells that fail to differentiate, and they represent the most common soft-tissue sarcoma of childhood (12). The two subtypes of RMS, embryonal and alveolar, exhibit different clinical manifestations. Although the cell of origin requires further clarification, RMS tumors are thought to arise from uncommitted mesodermal cells, mesenchymal and muscle progenitor cells, myoblasts, and adipocyte precursors (13, 14).
Early reports suggested that myoblasts often possess a cilium in interphase (15), and primary cilia were also observed in the mouse myoblast CO25 cell line (16). More recently, short, linear structures visualized with anti-detyrosinated tubulin in C2C12 myoblasts were described as cilia (17). Others have reported cilia assembly in nickel-sulfide-induced RMS in rats (18) and in a poorly differentiated human RMS (19). However, technical and reagent-associated limitations in earlier studies have precluded a comprehensive characterization of centrosomes and primary cilia in these muscle cell and RMS samples. Here we show that primary cilia and their associated Hh components are highly dynamic during myogenic differentiation. We found that depletion of three cilia-related proteins, centrosomal protein of 290 kDa (Cep290), intraflagellar transport protein 80 homolog (IFT80), and intraflagellar transport protein 88 homolog (IFT88), significantly inhibited primary cilia assembly in myoblasts. Our studies suggest that the assembly and subsequent disassembly of primary cilia are required to regulate proliferation, Hh signaling, and differentiation of skeletal muscle. Furthermore, our results showed that both cilia assembly and Hh pathways are deregulated in RMS and that cilia may contribute to the hyper-activation of Hh signaling in a subset of RMS populations. Together, we have revealed critical regulatory mechanisms for primary cilia assembly in normal muscle differentiation as well as the potential to therapeutically target cilia and Hh signaling in RMS.
Results and Discussion
Primary Cilia and Hh Components Are Dynamically Regulated During Skeletal Muscle Differentiation.
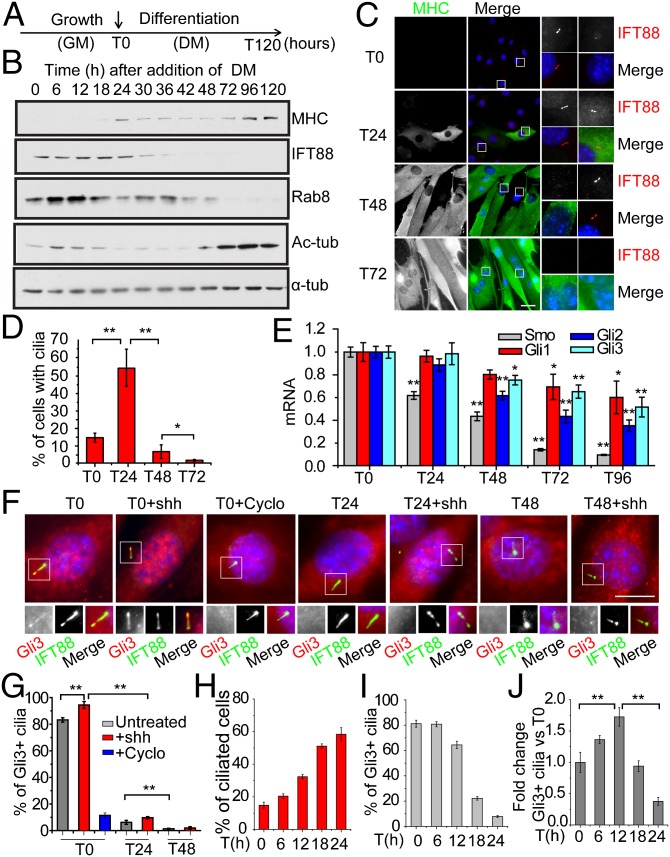
We investigated the occurrence of primary cilia and the abundance of centrosomal and ciliary proteins in primary mouse myoblasts before and after induction of differentiation (Fig. 1). To induce myoblast differentiation, growth medium was replaced with differentiation medium, a stage designated as T0 (Fig. 1A). The presence of primary cilia in myoblasts at T0 was observed with various cilia markers (Fig. S1 A and B). Next the expression of multiple centrosome and cilia markers, as well as differentiation markers (including myosin heavy chain, MHC), was analyzed at different time points from T0 to T120 (Fig. 1B). Expression of MHC increased over time, as expected, whereas the expression of proteins required to assemble the cilium, including IFT88 and Rab8, decreased and essentially disappeared by T48, suggesting that myogenic differentiation is accompanied by down-regulation of centrosomal and ciliary proteins. Primary cilia were also examined by immunofluorescence at each time point (Fig. 1 C and D). Remarkably, we found that cilia assembled and disassembled in an orchestrated and highly dynamic manner during differentiation: primary cilia were assembled during an early phase of differentiation (before T24) and subsequently disassembled during a later period (between T24 and T48). Interestingly, disassembly coincided with the start of myoblast fusion, because most cells with more than one nucleus at T48 lacked cilia, revealing an inverse correlation between the presence of cilia and the extent of myogenic differentiation. Furthermore, cilium disassembly also coincided with the dramatic down-regulation of IFT88 after T24 (Fig. 1B). Costaining with antibodies against two cilia markers, glutamylated tubulin (GT335) and IFT88, verified the timing of cilia assembly/disassembly during differentiation (Fig. S1C). Interestingly, GT335 staining along the axoneme seemed to significantly contract at T24, before completion of cilia disassembly at T48. To further elucidate this phenomenon, we quantitatively analyzed cilia with reduced GT335 staining from T0 to T24 (Fig. S1D), and the data showed a marked increase of cilia with truncated GT335 staining after T12, implicating loss of glutamylated tubulin, a modification that stabilizes microtubules, from cilia and potential structural reorganization of primary cilia during differentiation. Staining of centrin-1 and γ-tubulin in primary mouse myoblasts and C2C12 cells confirmed that centrosome reorganization occurs during differentiation, consistent with previous reports using C2C12 cells (20) (Fig. S1 E and F).
Fig. 1.
Primary cilia and Hh components are dynamically regulated during muscle differentiation. (A) Experimental setup for differentiation of primary mouse myoblasts. At T0, growth medium (GM) was replaced with differentiation medium (DM) to induce differentiation. (B) Western blotting of indicated proteins at different time points during the differentiation of primary mouse myoblasts. α-tub, α-tubulin; Ac-tub, acetylated-tubulin. (C) Representative immunofluorescence images of primary mouse myoblasts induced to differentiate and stained with indicated antibodies or DAPI (DNA) at different time points. Magnified views of the boxed Insets are shown on the right. (Scale bar, 30 μm.) (D) Quantification of the percentages of cilia as shown in C. Data are presented as mean ± SD. (E) qRT-PCR analysis of mRNA levels of Smo, Gli1, Gli2, and Gli3. Error bars, SEM. Student’s t test was performed by comparing expression at T0 with indicated times points. (F) Staining of Hh components during differentiation. Representative images are shown. Shh and cyclopamine (Cyclo) were used as indicated. Proteins and DNA were visualized as indicated, and magnified images are shown on the right. (Scale bar, 10 μm.) (G) Quantitative analysis of data in F, showing percentages of ciliated cells positive for Gli3 staining. Gli3 was considered positive when a clear axonemal staining was observed. Data are presented as mean ± SD. (H and I) Quantitative analysis of the percentage of cilia and Gli3-positive cilia. (J) Total number of ciliated cells positive for Gli3 staining was calculated. The data were divided by total number of cells and normalized against T0. In all cases, * and ** indicate P < 0.05 and P < 0.01, respectively, by Student t test.
The cilium concentrates Hh pathway components and coordinates signaling in response to this ligand (2). Expression of Smo, Gli1, Gli2, and Gli3 was assessed by quantitative real-time PCR (qRT-PCR) at different time points during differentiation, and this analysis indicated that Hh components were down-regulated (Fig. 1E). Gli3 processing is inhibited by Hh, which causes full-length, activated Gli3 to accumulate in primary cilia, while suppressing formation of a processed, repressive form of the protein (21). Therefore, we examined the localization of this protein during differentiation in the presence and absence of ligand (Fig. 1 F and G). Treatment with Shh, an Hh agonist, increased the localization of Gli3 to cilia, whereas the Hh antagonist cyclopamine (22) reduced its recruitment, suggesting that myoblast cilia can transduce Hh-initiated signals. The percentages of Gli3-positive cells vary between different cell types and tissues, and in myoblasts, even without addition of exogenous Shh, a large proportion of myoblasts exhibit Gli3 targeting to cilia at T0, indicating activation of the pathway. Interestingly, localization of Gli3 to cilia sharply diminished at T24 compared with T0, although cilia persisted in these cells. Down-regulation of Hh signaling in cilia continued through T48, the period during which cilia were most rapidly lost (Fig. 1 F and G). To further investigate this process, we examined Gli3 localization to cilia during the initial stages of differentiation (T0–T24) (Fig. 1H). As cells differentiate, primary cilia are gradually assembled, and the percentage of Gli3-positive cilia is essentially the same at T0 and T6. However, the removal of Gli3 from cilia primarily occurred precipitously after T12 and coincided with the loss of glutamylated tubulin from cilia (Fig. S1D). Notably, the number of Gli3-positive cilia increased initially owing to an increase in cilia percentage and then decreased due to Gli3 removal (Fig. 1 I and J). Taken together, these data suggest that assembly of primary cilia and Hh signaling are highly dynamic and may be functionally linked during myoblast differentiation, and Hh signaling transduced through cilia could function in the very early stages of myogenic differentiation.
Primary Cilia Regulate Proliferation of Myoblasts.
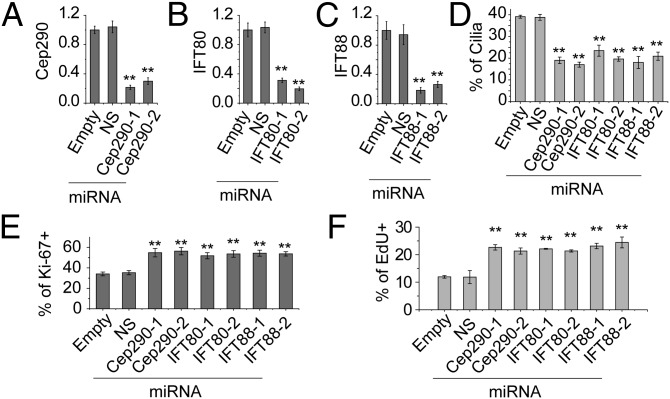
To understand the role of cilia in myogenesis, we investigated their functional role by inhibiting cilia assembly in C2C12 myoblasts. We expressed microRNAs (miRNAs) to individually silence three genes, CEP290, IFT80, and IFT88, each of which is required for cilia assembly (23). We expressed two different miRNAs per gene, each of which resulted in ∼70–80% suppression of mRNA levels (Fig. 2 A–C), and quantified cilia percentages in confluent C2C12 cells at T0 (Fig. 2D and Fig. S2A). Suppression of each protein significantly inhibited cilia assembly compared with controls. Cilium assembly is known to be linked with proliferation in many cell types (24, 25), and therefore we evaluated proliferation of myoblasts by Ki-67 staining at T0. As shown in Fig. 2E and Fig. S2A, depletion of Cep290, IFT80, and IFT88 led to an increase in cell proliferation, suggesting that cilia assembly can restrain cell cycle progression. Moreover, cells depleted of ciliary proteins consistently exhibited markedly increased 5-ethynyl-2′-deoxyuridine (EdU) incorporation (Fig. 2F and Fig. S2B). In agreement, FACS analysis indicated a moderate decrease in the percentage of G0/G1 cells, with a corresponding increase in the S phase fraction (Fig. S2C), although the mitotic index was not significantly affected (Fig. S2D). Given that Cep290, IFT80, and IFT88 are thought to play critical roles in cilium assembly and function, we propose that assembly of this structure in myoblasts facilitates, or results from, cell cycle exit before differentiation.
Fig. 2.
Assembly of primary cilia regulates proliferation of myoblasts. (A–D) qRT-PCR detection of knockdown efficiency in C2C12 myoblasts using miRNAs. Two different miRNAs targeting Cep290 (A), IFT80 (B), and IFT88 (C) were analyzed. Empty vector and a nonspecific (NS) miRNA were used as controls. Relative expression values are shown; error bars, SEM. (D) Quantification of cilia percentages in C2C12 cells expressing different miRNAs. (E) Quantification of percentages of Ki-67-positive cells. (F) Quantification of percentages of EdU-positive cells. Data are shown as mean ± SD. In each case, **P < 0.01.
Primary Cilia Regulate Myoblast Differentiation and Hh Signaling.
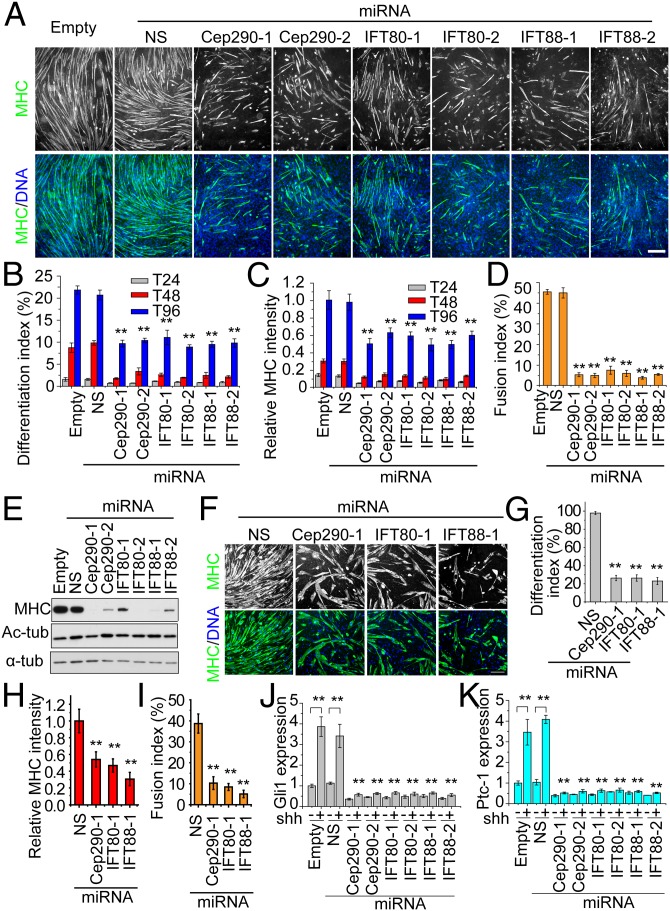
To test whether primary cilia are required for muscle differentiation, we first investigated the differentiation potential of C2C12 cell lines after stable silencing of all three cilia-related genes (CEP290, IFT80, and IFT88). Myoblast differentiation was investigated at T24, T48, and T96 by MHC staining (Fig. 3A and Fig. S3). In control groups, myotubes were well formed and normally aligned at T96 (Fig. 3A). However, cells depleted of Cep290, IFT80, or IFT88 formed significantly fewer myotubes by T96, and myotube length (reflecting the number of nuclei incorporated or extent of fusion) was considerably reduced. The extent of differentiation was also significantly reduced in cells with diminished cilia assembly at T24 and T48 (Fig. S3). To further elucidate the function of cilia in differentiation, we assessed the extent of differentiation in each miRNA-expressing population (Fig. 3 B–D). We estimated the differentiation index as the percentage of nuclei incorporated into MHC-positive cells (Fig. 3B), the relative MHC intensity per microscopic field (with 10× objective; Fig. 3C) at T24, T48, and T96, and the percentage of cells that have fused at T96 (the fusion index; Fig. 3D). Each of these criteria confirmed our assessment of the impact of depleting each protein on differentiation. Moreover, Western blotting showed that the abundance of MHC at T96 was significantly diminished in populations lacking cilia (Fig. 3E). To further validate the specificity of IFT88 depletion, we infected cells with viruses expressing two distinct RNAi-resistant IFT88 (R1 and R2) cDNAs, which restored normal levels of IFT88 in their respective targeted populations (Fig. S4A). Importantly, defective cilia assembly (Fig. S4 B and C) and differentiation (Fig. S4 D–G) were fully rescued by R1 and R2 but not by the control vector. To confirm our findings, we repeated the silencing experiments in primary mouse myoblasts. In agreement with results from C2C12 cells, depletion of Cep290, IFT80, and IFT88 led to significant reductions in the extent of differentiation (Fig. 3 F–I).
Fig. 3.
Inhibition of primary cilia assembly abolishes myoblast differentiation. (A) Inhibition of primary cilia in C2C12 cells suppresses differentiation. The differentiation efficiency of C2C12 cells expressing distinct miRNAs was examined at T24, T48, and T96. The empty and NS miRNAs were used as controls, and six miRNAs targeting Cep290, IFT80, and IFT88 were analyzed as indicated. Representative images of cells, visualized as indicated at T96, are shown. (B) Quantitative analysis of the differentiation index of the C2C12 cells expressing miRNAs at indicated time points. The differentiation index was calculated as the percentage of nuclei in MHC-positive cells. The differentiation indexes at T24, T48, and T96 are depicted by gray, red, and blue bars, respectively. Error bars, SD. (C) Quantification of the relative MHC intensity in each group as in A. Error bars, ±SD. (D) Quantification of fusion index at T96 as in A. Error bars, ±SD. (E) Western blots of extracts of C2C12 cells expressing individual miRNAs (Upper) and probed with indicated antibodies. (F) Inhibition of cilia assembly in primary mouse myoblasts suppresses differentiation. Differentiation efficiencies of primary mouse myoblasts expressing indicated miRNAs were examined by staining MHC (green) and DNA (blue). (G–I) Quantitative analysis of primary mouse myoblasts expressing different miRNAs shown in F. The differentiation index (G), relative MHC intensity (H), and fusion index (I) were measured. Error bars, ±SD. In each case, **P < 0.01. (J and K) Hh signaling is significantly diminished in C2C12 cells depleted of ciliary/centrosome components. The expression of Gli1 (J) and Patched-1 (K) was analyzed by qRT-PCR. Error bars, ±SEM. (Scale bars, 200 μm.)
As a complementary approach, we treated cells with ciliobrevin D (26), a small molecule that suppresses cilia assembly by inhibiting the activity of both cytoplasmic dynein 1 and 2 in a dose-dependent manner. Treatment of C2C12 cells with this inhibitor dramatically reduced both cilia assembly and the extent of differentiation (Fig. S5 A–E). By varying both the dosage and treatment intervals, we found that the extent of differentiation depended on both parameters. Together, these results demonstrate that cilia ablation by either depletion of essential ciliary proteins or chemical inhibition suppresses myogenesis both at early (T0–T24) and later (after T24) stages of differentiation.
Our studies indicated that Hh components localize to cilia in myoblasts and that appropriate responses to agonists (Shh ligand) and antagonists (cyclopamine) could be elicited (Fig. 1 E and F). Therefore, we hypothesized that Hh signaling might be attenuated as a result of loss of cilia. To investigate signaling in the absence of cilia, we examined the expression of two Hh response genes, Gli1 and Ptc1, in C2C12 cells depleted of Cep290, IFT80, or IFT88 in the presence and absence of Shh ligand (Fig. 3 J and K). Expression of both Gli1 and Ptc1 at T0 was strongly inhibited in the presence of Shh when cilia assembly was blocked, although control cells responded robustly, as expected. These data confirm that Hh components localize to cilia and are essential for transducing signals in response to this ligand in myoblasts.
Next, we investigated whether MRFs were also impacted by the loss of cilia. In control C2C12 and primary mouse myoblasts, myogenin (Myog) expression was induced between T0 and T24 (Fig. S6A), as expected (27). However, Myog expression was markedly reduced after cilia inhibition (Fig. S6B), and expression of two other MRFs (MyoD and Myf5), which are transcribed in myoblasts and at T0, was also reduced in cells depleted of ciliary proteins (Fig. S6C). Furthermore, stimulation of the Hh pathway with Shh modestly enhanced expression of MRFs, whereas pathway inhibition by cyclopamine reduced their expression (Fig. S6D). These data are consistent with observations in zebrafish and mice, wherein Shh signaling was required for induction of Myf5 and MyoD (28, 29). Taken together, these data suggest that assembly of cilia and a functional Hh pathway are essential for induction or sustained expression of MRFs and reinforce our conclusion that primary cilia are required for both Hh signaling and early myogenic differentiation.
Primary Cilia Assembly and Hh Signaling Are Deregulated in RMS.
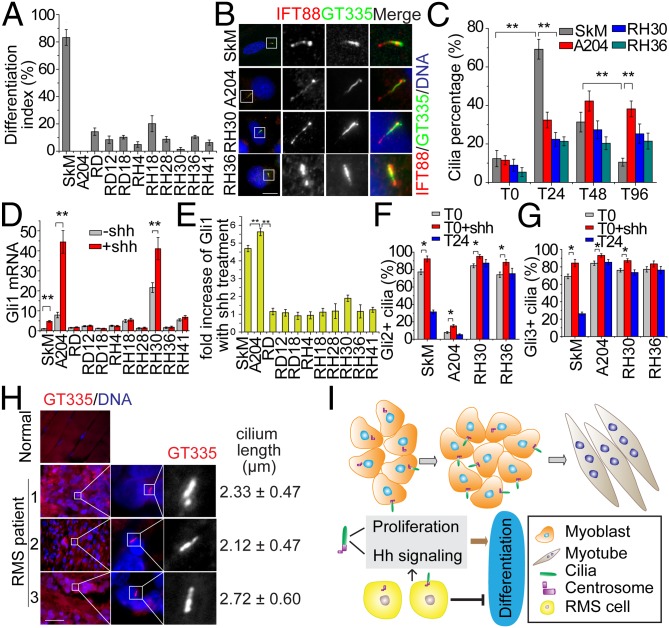
During normal myogenesis, Hh signaling is thought to act early to promote initiation of the myogenic program (30), whereas aberrant activation of Hh signaling through Shh addition or Gli overexpression can block differentiation (31, 32). Indeed, previous studies have shown that RMS formation is associated with Patched1 mutations, and different degrees of Hh activation have been observed in RMS, suggesting that Hh signaling plays an important role in RMS oncogenicity (33, 34). However, whether abnormal ciliogenesis contributes to aberrant Hh signaling in muscle and RMS has not been explored. We investigated ciliogenesis in five embryonal RMS (A204, RD, RD12, RD18, and RH36) and five alveolar RMS (RH4, RH18, RH28, RH30, and RH41) cell lines as well as normal primary human skeletal myoblasts (SkM). First, we confirmed that RMS cells differentiate poorly, compared with the SkM control (Fig. 4A and Fig. S7A). Next, we examined cilia assembly during differentiation of SkM and RMS cells (Fig. 4 B and C and Fig. S7 B and C). Cilium assembly in SkM cells was maximal at T24 and then decreased as differentiation proceeded (Fig. 4 B and C), reminiscent of primary mouse myoblasts. In striking contrast, cilia were observed in only 3 of 10 RMS cell lines (A204, RH30, and RH36); all other RMS cell lines were devoid of cilia altogether. Indeed, while our work was in progress, another group also reported the presence of cilia in RH30 (35). Interestingly, however, ciliation in all three cell lines was abnormal. First, cilia were not efficiently assembled in RMS cells during the first 24 h, and second, cilia did not disappear at later phases of differentiation, from T48 and T96 (Fig. 4C). To explore potential explanations for cilia deregulation in RMS cells, we analyzed the expression of multiple cilia and centrosomal proteins (Fig. S7D) and found that several proteins required for cilia assembly were deregulated in a few RMS cell lines, including IFT88 and Cep290, although many cell lines expressed normal levels of these proteins. We conclude that ciliogenesis is subverted in all RMS cell lines that we have investigated and that defects—both in the regulated assembly and disassembly of the organelle—could, in part, explain their inability to differentiate into skeletal muscle. Future studies will be required to determine the extent to which aberrant expression of key ciliary proteins could contribute to the defects observed in RMS cell lines.
Fig. 4.
Ciliogenesis and Hh pathway are deregulated in RMS. (A) SkM and 10 RMS cell lines were induced to differentiate for 96 h, and the extent of differentiation indexes was assessed. Error bars, ±SD. (B) Representative images of primary cilia in SkM and three RMS cell lines. Magnified images of boxed Insets are shown on the right. (Scale bar, 10 μm.) (C) Quantification of cilia percentage at indicated stages of differentiation of SkM and RMS cells. Error bars, ±SD. (D and E) Analysis of Hh responsiveness in RMS cell lines. Gli1 expression was detected by qRT-PCR in SkM and RMS cells with or without Shh addition. The relative expression of Gli1 (D) and fold increase of Gli1 expression with Shh treatment (E) are shown. Error bars, ±SEM. (F and G) Quantitative analysis of the percentages of ciliated cells positive for Gli2 and Gli3 staining. (H) Staining of cilia and nuclei in normal muscle and in three RMS patients. Length of cilia is indicated for three patients. The lengths are indicated as mean ± SD. (Scale bar, 200 μm.) In each case, *P < 0.05; **P < 0.01. (I) The role of primary cilia in normal muscle differentiation and RMS. In myoblasts, primary cilia regulate cell proliferation and transduce Hh signaling. Dynamic assembly and loss of cilia occurs during myogenic differentiation, and this may limit the period of Hh signaling to a narrow window. See text for details.
Next, we investigated whether cilia and Hh signaling are correlated in RMS. We examined the expression of Gli2 by Western blotting and found that Gli2 was aberrantly expressed in the majority of RMS cell lines (Fig. S7D). Furthermore, we examined Hh responsiveness in RMS and SkM by measuring expression of Gli1 in response to Shh treatment. Under basal conditions, RMS cells expressed Gli1 at similar or higher levels than the control (Fig. 4D). Interestingly, Gli1 expression was significantly up-regulated in SkM, A204, and RH30 cells compared with other RMS lines after Shh treatment (Fig. 4 D and E). These cell lines also assembled cilia more efficiently than other RMS lines (Fig. 4C), affirming the link between cilia and Hh signaling. Remarkably, both A204 and RH30 cells exhibited constitutive elevation of Gli1 levels in the absence of ligand (at levels 7.9-fold and 21.6-fold higher than SkM, respectively) and significantly higher overall induction of Gli1 after Shh treatment, suggesting that the Hh pathway in these cells was hyperactivated. Interestingly, however, RH36 cells assembled cilia to a degree comparable to RH30, but these cells did not respond to Shh robustly, suggesting additional aberrations. Furthermore, we examined the localization of Gli2 and Gli3 in the three above-mentioned ciliated RMS cells as well as SkM cells. Gli2 and Gli3 strongly localized to the distal tip of cilia upon Shh treatment (Fig. 4 F and G and Fig. S8), as observed in other cell types (21). We found that the bulk of Gli2 and Gli3 disappeared from cilia in normal myoblasts with the onset of differentiation (T24), whereas in RMS cells these Hh components largely persisted in the cilia, suggesting continuous activation of cilia-dependent Hh signaling. Remarkably, in A204 cells, Gli2 levels are very low (Fig. S7D) and most cilia are Gli2-negative (Fig. 4F), yet there is an increase in the percentage of Gli2-positive cilia, as well as a very robust increase in Gli1 expression (Fig. 4 D and E), after Shh treatment, indicating the existence of a functional, highly sensitive Hh response in this RMS line. Despite their ability to ciliate and respond to Shh, both A204 and RH30 cells were among the most poorly differentiated of all RMS cell lines (Fig. 4A), suggesting that aberrant persistence of cilia could also counteract differentiation. Altogether, our data indicate that defective Hh signaling in ciliated RMS cell lines is associated with cilia deregulation.
To investigate whether cilia are deregulated in muscle tissue from RMS patients, we stained paraffin-embedded sections with GT335 antibody and identified a low but readily detectable occurrence of ciliated cells in a small proportion (3 of 33 or 9.1%) of RMS patients (Fig. 4H and Table S1). Further, we found no evidence for the existence of cilia in normal muscle fibers (Fig. 4H). The average length of cilia in three RMS patients was ∼2–3 μm, similar to cilia in cultured normal skeletal myoblasts. Thus, unlike differentiation of normal muscle, cilia in three RMS patients persisted during development. Taken together, these data suggested that cilia are deregulated in a subset of RMS patients. Future studies will be required, however, to determine the extent to which aberrant ciliogenesis and defects in signaling pathways transduced by cilia contribute to defective differentiation and RMS.
In summary, our work leads us to hypothesize a unique model to illustrate the functions of primary cilia in normal muscle differentiation and its subversion in RMS. During the differentiation of normal myoblasts (Fig. 4I), cilia are dynamically assembled and disassembled before formation of mature myotubes, and Hh components are concomitantly down-regulated as differentiation proceeds. In myoblasts, cilia regulate proliferation and Hh signaling in preparation for differentiation. When cilia assembly is perturbed by depletion of proteins required for this process, cell proliferation is enhanced, most likely as a result of a block to cell cycle exit. In addition, Hh signaling is sharply diminished upon ablation of cilia, and this effect, combined with the block to cell cycle exit, could abolish differentiation. In RMS cells (Fig. 4I), cilia assembly is abrogated or deregulated, perhaps owing to deregulation of centrosomal and ciliary proteins. Deregulation of cilia assembly and disassembly in RMS could promote proliferation at the expense of differentiation in myoblasts. Moreover, primary cilia in a subset of RMS cell lines can respond to Hh ligand, which could further contribute to hyperactivation of Hh, leading to overproliferation and failure to differentiate. Because some RMS cell populations exhibit enhanced Hh activity in the face of low levels of ciliation, cilia-independent Hh activation might also play a role in RMS.
Given that normal mouse and human skeletal muscle cells assemble cilia before myoblast fusion (i.e., in the early phase of differentiation), we propose that cilia and Hh signaling could contribute to the maintenance of the myogenic fate and the initiation of differentiation. These data are consistent with our observation that cells depleted of ciliary proteins exhibit differentiation defects as early as T24 (Fig. 3 B–D and Fig. S3). It is well established that Hh signaling is essential for muscle differentiation, although our understanding of the role of this pathway in muscle is incomplete and has been confounded by contradictory observations. On one hand, Hh prevents differentiation of satellite cells into multinucleated myotubes (31), and Gli1 and Gli2 have been shown to block myoblast differentiation by inhibiting MyoD-mediated transcriptional activation (32). On the other hand, it has been shown that Hh enhances myotube formation by inducing proliferation of committed muscle cells (36) and that this pathway can drive terminal differentiation of slow skeletal muscle (37) and specify muscle cell fate (38). Together with findings that high Hh activity and low levels of differentiation exist in many RMS, our data suggest that cilia are required to fine-tune the response to Hh signaling, and either loss of ciliation or persistent ciliation (with attendant diminution or hyperactivation of Hh signaling, respectively) might abrogate normal differentiation. We propose that the primary cilium, by acting as a conduit for Hh signaling and regulation of proliferation, may determine the differentiation and fate of muscle cells. We note that cells of other mesodermal lineages, including osteoblasts, adipocytes, and chondrocytes, also assemble primary cilia (8–10), and together with our observations, we postulate that all mesodermal lineages could require this organelle to implement their respective differentiation programs.
It has been shown that centrosomal proteins dramatically relocalize in differentiating muscle cells (20). Taken together with our studies, it is intriguing to speculate that the destruction of centrosomes is an obligatory event that (i) eliminates the assembly of mitotic spindles, which are no longer needed after terminal differentiation, and (ii) precludes the ability to form a primary cilium able to promote Hh pathway activation and proliferation.
The first study that linked Hh with RMS showed that mice heterozygous for Patched exhibited elevated Gli1 and Patched transcript levels and an elevated incidence of RMS (33). It has been proposed that pathogenesis of RMS involves deregulation of Hh signaling pathways (39), and up-regulation of Hh signaling has been implicated in a subset of RMS cases (40). Hh pathway components are considered to be potential targets for RMS treatment (41). However, we have shown that ciliated and nonciliated RMS populations respond differently to Hh signaling, suggesting that RMS patients could be further stratified accordingly. Moreover, because the cilium is deregulated in RMS and is also required for normal differentiation, causal links between proper cilium assembly and RMS should be investigated in the future. Given that cilia can both mediate and suppress Hh pathway-dependent tumorigenesis (42), and because certain Hh inhibitors may be ineffective toward nonciliated cancer cells (43), this organelle might also represent a viable drug target. Further work is needed to maximize the potential of these therapeutic targets in RMS.
Materials and Methods
To study myoblast differentiation, primary mouse myoblasts, mouse C2C12, and primary human skeletal myoblasts were used. Five embryonal RMS cell lines (A204, RD, RD12, RD18, and RH36) and five alveolar RMS (RH4, RH18, RH28, RH30, and RH41) were also studied. RNA interference in myoblasts was performed using a modified version of the miRNA retroviral expression system as previously described (44). Details of cell culture and differentiation, RNA interference, retroviral infection, RNA isolation, qRT-PCR, antibodies, immunofluorescence, microscopic analysis, and statistical analysis are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank D. Kopinke and J. Reiter for sharing unpublished data; Drs. G. Germain and M. Hatley (St. Jude Research Hospital), P.-L. Lollini and L. Landuzzi (University of Bologna), S. Scales (Genetech), and C. Desdouets (Faculté de médecine R. Descartes) for cell lines, antibodies, and plasmids; and Drs. S. Kim and K. Lee and other members of the B.D.D. laboratory for constructive advice. Work in the B.D.D. laboratory was supported by the Alex Lemonade Stand Foundation and National Institutes of Health Grant R01 HD069647, for whose support we are grateful.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1323265111/-/DCSupplemental.
References
- 1.Kim S, Dynlacht BD. Assembling a primary cilium. Curr Opin Cell Biol. 2013;25(4):506–511. doi: 10.1016/j.ceb.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goetz SC, Anderson KV. The primary cilium: A signalling centre during vertebrate development. Nat Rev Genet. 2010;11(5):331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317(5836):372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- 4.Hu MC, et al. GLI3-dependent transcriptional repression of Gli1, Gli2 and kidney patterning genes disrupts renal morphogenesis. Development. 2006;133(3):569–578. doi: 10.1242/dev.02220. [DOI] [PubMed] [Google Scholar]
- 5.Ezratty EJ, et al. A role for the primary cilium in Notch signaling and epidermal differentiation during skin development. Cell. 2011;145(7):1129–1141. doi: 10.1016/j.cell.2011.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou J. Polycystins and primary cilia: Primers for cell cycle progression. Annu Rev Physiol. 2009;71:83–113. doi: 10.1146/annurev.physiol.70.113006.100621. [DOI] [PubMed] [Google Scholar]
- 7.Clement CA, et al. The primary cilium coordinates early cardiogenesis and hedgehog signaling in cardiomyocyte differentiation. J Cell Sci. 2009;122(Pt 17):3070–3082. doi: 10.1242/jcs.049676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao ZS, Quarles LD. Role of the polycytin-primary cilia complex in bone development and mechanosensing. Ann N Y Acad Sci. 2010;1192:410–421. doi: 10.1111/j.1749-6632.2009.05239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marion V, et al. Transient ciliogenesis involving Bardet-Biedl syndrome proteins is a fundamental characteristic of adipogenic differentiation. Proc Natl Acad Sci USA. 2009;106(6):1820–1825. doi: 10.1073/pnas.0812518106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wann AKT, et al. Primary cilia mediate mechanotransduction through control of ATP-induced Ca2+ signaling in compressed chondrocytes. FASEB J. 2012;26(4):1663–1671. doi: 10.1096/fj.11-193649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tummala P, Arnsdorf EJ, Jacobs CR. The role of primary cilia in mesenchymal stem cell differentiation: A pivotal switch in guiding lineage commitment. Cell Mol Bioeng. 2010;3(3):207–212. doi: 10.1007/s12195-010-0127-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dagher R, Helman L. Rhabdomyosarcoma: An overview. Oncologist. 1999;4(1):34–44. [PubMed] [Google Scholar]
- 13.Hettmer S, Wagers AJ. Muscling in: Uncovering the origins of rhabdomyosarcoma. Nat Med. 2010;16(2):171–173. doi: 10.1038/nm0210-171. [DOI] [PubMed] [Google Scholar]
- 14.Hatley ME, et al. A mouse model of rhabdomyosarcoma originating from the adipocyte lineage. Cancer Cell. 2012;22(4):536–546. doi: 10.1016/j.ccr.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Przybylski RJ. Occurrence of centrioles during skeletal and cardiac myogenesis. J Cell Biol. 1971;49(1):214–221. doi: 10.1083/jcb.49.1.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeytinoglu M, Ritter J, Wheatley DN, Warn RM. Presence of multiple centrioles and primary cilia during growth and early differentiation in the myoblast CO25 cell line. Cell Biol Int. 1996;20(12):799–807. doi: 10.1006/cbir.1996.0103. [DOI] [PubMed] [Google Scholar]
- 17.Mian I, et al. LKB1 destabilizes microtubules in myoblasts and contributes to myoblast differentiation. PLoS ONE. 2012;7(2):e31583. doi: 10.1371/journal.pone.0031583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hildebrand HF, Biserte G. Ultrastructural investigation of NI3S2-induced rhabdomyosarcoma in Wistar rat: Comparative study with emphasis on myofibrillar differentiation and ciliar formation. Cancer. 1978;42(2):528–554. doi: 10.1002/1097-0142(197808)42:2<528::aid-cncr2820420222>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 19.Bundtzen JL, Norback DH. The ultrastructure of poorly differentiated rhabdomyosarcomas: A case report and literature review. Hum Pathol. 1982;13(4):301–313. doi: 10.1016/s0046-8177(82)80220-7. [DOI] [PubMed] [Google Scholar]
- 20.Srsen V, Fant X, Heald R, Rabouille C, Merdes A. Centrosome proteins form an insoluble perinuclear matrix during muscle cell differentiation. BMC Cell Biol. 2009;10:28. doi: 10.1186/1471-2121-10-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wen X, et al. Kinetics of hedgehog-dependent full-length Gli3 accumulation in primary cilia and subsequent degradation. Mol Cell Biol. 2010;30(8):1910–1922. doi: 10.1128/MCB.01089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen JK, Taipale J, Cooper MK, Beachy PA. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev. 2002;16(21):2743–2748. doi: 10.1101/gad.1025302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsang WY, et al. CP110 suppresses primary cilia formation through its interaction with CEP290, a protein deficient in human ciliary disease. Dev Cell. 2008;15(2):187–197. doi: 10.1016/j.devcel.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackson PK. Do cilia put brakes on the cell cycle? Nat Cell Biol. 2011;13(4):340–342. doi: 10.1038/ncb0411-340. [DOI] [PubMed] [Google Scholar]
- 25.Yang Y, Roine N, Mäkelä TP. CCRK depletion inhibits glioblastoma cell proliferation in a cilium-dependent manner. EMBO Rep. 2013;14(8):741–747. doi: 10.1038/embor.2013.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Firestone AJ, et al. Small-molecule inhibitors of the AAA+ ATPase motor cytoplasmic dynein. Nature. 2012;484(7392):125–129. doi: 10.1038/nature10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blais A, et al. An initial blueprint for myogenic differentiation. Genes Dev. 2005;19(5):553–569. doi: 10.1101/gad.1281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coutelle O, et al. Hedgehog signalling is required for maintenance of myf5 and myoD expression and timely terminal differentiation in zebrafish adaxial myogenesis. Dev Biol. 2001;236(1):136–150. doi: 10.1006/dbio.2001.0193. [DOI] [PubMed] [Google Scholar]
- 29.Borycki AG, et al. Sonic hedgehog controls epaxial muscle determination through Myf5 activation. Development. 1999;126(18):4053–4063. doi: 10.1242/dev.126.18.4053. [DOI] [PubMed] [Google Scholar]
- 30.Hu JK, McGlinn E, Harfe BD, Kardon G, Tabin CJ. Autonomous and nonautonomous roles of Hedgehog signaling in regulating limb muscle formation. Genes Dev. 2012;26(18):2088–2102. doi: 10.1101/gad.187385.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koleva M, et al. Pleiotropic effects of sonic hedgehog on muscle satellite cells. Cell Mol Life Sci. 2005;62(16):1863–1870. doi: 10.1007/s00018-005-5072-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerber AN, Wilson CW, Li YJ, Chuang PT. The hedgehog regulated oncogenes Gli1 and Gli2 block myoblast differentiation by inhibiting MyoD-mediated transcriptional activation. Oncogene. 2007;26(8):1122–1136. doi: 10.1038/sj.onc.1209891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hahn H, et al. Rhabdomyosarcomas and radiation hypersensitivity in a mouse model of Gorlin syndrome. Nat Med. 1998;4(5):619–622. doi: 10.1038/nm0598-619. [DOI] [PubMed] [Google Scholar]
- 34.Roma J, Almazán-Moga A, Sánchez de Toledo J, Gallego S. Notch, wnt, and hedgehog pathways in rhabdomyosarcoma: From single pathways to an integrated network. Sarcoma. 2012;2012:695603. doi: 10.1155/2012/695603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li F, Shi W, Capurro M, Filmus J. Glypican-5 stimulates rhabdomyosarcoma cell proliferation by activating Hedgehog signaling. J Cell Biol. 2011;192(4):691–704. doi: 10.1083/jcb.201008087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duprez D, Fournier-Thibault C, Le Douarin N. Sonic Hedgehog induces proliferation of committed skeletal muscle cells in the chick limb. Development. 1998;125(3):495–505. doi: 10.1242/dev.125.3.495. [DOI] [PubMed] [Google Scholar]
- 37.Li X, et al. Hedgehog can drive terminal differentiation of amniote slow skeletal muscle. BMC Dev Biol. 2004;4:9. doi: 10.1186/1471-213X-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Flynt AS, Li N, Thatcher EJ, Solnica-Krezel L, Patton JG. Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate. Nat Genet. 2007;39(2):259–263. doi: 10.1038/ng1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tostar U, et al. Deregulation of the hedgehog signalling pathway: A possible role for the PTCH and SUFU genes in human rhabdomyoma and rhabdomyosarcoma development. J Pathol. 2006;208(1):17–25. doi: 10.1002/path.1882. [DOI] [PubMed] [Google Scholar]
- 40.Zibat A, et al. Activation of the hedgehog pathway confers a poor prognosis in embryonal and fusion gene-negative alveolar rhabdomyosarcoma. Oncogene. 2010;29(48):6323–6330. doi: 10.1038/onc.2010.368. [DOI] [PubMed] [Google Scholar]
- 41.Kawabata N, et al. Pharmacological inhibition of the Hedgehog pathway prevents human rhabdomyosarcoma cell growth. Int J Oncol. 2011;39(4):899–906. doi: 10.3892/ijo.2011.1076. [DOI] [PubMed] [Google Scholar]
- 42.Wong SY, et al. Primary cilia can both mediate and suppress Hedgehog pathway-dependent tumorigenesis. Nat Med. 2009;15(9):1055–1061. doi: 10.1038/nm.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hassounah NB, Bunch TA, McDermott KM. Molecular pathways: The role of primary cilia in cancer progression and therapeutics with a focus on Hedgehog signaling. Clin Cancer Res. 2012;18(9):2429–2435. doi: 10.1158/1078-0432.CCR-11-0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Asp P, Acosta-Alvear D, Tsikitis M, van Oevelen C, Dynlacht BD. E2f3b plays an essential role in myogenic differentiation through isoform-specific gene regulation. Genes Dev. 2009;23(1):37–53. doi: 10.1101/gad.1727309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.