Significance
Using crystallographic and biochemical studies, we explored how a viral protein, the matrix (M) protein of vesicular stomatitis virus (VSV), affects a pair of cellular proteins: mRNA export factor 1 (Rae1) and nucleoporin Nup98. We identified Rae1 as a nucleic acid-binding protein and showed that the viral M protein mimics the phosphate backbone of any nucleic acid. This mimicry enables VSV M protein to occupy the nucleic acid binding site of Rae1. Because this site is also used as a temporary stopover by host cell mRNA on its passage through the nuclear pore, the viral M protein sabotages host cell mRNA export, thereby paving the way for exclusively cytoplasm-synthesized VSV mRNAs to take over the cell’s protein biosynthesis machineries.
Keywords: phosphate backbone binding, M protein mimicry, mRNP export
Abstract
mRNA export factor 1 (Rae1) and nucleoporin 98 (Nup98) are host cell targets for the matrix (M) protein of vesicular stomatitis virus (VSV). How Rae1 functions in mRNA export and how M protein targets both Rae1 and Nup98 are not understood at the molecular level. To obtain structural insights, we assembled a 1:1:1 complex of M•Rae1•Nup98 and established a crystal structure at 3.15-Å resolution. We found that the M protein contacts the Rae1•Nup98 heterodimer principally by two protrusions projecting from the globular domain of M like a finger and thumb. Both projections clamp to the side of the β-propeller of Rae1, with the finger also contacting Nup98. The most prominent feature of the finger is highly conserved Methionine 51 (Met51) with upstream and downstream acidic residues. The complementary surface on Rae1 displays a deep hydrophobic pocket, into which Met51 fastens like a bolt, and a groove of basic residues on either side, which bond to the acidic residues of the finger. Notably, the M protein competed for in vitro binding of various oligonucleotides to Rae1•Nup98. We localized this competing activity of M to its finger using a synthetic peptide. Collectively, our data suggest that Rae1 serves as a binding protein for the phosphate backbone of any nucleic acid and that the finger of M mimics this ligand. In the context of mRNA export, we propose that a given mRNA segment, after having been deproteinated by helicase, is transiently reproteinated by Nup98-tethered Rae1. We suggest that such repetitive cycles provide cytoplasmic stopover sites required for ratcheting mRNA across the nuclear pore.
Viruses have long been used as tools for probing and elucidating pathways of host cells. A prominent example is the membrane-enclosed vesicular stomatitis virus (VSV). On entry into the host cell cytoplasm, its negative ssRNA genome is transcribed by virus-associated RNA-dependent RNA polymerase. The positive RNA transcripts serve as mRNAs for viral proteins and templates for synthesizing more negative RNA transcripts. Each of the negative RNA transcripts can then be packaged by viral proteins to give rise to a new viral particle. Packaging is likely to begin with cotranscriptional coverage of the entire phosphate backbone of viral negative RNA using more than 1,000 copies of nucleocapsid protein in a density of about 1 nucleocapsid protein per 10 nucleotides of viral RNA (1). It continues with the resulting viral ribonucleoprotein (RNP) spiral being encased into a bullet-shaped particle by another protein shell consisting of more than 1,000 copies of matrix (M) protein, and it ends with budding of this particle through a specific region of the host cell plasma membrane containing the viral membrane glycoprotein.
Other than serving as one of the principal construction materials of VSV, the M protein also evolved mimicry capabilities to affect several host cell pathways in favor of virus reproduction. The principal host cell targets of the viral M protein have been identified. They are the mRNA export factor 1 (Rae1; also known as Gle2 or mrnp41) (2), and the nucleoporin Nup98 (3). As for their cellular location, Rae1 and Nup98 home not only at the cytoplasmic side of the nuclear pore complex (NPC) (reviewed in ref. 4) but also, are kinetically partitioned among other components in both cytoplasmic and nucleoplasmic compartments (5, 6). Not surprisingly, M protein, therefore, affects multiple cellular pathways in both interphase and mitotic cells, such as mRNA export across the NPC (2, 3, 7), transcription or translation (8, 9) and assembly of spindle poles (10), respectively. Although identification of Rae1 and Nup98 as host cell targets for the M protein provided the first clues in the search for a common denominator in the bewilderingly large spectrum of M-protein effects, the function of these two host cell proteins remains only partially understood. As for Nup98, it not only contains numerous Phenylalanine-Glycine (FG) modules serving as ligands for nucleocytoplasmic transport factors (6, 11), but more tellingly, Nup98 also features a binding module for Rae1 (2, 12). As for Rae1, it displays an RNA binding activity that was first detected by cross-linking experiments (5). We recently determined a crystal structure for Rae1 in complex with Nup98 (12). Consistent with the presence of the RNA binding activity of Rae1, we found a surface groove of highly conserved positively charged residues along the side of the Rae1 β-propeller (12). Indeed, in vitro experiments detected binding of single-stranded oligoribonucleotides (12), suggesting that this surface feature of Rae1 may represent its RNA binding site. As a canonical seven-bladed β-propeller, Rae1 is a likely binding platform for other proteins; however, none of these putative interactomes have so far been characterized at the molecular level.
As a logical next step in shedding additional light on the dialogue between M protein, Rae1, and Nup98, we followed up our previous work on the crystal structure of the Rae1•Nup98 heterodimer. We succeeded in assembling a higher-order structure consisting of a 1:1:1 heterotrimeric complex of M•Rae1•Nup98 and solved its crystal structure at 3.15-Å resolution. Our crystal data revealed that M protein attaches to both Rae1 and Nup98 of the Rae1•Nup98 heterodimer. Binding occurs essentially through two prominent protrusions, designated finger (residues 49–61) and thumb (residues 213–223), which project from a fist-like globular domain of M protein. Notably, the finger region of M attaches to a previously identified groove of positively charged residues on the side of the Rae1 β-propeller, which we previously noted as a candidate for the binding site of Rae1 for ssRNA (12). To further test this notion, we carried out in vitro binding assays using a larger variety of nucleic acid proxies than previously used, including single- and double-stranded oligomers of ribonucleotides or deoxyribonucleotides or tRNA. We found that M protein was able to displace all of them from Rae1•Nup98. These data suggest that (i) Rae1 not only binds phosphate backbone of ssRNA but recognizes the phosphate backbone of any nucleic acid and (ii) M protein behaves like a mimic for phosphate backbone and competes for nucleic acid binding to Rae1. Our structural interactome data strongly suggested that the activity of M protein is localized to (or overlaps with) its finger region. Indeed, a synthetic peptide representing the finger of the M protein (residues 49–61) competed for nucleic acid phosphate backbone binding, whereas a peptide representing its thumb (residues 213–223) did not. Collectively, our data assist in rationalizing the large spectrum of effects of M protein. One of these effects, namely its inhibition of mRNA export, is discussed in the context of a Brownian ratchet model (13, 14) for transport of mRNA across a ligand-gated channel of the nuclear pore complex (15, 16).
Results
Assembly and Structure Determination of M•Rae1•Nup98 Complex.
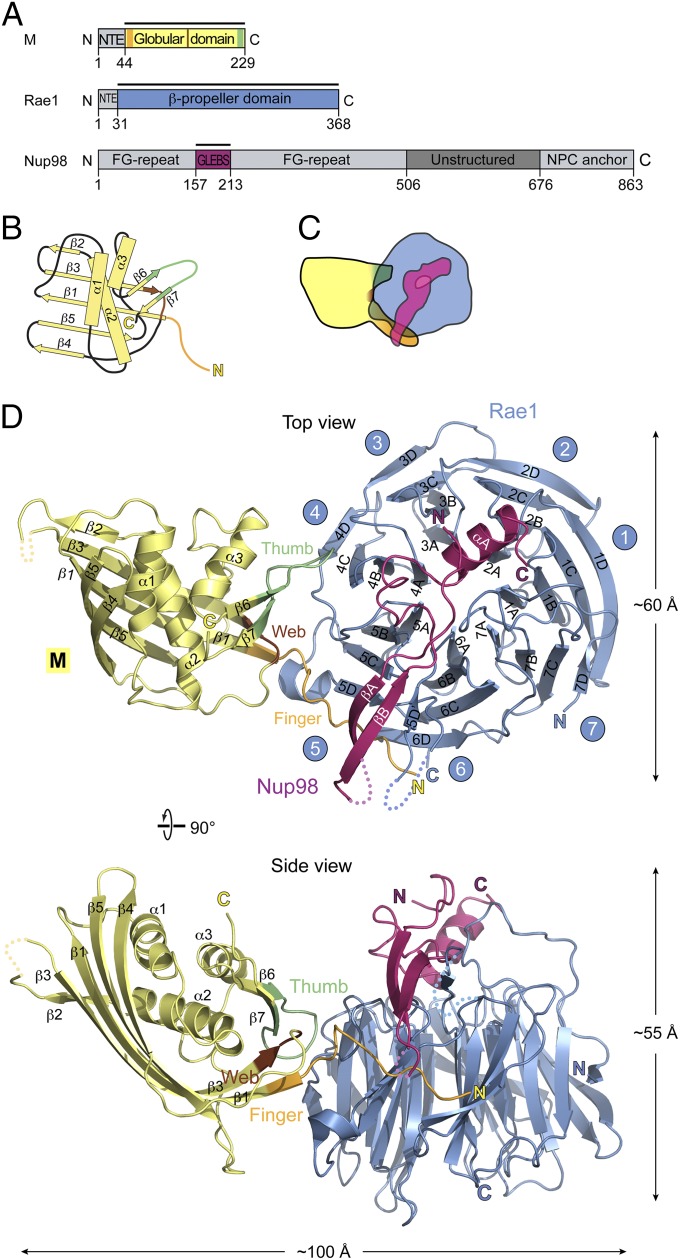
Relevant annotated primary and domain structures for M protein of VSV, Rae1, and Nup98 are shown in Fig. 1A and Figs. S1–S3. In the assembled virus, M protein multimerizes through contacts between N-terminal and central regions of neighboring M proteins (Fig. S4) (17, 18). These repetitive contacts yield an M protein spiral of over 1,000 molecules wrapping up viral RNP (17). To prevent multimerization, we mutated the central region of M protein (residues 121–124) from AVLA to GGSG (Figs. S1 and S4) and designated this mutant as *M protein. Moreover, deletion of an N-terminal extension (NTE; residues 1–43) of full-length *M protein yielded *M44–229 (Fig. S1). By the criterion of size-exclusion chromatography and multiangle light scattering, *M44–229 behaved as a monomer (Fig. S5). Importantly, *M44–229 remained competent to associate with an Rae1•Nup98 1:1 heterodimer to form a 1:1:1 heterotrimeric complex of *M44–229•Rae1•Nup98 (Fig. S5). This heterotrimer only yielded poorly diffracting crystals. An important clue for obtaining better diffracting crystals came from our previous structure determination of Rae1•Nup98 showing that full-length Rae1 (denoted by Rae1FL) contains an unstructured N-terminal extension (NTE; residues 1–30) that mediates Rae1 tetramer formation in crystallo (12). Notably, this Rae1 tetramer formation involves two of three FG motifs, which are present in the NTE of Rae1; these two FG motifs attached to respective hydrophobic pockets in the β-propeller region of a neighboring Rae1 molecule (see below) (12). Because Rae1 tetramerization might interfere with crystallization of an *M44–229•Rae1•Nup98 heterotrimer, we replaced Rae1FL by N-terminally truncated Rae1 (Fig. 1A). We refer to this complex in the text as M•Rae1•Nup98 heterotrimer. The heterotrimer formed crystals in the primitive tetragonal space group P4212, with one trimer in the asymmetric unit. The structure was solved by molecular replacement and refined to a 3.15-Å resolution with Rwork and Rfree values of 21.3% and 28.9%, respectively. Details of crystallographic statistics are in Table S1.
Fig. 1.
Structure of a 1:1:1 complex of M•Rae1•Nup98. (A) Domains of each full-length polypeptide are annotated (Figs. S1–S3). Recombinant protein fragments that were used for assembling the M•Rae1•Nup98 trimeric complex for crystallization (in the text) are indicated by bars and color-coded: blue, Rae1; magenta, Nup98; yellow, M protein. Moreover, the three surface features of M protein interacting with Rae1•Nup98 (see below) were designated and color-coded: brown, web (residues 142–144); green, thumb (residues 213–223); orange, finger (residues 49–61). Nup98 features numerous FG repeats, a predicted unstructured region, and binding sites for Rae1 (also referred to as GLEBS for Gle2 binding sequence) and Nup88 (denoted by NPC anchor) (Fig. 5). (B) Schematic topology diagram: location of secondary structural elements of the globular portion of M protein with respect to finger, web, and thumb. (C) Schematic contour diagram of the trimeric complex colored as in A and viewed as in D, Upper. (D) Ribbon presentation of the trimeric complex, colored as in A; Upper and Lower represent top and side views, respectively, with regard to the β-propeller domain of Rae1 and include annotations for secondary and tertiary structures of the three moieties of the trimeric complex (Figs. S1–S3). Note that the finger of M protein contacts both Rae1 and Nup98 moieties, whereas the web and thumb contact only Rae1.
Structural Overview.
The previously reported structures of either M protein (19) or the 1:1 Rae•Nup98 heterodimer (12) are not significantly altered when these reactants associate with each other and form a heterotrimeric M•Rae1•Nup98 complex (Fig. 1). The M protein contacts the Rae1•Nup98 heterodimer primarily through three prominent surface features designated finger (residues 49–61), thumb (residues 213–223), and web (a small region in between; residues 142–144) (Fig. 1 and Fig. S1). Notably, the finger contacts both Rae1 and Nup98 of the Rae•Nup98 heterodimer, whereas the web and thumb contact only Rae1 (Figs. 1 and 2). The complementary contact sites on the Rae1•Nup98 heterodimer extend alongside blades 4–6 of the Rae1 β-propeller and include the β-tongue of the Nup98 hairpin overlying the top of the Rae1 β-propeller (Fig. 1) (12). The dimensions of the complex are ∼100 × 55 × 60 Å. Surface representations of the heterotrimeric complex and its various interfaces show patches of evolutionary conservation and distinct surface potentials (Fig. S6) (12).
Fig. 2.
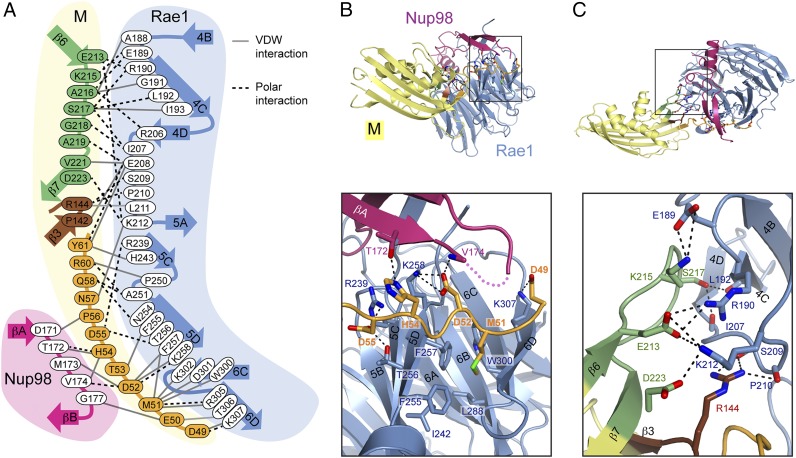
Interface between M protein and Rae1•Nup98. (A) All residues involved at the interface between M and Rae1•Nup98 are shown. Black dotted lines indicate polar interactions, including salt bridges and hydrogen bonds. Gray lines indicate van der Waals (VDW) interactions. (B and C) Ribbon representation is colored as in Fig. 1. B, Inset and C, Inset illustrate the positions of (B) index finger and (C) thumb and web of the M protein, respectively and are expanded B, Lower and C, Lower. Polar interaction network at the interfaces is indicated by black dashes.
Interface Between M Protein and Rae1•Nup98 Heterodimer.
A residue-by-residue interactome of the finger and thumb region of M protein with Rae1•Nup98 is shown in Fig. 2 (corresponding annotations are in Figs. S1–S3). The finger of M protein contains several key residues. Of them, a highly conserved Methionine (Met; Met51) (Fig. S1) together with acidic residues (D49, E50, D52, and D55) on either side of Met51 provide the majority of bonds to Rae1. Met51 of the finger region bonds to five residues (F257, W300, D301, K302, and R305) lining a deep hydrophobic pocket along blade 5 of the Rae1 β-propeller. Curiously, a completely unrelated Met residue, Met17 of the NTE of Rae1, bonds to identical residues on that same hydrophobic pocket on blade 5 of a collateral Rae1 (Fig. S7) (12), suggesting mimicry of these two Met residues (Discussion). Of the other residues of the finger of M protein, His54 stands out by providing several bonds to β-tongue of Nup98 (Fig. 2 A and B). However, by far the most revealing aspect of the interactome of M protein with Rae1•Nup98 is the location of the finger of M protein in the previously noted groove along blades 5 and 6 of the β-propeller of Rae1. This groove is lined by highly conserved basic residues (R239, K258, K302, R305, and K307) and for this reason, has previously been surmised to serve as RNA binding site (see below) (12). The bonding of the web and thumb of M protein is accomplished by polar and van der Waals interactions (Figs. 1 and 2 and Figs. S1–S3). These interactomes seem to serve as downstream reinforcements for the interaction of the finger of M protein with Rae1•Nup98.
Binding Sites on Rae1 for the Finger of M Protein or Phosphate Backbone of Nucleic Acids Overlap.
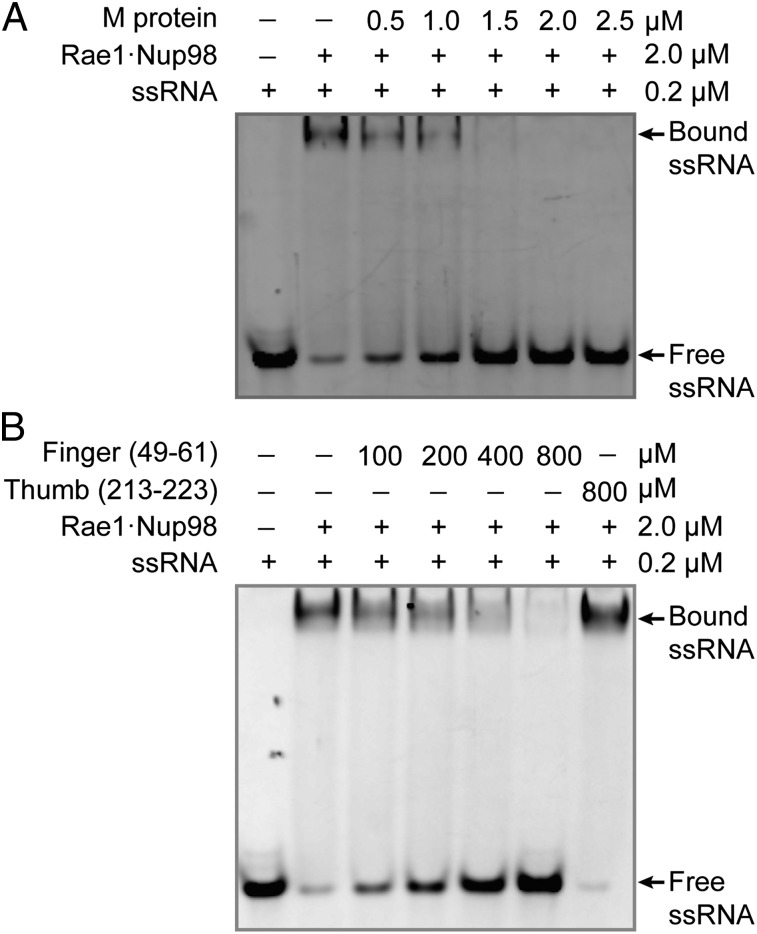
Using an EMSA, we found that M protein competed with ssRNA for binding to Rae1•Nup98 in a concentration-dependent manner. Complete inhibition of RNA binding was observed at an about equimolar concentration of M protein with respect to Rae1•Nup98 (Fig. 3A).
Fig. 3.
VSV M protein displaces ssRNA from Rae1FL•Nup98. (A) After preincubation of Alexa-488–labeled 10-mer poly(U) ssRNA and Rae1FL•Nup98157–213, the mixture was further incubated with increasing amounts of *M44–229 and analyzed by an EMSA. The final concentrations of the mixture are indicated. Presence of RNA was detected by fluorescence of Alexa-488. (B) Increasing amounts of the 13-mer finger peptide (residues 49–61) as indicated were added to the preformed ssRNA•Rae1FL•Nup98157–213 complex and analyzed as in A. The 11-mer thumb peptide (residues 213–223) was tested at 800 μM.
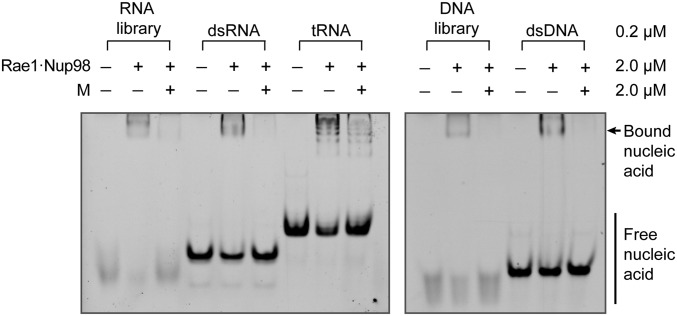
To biochemically localize the inhibitory activity of M protein, we tested two synthetic peptides: one representing the finger (residues 49–61) and one representing the thumb (residues 213–223). Strikingly, the finger peptide but not the thumb peptide inhibited RNA binding to Rae1•Nup98 (Fig. 3B), although inhibition required considerably higher concentration of the finger peptide than M protein. We conclude that the inhibition of RNA binding to Rae1•Nup98 by M protein is localized to its finger, whereas the web and thumb serve to merely reinforce the binding activity of the finger. Binding of random sequences of ssRNA suggested that recognition is not base-specific but only involves the phosphate backbone. This conclusion was further supported by showing that a variety of other synthetic oligomers, both of ribo- and deoxyribonucleotides, single or double stranded, as well as a mixture of yeast tRNAs were all outcompeted by M protein (Fig. 4). These data suggest that Rae1 is a member of the large family of proteins that binds nucleic acid phosphate backbones.
Fig. 4.
VSV M protein displaces different types of nucleic acids from the Rae1•Nup98 complex. After preincubation with nucleic acid and the Rae1FL•Nup98157–213 complex, the mixture was further incubated in the absence or presence of *M44–229. All final concentrations are indicated on the right. Representative nucleic acids include 21-mer RNA and DNA libraries containing random sequences, 21-mer dsRNA and dsDNA of nonrandom sequence (28), and yeast tRNAs (Roche). After EMSA, nucleic acid binding was visualized by SYBR-GOLD nucleic acid staining. Note that *M44–229 displaced each of the tested nucleic acids from the Rae1FL•Nup98157–213 complex.
Discussion
Our crystallographic data here on 1:1:1 M•Rae1•Mup98 heterotrimer showed how M protein of VSV interacts with Rae1 and Nup98. The M protein contacts the Rae1•Nup98 heterodimer through three distinct regions that project from a fist-like globular domain: a finger (residues 49–61), a thumb (residues 213–223), and a small region in between termed the web (residues 142–144) (Figs. 1 and 2). Additional biochemical studies revealed binding of phosphate backbones of any of the tested nucleic acid to Rae1•Nup98 heterodimer and showed that this binding is mutually exclusive with that of M protein.
The finger attaches to both the Rae1 and Nup98 moieties of Rae1•Nup98 heterodimer, whereas the thumb and web contact only Rae1 (Fig. 2). Notably, the finger’s Met51 interacts with five residues (F257, W300, D301, K302, and R305) that line a deep pocket on the side of the Rae1 β-propeller (Fig. 2B); this extensive bonding of Met51 is consistent with previously reported mutational studies that pointed to its critical role in M-protein function (20). Remarkably, the identical Rae1 pocket can be alternatively occupied by a completely unrelated Met residue, namely Met17 of a neighboring Rae1, which was recently described in the Rae1•Nup98 crystal structure (Fig. S7) (12); this observation suggests that M protein’s Met51 mimics Rae1’s Met17 (see below for discussion of this mimicry). Strikingly, the acidic residues of the finger (D49, D52, and D55) on either side of Met51 form polar bonds with highly conserved residues on the side of Rae1 β-propeller (R239, K258, and K307) (Fig. 2 and Figs. S1 and S2). Notably, the finger also latches on to Nup98 (Fig. 2 and Fig. S3).
To extend previous biochemical data showing binding of synthetic single-stranded oligoribonucleotides to Rae1•Nup98 heterodimer, we also tested oligomers of dsRNA, oligomers of single- and double-stranded deoxyribonucleotides, and a mixture of yeast tRNAs. All of these nucleic acid proxies acted as ligands and bound to Rae1•Nup98, suggesting that Rae1•Nup98 recognizes the phosphate backbone of nucleic acids. Strikingly, M protein outcompeted these ligands when present in the incubation mixture at equimolar concentration relative to Rae1•Nup98. We localized the inhibitory activity of the M protein to the finger region (residues 49–61) using a synthetic peptide. Collectively, these data suggest that Rae1 is a member of the large family of proteins that binds to nucleic acid phosphate backbone and that the finger of M protein inhibits this activity of Rae1.
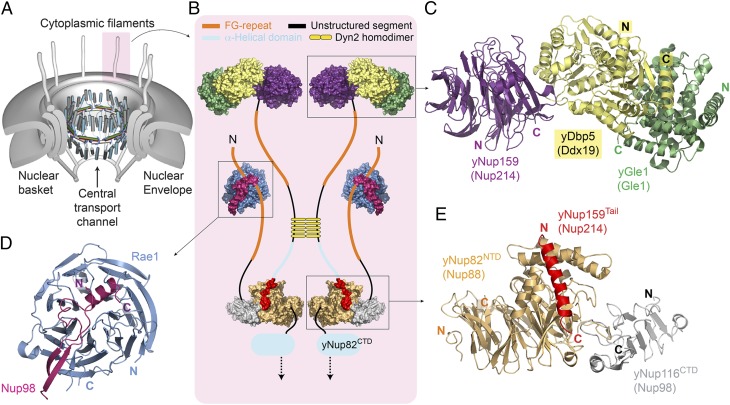
To relate our data to M-protein inhibition of mRNA export across the NPC (2, 7), we schematically summarized our current level of understanding of molecular interactomes of various filament nucleoporins anchored to the cytoplasmic side of the NPC (Fig. 5). This schematic model is based on crystallographic data derived from interactome analyses of yeast (y) or homologous vertebrate nucleoporins. Our model in Fig. 5 omits two additional vertebrate nucleoporins on the cytoplasmic side of the NPC, Nup358 and hCG1 (yNup42), because structural analyses regarding anchorage of these two nucleoporins to the NPC are presently not available (the nucleoporin inventory is given in ref. 4). The symmetric core of NPC (twofold axis in midplane and an eightfold rotational axis in the nucleocytoplasmic direction) is schematically depicted as a series of concentric cylinders (Fig. 5A). Principal structural elements of a recently proposed model for the architecture of a central transport channel, shown in its dilated state, are depicted in the center of the symmetric core (15, 16) (Fig. 5A).
Fig. 5.
Neighborhood of Rae1•Nup98 on the cytoplasmic side of the nuclear pore. (A) An overall schematic model for NPC architecture. It shows a series of concentric cylinders (anchored to the pore membrane domain of the nuclear envelope) and the attached cytoplasmic filaments and nuclear basket. A recently proposed structural model for the central transport channel with a midplane ring (shown in the open state) and attached structures on either side (16) are indicated. Modified from ref. 15. (B–E) Close-up views of components of one cytoplasmic filament (shaded in pink in A): the crystal structures are composites representing interactomes of either yeast or vertebrate (in parentheses) nucleoporins, color-coded, and shown in (B) space-filling or (C–E) ribbon representation. (C) yGle1•Dbp5•Nup159 [Protein Data Bank (PDB) ID code 3RRM] (24). (D) Human Rae1•Nup98 (PDB ID code 4OWR). (E) yNup82•Nup159•Nup116 (PDB ID code 3PBP) (21). At the cytoplasmic filament base, yNup82 (Nup88) serves as a node. (B and E) Its N-terminal domain interactome encompasses the C-terminal tail of yNup159 and the C-terminal domain (CTD) of yNup116 (Nup98). (B) The yNup82 (Nup88)-CTD interactome with nucleoporin(s) of the cylindrical core still awaits analyses at the molecular level. The farther course of yNup159 (Nup214) and yNup116 (Nup98) with (B–E) upstream interactomes and various unstructured regions with and without FG repeats is annotated in B (in the text); the length of the color-coded lines is roughly proportional to the predicted length of unstructured regions. Note that five homodimers of yeast dynein light chain (yDyn2) link two yNup159 molecules and thereby, dimerize two yNup82 modules. Because interactomes of vertebrate Nup358 and hCG1 (yNup42) with other nucleoporins remain uncharacterized at the molecular level, they have been omitted in this neighborhood model of Rae1•Nup98.
A close-up model of a cytoplasmic filament (Fig. 5 A and B) shows yNup82 at the base of the cytoplasmic filaments, where it is circularly anchored to the cytoplasmic side of the cylindrical NPC core. To date, the binding of yNup82 to nucleoporin(s) of the cylindrical symmetric core remains uncharacterized at the molecular level (Fig. 5B). In contrast, the binding site of yNup82 to two filament nucleoporins, yNup116 (vertebrate Nup98) and yNup159 (vertebrate Nup214), has been elucidated at the molecular level (21). Both yNup116 and yNup159 are anchored to yNup82 by their C-terminal domains. For yNup159, additional upstream regions of two yNup159 molecules are dimerized by five homodimers of yeast dynein light chain (22); the yNup159 polypeptide then continues in a natively unfolded form with numerous interspersed FG motifs (serving as ligands for transport factors) before folding into a β-propeller that reversibly binds helicase yDbp5 and its associated proteins (Fig. 5B) (23–25). After its C-terminal anchorage to yNup82, yNup116 (Nup98) also continues as a natively unfolded chain with interspersed FG motifs and then folds into a hairpin structure that binds to the top of the Rae1 β-propeller before ending with another natively unfolded stretch with FG motifs (Fig. 5B) (12, 21).
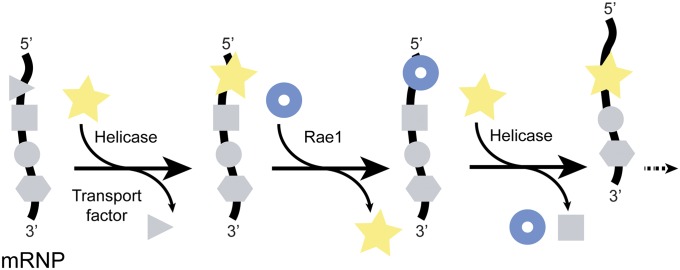
Extensive remodeling of mRNP occurs at the cytoplasmic filaments of the NPC. Here, a given mRNA-bound protein is replaced during an ATPase cycle during which helicase yDbp5 (vertebrate Ddx19) binds and unbinds from its cognate yNup159 (Nup214) (reviewed in ref. 26). The resulting deproteinated stretch of mRNA is likely to be rapidly reproteinated. We propose that the nearby Nup98-anchored Rae1 containing a binding site for nucleic acid phosphate backbone serves as a proxy for an mRNA-binding protein, rapidly saturating a transiently vacated protein binding site on mRNA (Fig. 6). Alternatively, other nearby stopover sites might be available (e.g., in the form of the recently identified RNA binding site on Nup358) (27) to assure that ratcheting of mRNA (13, 14) across the central channel of the NPC does not fail.
Fig. 6.
Rae1•Nup98 functions as a phosphate backbone-binding protein in a ratchet model for mRNA export across the central transport channel of the nuclear pore. At cytoplasmic filaments of the nuclear pore, mRNA-bound proteins (gray symbols) are sequentially removed, one by one, in the 5′ to 3′ direction (29). The removal of each protein requires one ATPase cycle of helicase (yellow star) (24, 30, 31). The protein-vacated stretch of the mRNA phosphate backbone is immediately occupied by the nearby Rae1 (blue ring) (in the text and Fig. 5). The number of these repeat cycles is predicted to be proportional to the length of mRNA.
In the context of a central channel of the nuclear pore that dilates in response to cellular needs for nucleocytoplasmic transport (15, 16), one can surmise numerous reactions to cooperate in accomplishing flexibility in the overall structure of the NPC. One of these adjuvant reactions could be dimerization (or tetramerization) of Nup98-attached Rae1 (12), conceivably to reinforce a constricted configuration of the central transport channel on its cytoplasmic side. Because Met51 of the finger region of M protein might prevent this oligomerization by mimicking Met17 of Rae1 (Fig. S7), it could abolish the contribution of Rae1 to a constricted state on the cytoplasmic side. By affecting the central channel dynamics, the M protein could make the NPC more independent of cellular controls for nucleocytoplasmic traffic.
Materials and Methods
The details of protein expression and purification, crystallization, structure determination, multiangle light scattering, and EMSA are described in SI Materials and Methods. In short, the dimeric Rae131–368·Nup98157–213 complex was coexpressed in High Five insect cells (Invitrogen), and *M44–229 was expressed in Escherichia coli. Rae131–368·Nup98157–213 and *M44–229 were purified separately, and the trimeric 1:1:1 complex of *M44–229·Rae131–368·Nup98157–213 was purified by size-exclusion chromatography. X-ray diffraction data were collected at Beamline 24ID-C at the Advanced Photon Source (Argonne National Laboratory). The structure was solved by molecular replacement. Data collection and refinement statistics are summarized in Table S1.
Supplementary Material
Acknowledgments
We thank Sozanne Solmaz for help with multiangle light-scattering experiment and Deena Oren for help with initial screening of crystals using an in-house X-ray generator. We also thank Beatriz Fontoura for reagents and the staffs at Beamline 24ID-C at Advanced Photon Source for help with data collection.
Footnotes
The authors declare no conflict of interest.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4OWR).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1409076111/-/DCSupplemental.
References
- 1.Thomas D, et al. Mass and molecular composition of vesicular stomatitis virus: A scanning transmission electron microscopy analysis. J Virol. 1985;54(2):598–607. doi: 10.1128/jvi.54.2.598-607.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Faria PA, et al. VSV disrupts the Rae1/mrnp41 mRNA nuclear export pathway. Mol Cell. 2005;17(1):93–102. doi: 10.1016/j.molcel.2004.11.023. [DOI] [PubMed] [Google Scholar]
- 3.von Kobbe C, et al. Vesicular stomatitis virus matrix protein inhibits host cell gene expression by targeting the nucleoporin Nup98. Mol Cell. 2000;6(5):1243–1252. doi: 10.1016/s1097-2765(00)00120-9. [DOI] [PubMed] [Google Scholar]
- 4.Hoelz A, Debler EW, Blobel G. The structure of the nuclear pore complex. Annu Rev Biochem. 2011;80:613–643. doi: 10.1146/annurev-biochem-060109-151030. [DOI] [PubMed] [Google Scholar]
- 5.Kraemer D, Blobel G. mRNA binding protein mrnp 41 localizes to both nucleus and cytoplasm. Proc Natl Acad Sci USA. 1997;94(17):9119–9124. doi: 10.1073/pnas.94.17.9119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Radu A, Moore MS, Blobel G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell. 1995;81(2):215–222. doi: 10.1016/0092-8674(95)90331-3. [DOI] [PubMed] [Google Scholar]
- 7.Her LS, Lund E, Dahlberg JE. Inhibition of Ran guanosine triphosphatase-dependent nuclear transport by the matrix protein of vesicular stomatitis virus. Science. 1997;276(5320):1845–1848. doi: 10.1126/science.276.5320.1845. [DOI] [PubMed] [Google Scholar]
- 8.Black BL, Lyles DS. Vesicular stomatitis virus matrix protein inhibits host cell-directed transcription of target genes in vivo. J Virol. 1992;66(7):4058–4064. doi: 10.1128/jvi.66.7.4058-4064.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Black BL, Brewer G, Lyles DS. Effect of vesicular stomatitis virus matrix protein on host-directed translation in vivo. J Virol. 1994;68(1):555–560. doi: 10.1128/jvi.68.1.555-560.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chakraborty P, et al. Vesicular stomatitis virus inhibits mitotic progression and triggers cell death. EMBO Rep. 2009;10(10):1154–1160. doi: 10.1038/embor.2009.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conti E, Müller CW, Stewart M. Karyopherin flexibility in nucleocytoplasmic transport. Curr Opin Struct Biol. 2006;16(2):237–244. doi: 10.1016/j.sbi.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 12.Ren Y, Seo H-S, Blobel G, Hoelz A. Structural and functional analysis of the interaction between the nucleoporin Nup98 and the mRNA export factor Rae1. Proc Natl Acad Sci USA. 2010;107(23):10406–10411. doi: 10.1073/pnas.1005389107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simon SM, Peskin CS, Oster GF. What drives the translocation of proteins? Proc Natl Acad Sci USA. 1992;89(9):3770–3774. doi: 10.1073/pnas.89.9.3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stewart M. Ratcheting mRNA out of the nucleus. Mol Cell. 2007;25(3):327–330. doi: 10.1016/j.molcel.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 15.Solmaz SR, Chauhan R, Blobel G, Melčák I. Molecular architecture of the transport channel of the nuclear pore complex. Cell. 2011;147(3):590–602. doi: 10.1016/j.cell.2011.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Solmaz SR, Blobel G, Melcák I. Ring cycle for dilating and constricting the nuclear pore. Proc Natl Acad Sci USA. 2013;110(15):5858–5863. doi: 10.1073/pnas.1302655110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ge P, et al. Cryo-EM model of the bullet-shaped vesicular stomatitis virus. Science. 2010;327(5966):689–693. doi: 10.1126/science.1181766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graham SC, et al. Rhabdovirus matrix protein structures reveal a novel mode of self-association. PLoS Pathog. 2008;4(12):e1000251. doi: 10.1371/journal.ppat.1000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaudier M, Gaudin Y, Knossow M. Crystal structure of vesicular stomatitis virus matrix protein. EMBO J. 2002;21(12):2886–2892. doi: 10.1093/emboj/cdf284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petersen JM, Her LS, Varvel V, Lund E, Dahlberg JE. The matrix protein of vesicular stomatitis virus inhibits nucleocytoplasmic transport when it is in the nucleus and associated with nuclear pore complexes. Mol Cell Biol. 2000;20(22):8590–8601. doi: 10.1128/mcb.20.22.8590-8601.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshida K, Seo H-S, Debler EW, Blobel G, Hoelz A. Structural and functional analysis of an essential nucleoporin heterotrimer on the cytoplasmic face of the nuclear pore complex. Proc Natl Acad Sci USA. 2011;108(40):16571–16576. doi: 10.1073/pnas.1112846108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stelter P, et al. Molecular basis for the functional interaction of dynein light chain with the nuclear-pore complex. Nat Cell Biol. 2007;9(7):788–796. doi: 10.1038/ncb1604. [DOI] [PubMed] [Google Scholar]
- 23.Napetschnig J, et al. Structural and functional analysis of the interaction between the nucleoporin Nup214 and the DEAD-box helicase Ddx19. Proc Natl Acad Sci USA. 2009;106(9):3089–3094. doi: 10.1073/pnas.0813267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montpetit B, et al. A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export. Nature. 2011;472(7342):238–242. doi: 10.1038/nature09862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.von Moeller H, Basquin C, Conti E. The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner. Nat Struct Mol Biol. 2009;16(3):247–254. doi: 10.1038/nsmb.1561. [DOI] [PubMed] [Google Scholar]
- 26.Valkov E, Dean JC, Jani D, Kuhlmann SI, Stewart M. Structural basis for the assembly and disassembly of mRNA nuclear export complexes. Biochim Biophys Acta. 2012;1819(6):578–592. doi: 10.1016/j.bbagrm.2012.02.017. [DOI] [PubMed] [Google Scholar]
- 27.Kassube SA, et al. Crystal structure of the N-terminal domain of Nup358/RanBP2. J Mol Biol. 2012;423(5):752–765. doi: 10.1016/j.jmb.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seo H-S, Blus BJ, Jankovic NZ, Blobel G. Structure and nucleic acid binding activity of the nucleoporin Nup157. Proc Natl Acad Sci USA. 2013;110(41):16450–16455. doi: 10.1073/pnas.1316607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mehlin H, Daneholt B, Skoglund U. Translocation of a specific premessenger ribonucleoprotein particle through the nuclear pore studied with electron microscope tomography. Cell. 1992;69(4):605–613. doi: 10.1016/0092-8674(92)90224-z. [DOI] [PubMed] [Google Scholar]
- 30.Hodge CA, et al. The Dbp5 cycle at the nuclear pore complex during mRNA export I: dbp5 mutants with defects in RNA binding and ATP hydrolysis define key steps for Nup159 and Gle1. Genes Dev. 2011;25(10):1052–1064. doi: 10.1101/gad.2041611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noble KN, et al. The Dbp5 cycle at the nuclear pore complex during mRNA export II: Nucleotide cycling and mRNP remodeling by Dbp5 are controlled by Nup159 and Gle1. Genes Dev. 2011;25(10):1065–1077. doi: 10.1101/gad.2040611. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.