Significance
Error-prone DNA repair of activation-induced cytidine deaminase (AID)-induced lesions in B cells is known to cause hypermutation of the antibody genes and, when coupled with selection mechanisms in germinal centers, leads to increased affinity of antibody. These lesions can also lead to substantial genomic instability that can promote lymphomagenesis, and the cause of error-prone repair is unknown. We have found that expression of an essential DNA repair protein, AP endonuclease (APE) 1, is dramatically reduced in mouse germinal center B cells where somatic hypermutation occurs. By contrast, the very inefficient homologue, APE2, becomes highly expressed. We show that APE2 promotes mutations and present a model proposing that differential expression of APE homologues in germinal centers is a major reason for error-prone repair of AID-induced lesions.
Abstract
Somatic hypermutation (SHM) of antibody variable region genes is initiated in germinal center B cells during an immune response by activation-induced cytidine deaminase (AID), which converts cytosines to uracils. During accurate repair in nonmutating cells, uracil is excised by uracil DNA glycosylase (UNG), leaving abasic sites that are incised by AP endonuclease (APE) to create single-strand breaks, and the correct nucleotide is reinserted by DNA polymerase β. During SHM, for unknown reasons, repair is error prone. There are two APE homologs in mammals and, surprisingly, APE1, in contrast to its high expression in both resting and in vitro-activated splenic B cells, is expressed at very low levels in mouse germinal center B cells where SHM occurs, and APE1 haploinsufficiency has very little effect on SHM. In contrast, the less efficient homolog, APE2, is highly expressed and contributes not only to the frequency of mutations, but also to the generation of mutations at A:T base pair (bp), insertions, and deletions. In the absence of both UNG and APE2, mutations at A:T bp are dramatically reduced. Single-strand breaks generated by APE2 could provide entry points for exonuclease recruited by the mismatch repair proteins Msh2–Msh6, and the known association of APE2 with proliferating cell nuclear antigen could recruit translesion polymerases to create mutations at AID-induced lesions and also at A:T bp. Our data provide new insight into error-prone repair of AID-induced lesions, which we propose is facilitated by down-regulation of APE1 and up-regulation of APE2 expression in germinal center B cells.
During humoral immune responses, the recombined antibody variable [V(D)J] region genes undergo somatic hypermutation (SHM), which, after selection, greatly increases the affinity of antibodies for the activating antigen. This process occurs in germinal centers (GCs) in the spleen, lymph nodes, and Peyer’s patches (PPs) and entirely depends on activation-induced cytidine deaminase (AID) (1, 2). AID initiates SHM by deamination of cytidine nucleotides in the variable region of antibody genes, converting the cytosine (dC) to uracil (dU) (1, 3, 4). Some AID-induced dUs are excised by the ubiquitous enzyme uracil DNA glycosylase (UNG), resulting in abasic (AP) sites that can be recognized by apurinic/apyrimidinic endonuclease (APE) (4, 5). APE cleaves the DNA backbone at AP sites to form a single-strand break (SSB) with a 3′ OH that can be extended by DNA polymerase (Pol) to replace the excised nucleotide (6). In most cells, DNA Pol β performs this extension with high fidelity, reinserting dC across from the template dG. In contrast, GC B cells undergoing SHM are rapidly proliferating, and some of the dUs are replicated over before they can be excised and are read as dT by replicative polymerases, resulting in dC to dT transition mutations. Unrepaired AP sites encountering replication lead to the nontemplated addition of any base opposite the site, causing transition and transversion mutations. However, it is not clear why dUs and AP sites escape accurate repair by the highly efficient enzymes UNG and APE1 and lead instead to mutations.
Instead of removal by UNG, some U:G mismatches created by AID activity are recognized by the mismatch repair proteins Msh2–Msh6, which recruit exonuclease 1 to initiate excision of one strand surrounding the mismatch (7–9). The excised region (estimated at ∼200 nt; ref. 10) is subsequently filled in by DNA Pols, including error-prone translesion Pols, which spreads mutations beyond the initiating AID-induced lesion. The combined, but noncompeting interaction of the UNG and MMR pathways in generating mutations at A:T base pairs (bp) has been described (10–12). This mismatch repair-dependent process has been termed phase II of SHM (3). Pol η and Msh2–Msh6 have been shown to be essential for nearly all mutations at A:T bp (13–15). During repair of the excision patch, additional C:G bp can be mutated by translesion Pols, but mutations at C:G bp due to AID activity can also be repaired back to the original sequence during this step (16).
Mammals express two known homologs of AP endonuclease (APE), APE1 and APE2. APE1 is the major APE; it is ubiquitously expressed and essential for early embryonic development in mice and for viability of human cell lines (17–19). APE1 has strong endonuclease activity and weaker 3′-5′ exonuclease (proofreading) and 3′-phosphodiesterase (end-cleaning) activities (20, 21). Recombinant purified human APE2 has much weaker AP endonuclease activity than APE1, but its 3′-5′ exonuclease activity is strong compared with APE1, although it is not processive (20). However, APE2 has been shown to interact with proliferating cell nuclear antigen (PCNA) (22), which can recruit error-prone translesion polymerases (23, 24), and PCNA also increases the processivity of APE2 exonuclease in vitro (25). Both APE1 and APE2 are expressed in splenic B cells activated in culture (26). APE2 is nonessential, but APE2-deficient mice show a slight growth defect, a twofold reduction of peripheral B and T cells (27), and impaired proliferation of B-cell progenitors in the bone marrow (28).
In this study we examine SHM in GC B cells isolated from the PPs of unimmunized apex1+/−, apex2Y/−, and apex1+/−apex2Y/− mice relative to WT mice. [Because the APE2 gene is located on the X chromosome, we used APE2-deficient male mice (apex2Y/−) in all experiments.] We demonstrate that not only is APE2 important for SHM frequency, as reported (29), but APE2 also contributes to the generation of A:T mutations. The proportion of mutations at A:T bp is reduced in apex2Y/− mice to the same extent as it is in ung−/− mice, consistent with APE2 acting as an endonuclease that incises AP sites generated by UNG. Surprisingly, in the absence of both UNG and APE2, mutations at A:T bp are greatly reduced. In addition, we find that expression of APE1 is dramatically reduced in GC B cells, and APE1 haploinsufficiency has very little effect on SHM. We propose a model in which APE2 promotes SHM through inefficient and error-prone repair, whereas APE1, which is known to interact with XRCC1 and Pol β to promote error-free SSB repair (30, 31), is suppressed in GC B cells.
Results
APE1 Is Down-Regulated and APE2 Is Up-Regulated in GC Cells.
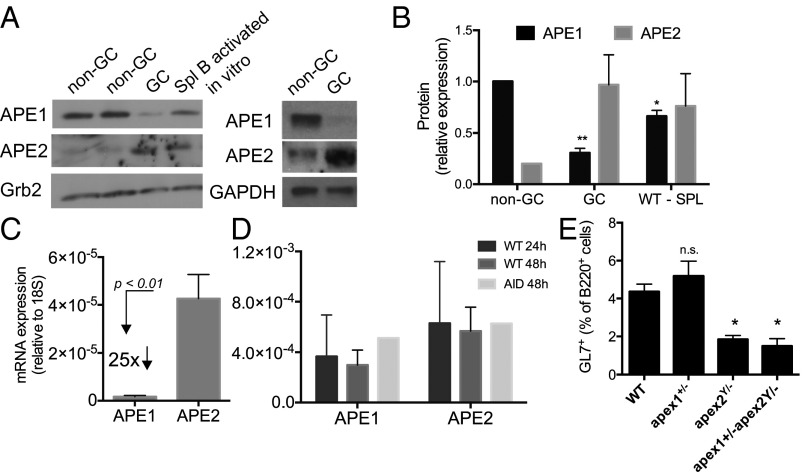
Previously we showed that APE1 is constitutively expressed and APE2 is inducibly expressed in mouse splenic B cells induced to undergo CSR in culture (26). Because these cultured B cells do not undergo SHM, which occurs only in vivo in GCs, we compared the expression of APE1 and APE2 in total cell extracts of GC and non-GC cells from PPs. Surprisingly, APE1 is expressed at dramatically lower levels in GC B cells than in non-GC B cells, whereas APE2 expression is greatly induced in GC cells (Fig. 1 A and B). APE1 expression is also lower in GC B cells than in spleen B cells activated in culture with LPS and IL-4 (P < 0.008). The expression of APE1 in human tonsil GCs is also very low, as shown by immunohistochemistry (32), The Human Protein Atlas (www.proteinatlas.org). In contrast, we showed that DNA Pol β is expressed equally in mouse GC and non-GC B cells, and in spleen B cells activated in culture (33). The low level of APE1 is reflected in its very low mRNA level relative to that of APE2 mRNA, using quantitative PCR primers that amplify equally efficiently (Fig. 1C). Recent reports have also shown low levels of APE1 transcripts in the GC (34, 35), especially in the rapidly proliferating centroblasts of the dark zone, relative to centrocytes in the light zone where selection occurs. Cultured spleen B cells express roughly equivalent amounts of APE1 and APE2 mRNA (Fig. 1D), similar to results for protein levels. APE2 is clearly important for the proliferation and/or survival of GC B cells, because APE2 deficiency causes a twofold reduction in the percent of B cells that are GL7+ GC cells (Fig. 1E). APE1 haploinsufficiency does not reduce the percent of GC cells, in keeping with the already very low levels of APE1 expressed.
Fig. 1.
APE1 expression is reduced and APE2 is increased in germinal center (GC) B cells. (A) Western blotting of 20 μg of whole-cell extract from FACS-purified PP GC and non-GC B cells, or spleen B cells activated in vitro for 48 h with LPS, IL-4, and BLyS. Anti-Grb2 or anti-GAPDH antibody was used as a loading control; two representative blots from three independent sorting experiments are shown. (B) Protein expression levels relative to non-GC B cells determined by densitometry analysis of Western blots (mean ± SEM, n = 3). Results were normalized to Grb2 levels. APE1 levels in non-GC B cells were arbitrarily set at 1.0; APE2 levels in non-GC B were set at 0.2, which represented the average ratio of APE2 to APE1 in non-GC B cells, relative to the loading control. **P < 0.004; *P < 0.03; paired t test analysis. APE1 in GC B cells is also reduced relative to spleen B cells activated in vitro (P < 0.008). (C and D) mRNA expression levels, relative to 18S rRNA, in GC B cells (C) (mean ± SEM, n = 4) and spleen B cells activated in vitro (D) (n = 2 for WT 24h and 48h, mean ± range, and n = 1 for AID 48h). (E) PP GC cells as a percent of B220+ cells (mean ± SEM, n = 5 except for apex1+/−apex2Y/−, n = 4); *P < 0.05.
Analysis of SHM in APE-Deficient Mice.
To analyze SHM in apex1+/−, apex2Y/−, and apex1+/−apex2Y/− mice in comparison with wild-type (WT) mice, we examined PP GC cells because they are undergoing persistent activation by numerous antigens, thus allowing examination of a large variety of B-cell clones. Similar to studies by other investigators, we analyzed SHM in a 493-bp segment located immediately 3′ to JH4 to avoid effects on the DNA sequences due to selection for antigen specificity (36). GC B cells were sorted from 7-wk to 9-mo-old unimmunized mice, and age did not appear to affect the results. APE1-heterozygous mice were analyzed because apex1−/− mice die during early embryogenesis, and APE1 heterozygotes are haploinsufficient and have DNA repair and CSR deficiencies (26, 37, 38).
Frequency of SHM Is Decreased in apex2Y/− Mice and Not Affected by APE1 Haploinsufficiency.
Table 1 presents the mutation frequency and base specificity data for the WT and APE-deficient mice. We analyzed an average of 30,000 nt per mouse, and the average of the individual mice is shown ± SEM. The data on individual mice, and the number of nts sequenced and mutations analyzed are reported in Table S1. Relative to WT mice, SHM in the JH4–3′ intron is essentially unaffected by apex1 haploinsufficiency. Although we cannot fully assess the impact of APE1 deficiency on SHM because we can only study apex1 heterozygotes, it is clear from the mutation frequencies and base specificity that APE1 has very little role in this process, consistent with its very low expression levels in GC B cells. In contrast, the mutation frequency is reduced by 50% in apex2Y/− mice, as shown (29), and also in apex1+/−apex2Y/− mice. The contribution of APE2 toward mutation frequency may be either direct, as discussed below, or indirect, because APE2 is important for B-cell proliferation and survival (28), and the percentage of GC B cells is reduced in its absence. However, because the nucleotide specificity of mutations is also altered in the absence of APE2, we conclude that APE2 has a direct role in SHM.
Table 1.
SHM analysis of the 3′JH4 flank in GC B cells deficient in APE and UNG
| Genotype | No. of mice | Mutation frequency (x 10−3)* | Percent of mutations at A:T bp† | Percent of C:G mutations that are transitions‡ | Percent of mutations at WRC/GYW hotspots§ | Frequency of indels (x 10−4)¶ |
| Wild type | 7 | 22.4 ± 3.5 | 58.9 ± 1.3 | 58.5 ± 2.9 | 17.7 ± 0.7 | 1.81 ± 0.24 |
| apex1+/− | 5 | 19.2 ± 2.6 | 55.1 ± 2.4 | 60.2 ± 1.8 | 20.3 ± 1.9 | 2.22 ± 1.02 |
| 0.516 | 0.154 | 0.675 | 0.161 | 0.660 | ||
| apex2Y/− | 5 | 11.1 ± 2.5 | 48.3 ± 1.8 | 63.3 ± 2.6 | 23.8 ± 1.1 | 0.77 ± 0.38 |
| 0.037 | <0.001 | 0.267 | <0.001 | 0.033 | ||
| apex1+/−apex2Y/− | 5 | 11.5 ± 1.1 | 46.2 ± 3.9 | 66.5 ± 4.2 | 22.2 ± 0.3 | 0.99 ± 0.36 |
| 0.031 | 0.005 | 0.135 | <0.001 | 0.072 | ||
| ung−/− | 3 | 20.0 ± 1.9 | 45.6 ± 1.4 | 90.7 ± 1.5 | 28.8 ± 1.2 | 0.71 ± 0.24 |
| 0.695 | <0.001 | <0.001 | <0.001 | 0.024 | ||
| ung−/−apex1+/− | 3 | 15.5 ± 2.7 | 41.5 ± 4.0 | 92.6 ± 1.2 | 29.3 ± 2.4 | 0.76 ± 0.23 |
| v. WT | 0.275 | <0.001 | <0.001 | <0.001 | 0.029 | |
| v. ung | 0.240 | 0.387 | 0.521 | 0.854 | 0.885 | |
| ung−/−apex2Y/− | 3 | 6.5 ± 0.6 | 24.4 ± 2.1 | 95.7 ± 1.5 | 45.6 ± 0.4 | 0.88 ± 0.64 |
| v. WT | 0.022 | <0.001 | <0.001 | <0.001 | 0.120 | |
| v. ung | 0.024 | 0.001 | 0.170 | <0.001 | 0.808 | |
| ung−/−apex1+/−apex2Y/− | 4 | 7.8 ± 1.9 | 25.0 ± 4.7 | 97.6 ± 1.1 | 38.5 ± 2.4 | 0.27 ± 0.16 |
| v. WT | 0.017 | <0.001 | <0.001 | <0.001 | 0.001 | |
| v. ung | 0.007 | 0.015 | 0.040 | 0.023 | 0.170 | |
| v. ung/apex2 | 0.595 | 0.567 | 0.349 | 0.056 | 0.330 |
Numbers in italics are P values.
Mean frequency of mutations/103 bp of individual mice ± SEM; P values, shown below each mean, are from t test of means relative to WT. For data on individual mice, see Table S1.
Corrected for sequence composition.
The calculations for base specificity exclude identical mutations found in sequences from the same mouse that have the same CDR3.
AID hotspots defined as WRC/GYW where Y = pyrimidine, W = A or T, R = purine. Mutations counted if they occur at the underlined C or G.
Insertions and deletions per 104 nt; mean of individual mice ± SEM.
Decreased Mutations at A:T Base Pairs in apex2Y/− Mice.
Although SHM is initiated by the deamination of dC by AID, 59% of the mutations (after correction for the base composition of the sequence) in WT mice are at A:T bp. A:T mutations are generated by the activities of Msh2-Msh6 and Pol η, which require SSBs as entry points to initiate excision and translesion synthesis (5, 39). The percent of mutations occurring at A:T bp is decreased to 48% (P < 0.001) in mice that lack APE2 and to 46% in both apex1+/−apex2Y/− mice and ung−/− mice (Table 1). Apex1+/− mice have only a very small decrease in A:T mutations, which is not statistically different from WT. The percent of mutations at AID-target hotspots (WRC/GYW) is also slightly, but significantly, increased from 18% in WT to 24% of mutations in the absence of APE2. Based on these data, we propose that APE2 functions as an endonuclease creating SSBs at AP sites generated by AID and UNG activities that allow mismatch repair and Pol η to generate mutations at surrounding A:T bp. However, because nearly half of the mutations are still at A:T bp, it is clear that there is another source of SSBs for creating mutations at A:T bp.
APE2 Function in the Absence of UNG.
Having proposed that APE2 and UNG act in the same pathway, we decided to test this by creating ung−/−apex2Y/− mice, which we expected would have the same phenotype with respect to A:T mutations as UNG deficiency. However, as shown in Table 1, ung−/−apex2Y/− mice have a further twofold reduction in the percent of mutations at A:T bp (24%; P < 0.001) compared with ung−/− mice and apex2Y/− mice, and also the proportion of mutations at AID hotspots is 2.6-fold greater in ung−/−apex2Y/− mice than in WT mice. These results are surprising because they indicate that APE2 can affect SHM in the absence of UNG, and suggest that another uracil DNA glycosylase might substitute for UNG during SHM, creating AP sites that can be incised by APE2. APE1 heterozygosity on an UNG-deficient background does not alter SHM relative to UNG deficiency alone. Similarly, APE-1 heterozygosity in ung−/−apex2Y/− mice does not alter SHM significantly relative to ung−/−apex2Y/− mice (Table 1). These data indicate that APE2 is the predominant APE for introducing SSBs at AP sites created by the AID-UNG pathway during SHM, and that backup activity, which may include SMUG1 (Discussion) and low levels of APE1, is very inefficient at spreading mutations beyond the initial lesion introduced by AID.
Insertions and Deletions Are Reduced.
SSBs made by APE during SHM could lead to insertion and deletion mutations (indels). Although this type of mutation is rare within the JH4 3′ inton during SHM (1.8 × 10−4 per bp in WT mice), we observed a 60% reduction in the frequency of indels in the absence of APE2 (Table 1), and also in the absence of UNG. The frequency of indels was reduced by 85% in the ung−/−apex1+/−apex2Y/− mice, suggesting that some SSBs made by the UNG-APE pathway could lead to double-strand breaks (DSBs) during SHM. However, indels can also be introduced by other mechanisms besides DSBs.
Transition Mutations at C:G bp Are Slightly Increased in the Absence of APE2.
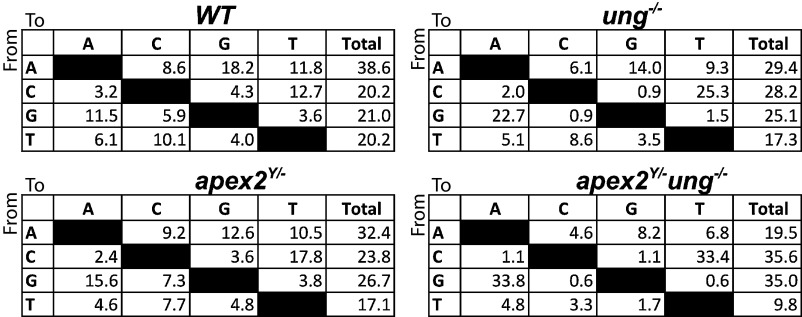
In WT B cells, 59% of the mutations at C:G bp are transition mutations, suggesting that many of the dUs introduced by AID are replicated over before excision by UNG. Although there is a trend toward increased transition mutations at C:G bp in the APE-deficient mice (Table 1 and Fig. 2), the difference is not significant, as might be expected because UNG is present. As reported by others (4), UNG-deficient mice have greatly increased transitions (91%), and we find that in the absence of both UNG and APE2, nearly all mutations at C:G are transitions (96% and 98% in ung−/−apex2Y/− and ung−/−apex1+/−apex2Y/−, respectively). An alternative uracil DNA glycosylase, acting in the absence of UNG, could explain the few transversions still observed in the ung−/− and ung−/−apex1+/−apex2Y/− mice.
Fig. 2.
Spectra of mutations in the J558VHFR3-JH4 3′ intron segments in pooled sequences from PP GC B cells from the indicated genotypes. The data are presented as percent of mutations, corrected for the base composition of the 493-bp segment. Base composition of the 493-bp segment is as follows: A, 26.4%; C, 14.4%; G, 27.2%; T, 31.8%.
To attempt to determine whether APE deficiency affects the activity of Pol η during SHM, we analyzed the ratio of A:T mutations, because Pol η shows strand bias with a twofold preference for mutating A relative to T (40). We observed no consistent changes in the A:T ratio of the mutations (Fig. 2 and Table S1). These data suggest that another translesion Pol does not substitute for Pol η in the absence of APE.
Discussion
We report here the surprising finding that expression of APE1, the highly efficient, essential, and otherwise ubiquitous AP endonuclease, is dramatically down-regulated in GC B cells. APE1 is effectively replaced in the GC by APE2, a very inefficient APE homolog whose expression is greatly induced. We found that APE2 not only increases the frequency of mutations, but also specifically increases mutations at A:T bp. We propose that the decrease in APE1 combined with up-regulation of APE2 in rapidly dividing GC cells explains a long-standing mystery as to why repair of AID-induced lesions is error prone.
The reduction in APE1 expression alone could be sufficient to explain a large portion of phase I mutations, which are generated when AID-induced lesions at C:G bp are encountered by DNA polymerase. It has not been clear why dUs and the resulting AP sites generated by UNG encounter polymerase and give rise to transitions and transversions in GC B cells, in contrast to their accurate repair in all other cell types. Reduced APE1 levels could explain both aspects of this error-prone repair. First, APE1 has been shown to increase the turnover rate of UNG by displacing it from double-stranded AP sites, thus increasing UNG activity (41). As such, decreased APE1 levels would lead to less UNG activity, leaving more dUs to encounter polymerase. APE2 might also have this activity, but might be much less efficient because it is an inefficient endonuclease. Second, it is unlikely that AP sites would go unrepaired and encounter DNA polymerase given normal levels of APE1 expression. The enzymatic activity of APE1 for AP-site repair has been reported to be from 70- to 450,000-fold more efficient than that of APE2 (20, 21, 42). In addition, APE1 interacts directly with XRCC1 (30, 43), a scaffolding protein that recruits Pol β and ligase to promote SSB repair. This interaction was mapped to the N terminus of APE1, a unique domain not found in APE2. XRCC1 is an essential gene, but xrcc1+/− mice have slightly increased SHM frequency (44), demonstrating that this APE1-mediated repair pathway could suppress SHM. Indeed, the fact that only a small effect on SHM is seen in xrcc1+/− mice could be due to the very low levels of APE1 expression in GC cells. Pol β deficiency does not affect the base specificity of mutations during SHM (33, 45), consistent with our hypothesis that the APE1-Pol β pathway has little or no direct effect on SHM. The lack of effect of Pol β likely results from the very low APE1 expression in GC B cells, as opposed to our previous interpretation that the BER pathway does not contribute to SSBs needed for A:T mutations (33). By contrast, it has recently been shown that mice expressing a mutant Pol β with very slow polymerization kinetics have increased SHM of the JH4 flank in GC B cells, with the largest effect on transversions at C:G bp (46). Because Pol β interacts with APE1 and XRCC1 during repair of AP sites (31, 47), this mutant Pol β might reduce the turnover of APE1, further decreasing the effective APE1 levels and allowing increased access of APE2 to AP sites.
In this study, we found that the alternative use of APE2 contributes to SHM in GC B cells. APE2-deficient GC cells are reduced in number and may proliferate fewer times because of increased levels of DNA damage, which could contribute indirectly to the reduced frequency of mutations we and others (29) observed. However, the fact that the base specificity of the mutations is also changed indicates that APE2 plays a direct role in SHM in addition to supporting GC cell proliferation. A:T mutations, which require SSBs as entry points for recruitment of exonuclease by Msh2–Msh6 and for Pol η to perform translesion synthesis, are reduced in apex2Y/− B cells to the exact same extent as they are in ung−/− cells. This reduction is consistent with APE2 endonuclease incision activity at AP sites generated by UNG. Although the incision activity of APE2 is weak in biochemical studies, APE2 is highly expressed in GC B cells and could be specifically recruited to increase its efficacy. Our previous data on the role of APE2 in CSR also supports the conclusion that APE2 acts as an endonuclease at AP sites made by UNG (26). We showed previously that DSBs occurring at AID hotspots are near background levels in apex1+/−apex2Y/− B cells, even with one allele of apex1 intact. Furthermore, A:T mutations induced in the unrearranged (germ-line) 5′ Sμ segment in cells undergoing Ig class switching are also reduced in apex1+/−apex2Y/− B cells relative to WT cells (26).
In addition to the inefficient endonuclease activity of APE2, we hypothesize that an interaction between APE2 and PCNA also promotes error-prone repair in GCs. Unlike APE1, APE2 has a functional PCNA-interacting domain (48). PCNA monoubiquitinated at lysine 164 is known to recruit the translesional polymerases Pol η and Rev1 (23), and SHM analyses in mice expressing a knock-in PCNAK164R mutation show a reduction in A:T mutations (49, 50). The 3′ to 5′ exonuclease processivity of APE2 is also stimulated by PCNA (25) and could contribute to mutations by excising a few nucleotides at a SSB, exposing a short single-strand patch 5′ to the AP site that could be filled in by Pol η, also resulting in mutations at A:T bp. This activity could explain the fact that during SHM, there are some A:T mutations in the absence of Msh2–Msh6, although Msh2–Msh6 is required for the vast majority of mutations at A:T bp (7). However, because the exonuclease activities of APE1 and APE2 are associated with proofreading functions, it is possible that their activity also correctly repairs some AID-induced lesions (20, 51).
Because UNG excises the dU base, thus creating the substrate for APE2, we predicted that mice deficient in both UNG and APE2 would have the same SHM phenotype as that of UNG-deficient mice. However, they do not. We find that ung−/−ape2Y/− mice have a further twofold reduction in the percent of mutations at A:T bp, with almost half (46%) of the mutations occurring at AID hotspots. This represents a dramatic decrease in phase II mutations, demonstrating that the spreading of mutations beyond the initial lesion made by AID is very inefficient in the absence of both UNG and APE2. That APE2 and UNG deficiencies affect SHM additively suggests that UNG and APE2 can each act in alternative pathways. UNG works very efficiently with APE1 (52), and the low level of APE1 might partially substitute for APE2 in its absence. Another uracil DNA glycosylase, such as SMUG1, could substitute for UNG (53), although SMUG1 is much less efficient than UNG. It binds AP sites very tightly and depends more on APE for its turnover (41, 52). Therefore, in ung−/−ape2Y/− GC B cells, which have SMUG1 and only very low levels of APE1, this pathway would be very inefficient at generating SSBs that can lead to mutations at A:T bp. Alternatively, or in addition, our data are consistent with the possibility that APE2 might incise the DNA backbone at dU bases, in addition to AP sites, consistent with a recent report (54). Although APEs are thought to act specifically at AP sites, this study showed that APE1 can incise DNA at dU in duplex DNA, albeit inefficiently. APE2 has not been tested for this activity, which would not be expected to contribute to the process of SHM under normal conditions, but could impact mutations in the absence of UNG.
It will be interesting to see how our finding of very low APE1 expression in GC B cells impacts current models of CSR. Although the process of CSR is associated with the GC reaction, class switching also occurs in vivo before GC formation. CSR requires DSB intermediates and, therefore, might require the more robust enzymatic activity of APE1. The mechanism and timing of APE1 down-regulation is unknown, but our results indicate that it occurs at the RNA level. APE1 might be highly expressed in cells undergoing CSR in extrafollicular sites and in B-cell follicles before differentiation of GCs, just as it is in activated B cells in culture. Also, the recently reported association of APE1 with AID (55), dependent on phosphorylation of AIDSer38, could result in specific recruitment of APE1 in GC B cells to switch regions to make DSBs required for CSR, thus overcoming its low level of expression. Although we find that APE2 contributes to CSR in cultured splenic B cells (26), APE2 does not contribute to CSR in CH12F3 cells (56) possibly due to specific requirements or genetic alterations of this cell line. However, because almost all studies to date on the mechanism of CSR and switch region DSBs have been performed in cultured cells, more studies on CSR in B cells in vivo are needed. It is possible that the high levels of APE1 expressed in B cells induced in vitro to undergo CSR could promote accurate repair of AID-induced lesions in the V region and explain the fact that SHM has not been observed in cultured B cells.
Materials and Methods
Mice.
All mouse strains were backcrossed to C57BL/6 for more than eight generations and before interbreeding to create double- and triple-deficient mice. Apex1+/− mice were obtained from E. Friedberg (38) (University of Texas Southwestern Medical Center, Dallas, TX). APE2-deficient mice were described (27), and UNG-deficient mice were obtained from T. Lindahl and D. Barnes (London Research Institute, London, England). Because apex2 is on the X chromosome, we used male apex2Y/− mice in all experiments. The WT mice were littermates of either the apex2Y/− or apex1+/−apex2Y/− mice. Mice were housed in the Institutional Animal Care and Use Committee-approved specific pathogen-free facility at the University of Massachusetts Medical School. The mice were bred and used according to the guidelines from the University of Massachusetts Animal Care and Use Committee.
Analysis and Purification of GC B Cells.
PP cells from WT, apex1+/−, apex2Y/−, and DBL mice were mechanically dispersed and stained with anti–B220-APC, anti–CD95-PE, anti–GL7-FITC, and 7AAD. Viable (7AAD−), GC (B220+CD95+GL7+), and non-GC (B220+CD95−GL7−) B cells were isolated on a FACSAriaII (>92% purity), and frozen for subsequent DNA extraction, or lysed in RIPA buffer for Western blotting or in TRIzol (Life Technologies) for RNA extraction. WT control spleen cells were T-depleted with antibody and complement and cultured as described (19), with LPS, IL-4, and BLyS.
Western Blotting.
Pelleted cells from FACS-purified or cultured B cells were lysed in RIPA buffer, and 15–20 μg of whole-cell extracts were analyzed on 8% (wt/vol) polyacrylamide gels as described (19) with goat anti-APE1 (R&D Systems), rabbit anti-APE2 (AnaSpec), and rabbit anti-Grb2 (growth factor receptor bound protein 2) and anti-GAPDH (glyceraldehyde 3-phosphate dehydrogenase) (Santa Cruz) primary antibodies, with goat anti-rabbit and donkey anti-goat secondary antibodies couple to horseradish peroxidase (Santa Cruz). Densitometry was performed on a G:BOX Chemi XL (Syngene).
Quantitative RT-PCR.
RNA was prepared from cells in TRIzol, and cDNA was made by using oligo dT and SuperScript II reverse transcriptase (Invitrogen). Quantitative PCR was performed on a ViiA7 (Applied Bioscience) at 60 °C with primers for apex1 [Forward (F): 5′- CTCAAGATATGCTCCTGGAA and reverse (R): 5′- GGTATTCCAGTCTTACCAGA, apex2 (F: 5′- CTTACACAGCACAAGATCCG and R: AAAGCATTCCAGACTACTTGC], and 18S RNA (F: 5′- TGGTGGAGGGATTTGTCTGG and R: 5′- TCAATCTCGGGTGGCTGAAC. Primers did not amplify genomic DNA.
Amplification, Cloning, and Sequence Analysis of J558VHFR3-JH4 3′ Intron Segments.
To assay SHM, the VHJ558L framework 3 - JH4 3′ flanking region was amplified by a nested PCR by using Pfu Ultra II (Stratagene); primers were modified slightly from ref. 36. Primers for the first amplification were as follows: F: 5′AGCCTGACATCTGAGGAC and R: 5′ GTGTTCCTTTGAAAGCTGGAC. Nested primers for the second amplification were as follows: F: 5′ CCGGAATTCCTGACATCTGAGGACTCTGC and R: 5′ GATGCCTTTCTCCCTTGACTC. The reaction conditions for the first primer set were 95 °C for 30 s, 57° for 30 s, and 72° for 1 min for 30 cycles; and for the second primer set, they were 95° for 30 s, 57° for 30 s, and 72° for 1 min for 35 cycles. The PCR products were electrophoresed on 1% agarose gels; the 600-bp band was purified by using QIAquick Gel Extraction Kit (Qiagen), dA tails were added with Taq polymerase, cloned by using TOPO TA cloning kit (Invitrogen), and sequenced by Macrogen. Unmutated sequences were excluded from the mutation frequency calculations shown in Table 1 and Table S1, because individual mice within each genotype, including WT mice, varied in the proportion of entirely unmutated sequences they have. As such, we cannot distinguish between sequences that are unmutated because of APE deficiency, and those that are due to contamination by nongerminal center cells, which could vary between experiments. Because SHM occurs over several cell generations, sequences with identical CDR3′s will have both shared and unique mutations. For frequency analyses, we counted all mutations in nonidentical sequences, but for base specificity analyses, we did not include identical mutations if they were in sequences with identical CDR3's.
Supplementary Material
Acknowledgments
We thank the University of Massachusetts Medical School Flow Cytometry Core Facility for excellent technical assistance and Dr. Michael Volkert (UMass Medical School) for helpful discussions. The research was supported by National Institutes of Health Grants R01 AI23283 and R21 AI88578 (to J.S.) and R01 AI065639 and R03 AI092528 (to C.E.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1405590111/-/DCSupplemental.
References
- 1.Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102(5):553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 2.Revy P, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102(5):565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 3.Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418(6893):99–103. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- 4.Rada C, et al. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr Biol. 2002;12(20):1748–1755. doi: 10.1016/s0960-9822(02)01215-0. [DOI] [PubMed] [Google Scholar]
- 5.Maul RW, Gearhart PJ. AID and somatic hypermutation. Adv Immunol. 2010;105:159–191. doi: 10.1016/S0065-2776(10)05006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Almeida KH, Sobol RW. A unified view of base excision repair: Lesion-dependent protein complexes regulated by post-translational modification. DNA Repair (Amst) 2007;6(6):695–711. doi: 10.1016/j.dnarep.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peled JU, et al. The biochemistry of somatic hypermutation. Annu Rev Immunol. 2008;26:481–511. doi: 10.1146/annurev.immunol.26.021607.090236. [DOI] [PubMed] [Google Scholar]
- 8.Schaetzlein S, et al. Mammalian Exo1 encodes both structural and catalytic functions that play distinct roles in essential biological processes. Proc Natl Acad Sci USA. 2013;110(27):E2470–E2479. doi: 10.1073/pnas.1308512110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bardwell PD, et al. Altered somatic hypermutation and reduced class-switch recombination in exonuclease 1-mutant mice. Nat Immunol. 2004;5(2):224–229. doi: 10.1038/ni1031. [DOI] [PubMed] [Google Scholar]
- 10.Frieder D, Larijani M, Collins C, Shulman M, Martin A. The concerted action of Msh2 and UNG stimulates somatic hypermutation at A. T base pairs. Mol Cell Biol. 2009;29(18):5148–5157. doi: 10.1128/MCB.00647-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schanz S, Castor D, Fischer F, Jiricny J. Interference of mismatch and base excision repair during the processing of adjacent U/G mispairs may play a key role in somatic hypermutation. Proc Natl Acad Sci USA. 2009;106(14):5593–5598. doi: 10.1073/pnas.0901726106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krijger PH, Langerak P, van den Berk PC, Jacobs H. Dependence of nucleotide substitutions on Ung2, Msh2, and PCNA-Ub during somatic hypermutation. J Exp Med. 2009;206(12):2603–2611. doi: 10.1084/jem.20091707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delbos F, Aoufouchi S, Faili A, Weill JC, Reynaud CA. DNA polymerase eta is the sole contributor of A/T modifications during immunoglobulin gene hypermutation in the mouse. J Exp Med. 2007;204(1):17–23. doi: 10.1084/jem.20062131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rada C, Ehrenstein MR, Neuberger MS, Milstein C. Hot spot focusing of somatic hypermutation in MSH2-deficient mice suggests two stages of mutational targeting. Immunity. 1998;9(1):135–141. doi: 10.1016/s1074-7613(00)80595-6. [DOI] [PubMed] [Google Scholar]
- 15.Wiesendanger M, Kneitz B, Edelmann W, Scharff MD. Somatic hypermutation in MutS homologue (MSH)3-, MSH6-, and MSH3/MSH6-deficient mice reveals a role for the MSH2-MSH6 heterodimer in modulating the base substitution pattern. J Exp Med. 2000;191(3):579–584. doi: 10.1084/jem.191.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roa S, et al. MSH2/MSH6 complex promotes error-free repair of AID-induced dU:G mispairs as well as error-prone hypermutation of A:T sites. PLoS ONE. 2010;5(6):e11182. doi: 10.1371/journal.pone.0011182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fung H, Demple B. A vital role for Ape1/Ref1 protein in repairing spontaneous DNA damage in human cells. Mol Cell. 2005;17(3):463–470. doi: 10.1016/j.molcel.2004.12.029. [DOI] [PubMed] [Google Scholar]
- 18.Xanthoudakis S, Smeyne RJ, Wallace JD, Curran T. The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc Natl Acad Sci USA. 1996;93(17):8919–8923. doi: 10.1073/pnas.93.17.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Izumi T, et al. Two essential but distinct functions of the mammalian abasic endonuclease. Proc Natl Acad Sci USA. 2005;102(16):5739–5743. doi: 10.1073/pnas.0500986102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burkovics P, Szukacsov V, Unk I, Haracska L. Human Ape2 protein has a 3′-5′ exonuclease activity that acts preferentially on mismatched base pairs. Nucleic Acids Res. 2006;34(9):2508–2515. doi: 10.1093/nar/gkl259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hadi MZ, Ginalski K, Nguyen LH, Wilson DM., 3rd Determinants in nuclease specificity of Ape1 and Ape2, human homologues of Escherichia coli exonuclease III. J Mol Biol. 2002;316(3):853–866. doi: 10.1006/jmbi.2001.5382. [DOI] [PubMed] [Google Scholar]
- 22.Tsuchimoto D, et al. Human APE2 protein is mostly localized in the nuclei and to some extent in the mitochondria, while nuclear APE2 is partly associated with proliferating cell nuclear antigen. Nucleic Acids Res. 2001;29(11):2349–2360. doi: 10.1093/nar/29.11.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garg P, Burgers PM. Ubiquitinated proliferating cell nuclear antigen activates translesion DNA polymerases eta and REV1. Proc Natl Acad Sci USA. 2005;102(51):18361–18366. doi: 10.1073/pnas.0505949102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: A possible mechanism for the polymerase switch in response to DNA damage. Mol Cell. 2004;14(4):491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 25.Burkovics P, Hajdú I, Szukacsov V, Unk I, Haracska L. Role of PCNA-dependent stimulation of 3′-phosphodiesterase and 3′-5′ exonuclease activities of human Ape2 in repair of oxidative DNA damage. Nucleic Acids Res. 2009;37(13):4247–4255. doi: 10.1093/nar/gkp357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guikema JE, et al. APE1- and APE2-dependent DNA breaks in immunoglobulin class switch recombination. J Exp Med. 2007;204(12):3017–3026. doi: 10.1084/jem.20071289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ide Y, et al. Growth retardation and dyslymphopoiesis accompanied by G2/M arrest in APEX2-null mice. Blood. 2004;104(13):4097–4103. doi: 10.1182/blood-2004-04-1476. [DOI] [PubMed] [Google Scholar]
- 28.Guikema JE, et al. Apurinic/apyrimidinic endonuclease 2 is necessary for normal B cell development and recovery of lymphoid progenitors after chemotherapeutic challenge. J Immunol. 2011;186(4):1943–1950. doi: 10.4049/jimmunol.1002422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sabouri Z, et al. Apex2 is required for efficient somatic hypermutation but not for class switch recombination of immunoglobulin genes. Int Immunol. 2009;21(8):947–955. doi: 10.1093/intimm/dxp061. [DOI] [PubMed] [Google Scholar]
- 30.Vidal AE, Boiteux S, Hickson ID, Radicella JP. XRCC1 coordinates the initial and late stages of DNA abasic site repair through protein-protein interactions. EMBO J. 2001;20(22):6530–6539. doi: 10.1093/emboj/20.22.6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caldecott KW. XRCC1 and DNA strand break repair. DNA Repair (Amst) 2003;2(9):955–969. doi: 10.1016/s1568-7864(03)00118-6. [DOI] [PubMed] [Google Scholar]
- 32.Uhlen M, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28(12):1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 33.Schrader CE, Linehan EK, Ucher AJ, Bertocci B, Stavnezer J. DNA polymerases β and λ do not directly affect Ig variable region somatic hypermutation although their absence reduces the frequency of mutations. DNA Repair (Amst) 2013;12(12):1087–1093. doi: 10.1016/j.dnarep.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Victora GD, et al. Identification of human germinal center light and dark zone cells and their relationship to human B-cell lymphomas. Blood. 2012;120(11):2240–2248. doi: 10.1182/blood-2012-03-415380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bannard O, et al. Germinal center centroblasts transition to a centrocyte phenotype according to a timed program and depend on the dark zone for effective selection. Immunity. 2013;39(5):912–924. doi: 10.1016/j.immuni.2013.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McDonald JP, et al. 129-derived strains of mice are deficient in DNA polymerase ι and have normal immunoglobulin hypermutation. J Exp Med. 2003;198(4):635–643. doi: 10.1084/jem.20030767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raffoul JJ, et al. Apurinic/apyrimidinic endonuclease (APE/REF-1) haploinsufficient mice display tissue-specific differences in DNA polymerase beta-dependent base excision repair. J Biol Chem. 2004;279(18):18425–18433. doi: 10.1074/jbc.M313983200. [DOI] [PubMed] [Google Scholar]
- 38.Meira LB, et al. Heterozygosity for the mouse Apex gene results in phenotypes associated with oxidative stress. Cancer Res. 2001;61(14):5552–5557. [PubMed] [Google Scholar]
- 39.Weill JC, Reynaud CA. DNA polymerases in adaptive immunity. Nat Rev Immunol. 2008;8(4):302–312. doi: 10.1038/nri2281. [DOI] [PubMed] [Google Scholar]
- 40.Mayorov VI, Rogozin IB, Adkison LR, Gearhart PJ. DNA polymerase eta contributes to strand bias of mutations of A versus T in immunoglobulin genes. J Immunol. 2005;174(12):7781–7786. doi: 10.4049/jimmunol.174.12.7781. [DOI] [PubMed] [Google Scholar]
- 41.Kavli B, et al. hUNG2 is the major repair enzyme for removal of uracil from U:A matches, U:G mismatches, and U in single-stranded DNA, with hSMUG1 as a broad specificity backup. J Biol Chem. 2002;277(42):39926–39936. doi: 10.1074/jbc.M207107200. [DOI] [PubMed] [Google Scholar]
- 42.Hadi MZ, Wilson DM., 3rd Second human protein with homology to the Escherichia coli abasic endonuclease exonuclease III. Environ Mol Mutagen. 2000;36(4):312–324. [PubMed] [Google Scholar]
- 43.Yamamori T, et al. SIRT1 deacetylates APE1 and regulates cellular base excision repair. Nucleic Acids Res. 2010;38(3):832–845. doi: 10.1093/nar/gkp1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saribasak H, et al. XRCC1 suppresses somatic hypermutation and promotes alternative nonhomologous end joining in Igh genes. J Exp Med. 2011;208(11):2209–2216. doi: 10.1084/jem.20111135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Esposito G, et al. Mice reconstituted with DNA polymerase beta-deficient fetal liver cells are able to mount a T cell-dependent immune response and mutate their Ig genes normally. Proc Natl Acad Sci USA. 2000;97(3):1166–1171. doi: 10.1073/pnas.97.3.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Senejani AG, et al. Mutation of POLB causes lupus in mice. Cell Reports. 2014;6(1):1–8. doi: 10.1016/j.celrep.2013.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bennett RA, Wilson DM, 3rd, Wong D, Demple B. Interaction of human apurinic endonuclease and DNA polymerase beta in the base excision repair pathway. Proc Natl Acad Sci USA. 1997;94(14):7166–7169. doi: 10.1073/pnas.94.14.7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ide Y, Tsuchimoto D, Tominaga Y, Iwamoto Y, Nakabeppu Y. Characterization of the genomic structure and expression of the mouse Apex2 gene. Genomics. 2003;81(1):47–57. doi: 10.1016/s0888-7543(02)00009-5. [DOI] [PubMed] [Google Scholar]
- 49.Roa S, et al. Ubiquitylated PCNA plays a role in somatic hypermutation and class-switch recombination and is required for meiotic progression. Proc Natl Acad Sci USA. 2008;105(42):16248–16253. doi: 10.1073/pnas.0808182105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langerak P, Nygren AO, Krijger PH, van den Berk PC, Jacobs H. A/T mutagenesis in hypermutated immunoglobulin genes strongly depends on PCNAK164 modification. J Exp Med. 2007;204(8):1989–1998. doi: 10.1084/jem.20070902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chou KM, Cheng YC. An exonucleolytic activity of human apurinic/apyrimidinic endonuclease on 3′ mispaired DNA. Nature. 2002;415(6872):655–659. doi: 10.1038/415655a. [DOI] [PubMed] [Google Scholar]
- 52.Pettersen HS, et al. Uracil-DNA glycosylases SMUG1 and UNG2 coordinate the initial steps of base excision repair by distinct mechanisms. Nucleic Acids Res. 2007;35(12):3879–3892. doi: 10.1093/nar/gkm372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dingler FA, Kemmerich K, Neuberger MS, Rada C. Uracil excision by endogenous SMUG1 glycosylase promotes efficient Ig class switching and impacts on A:T substitutions during somatic mutation. Eur J Immunol. 2014 doi: 10.1002/eji.201444482. 10.1002/eji.201444482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prorok P, et al. Uracil in duplex DNA is a substrate for the nucleotide incision repair pathway in human cells. Proc Natl Acad Sci USA. 2013;110(39):E3695–E3703. doi: 10.1073/pnas.1305624110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vuong BQ, et al. A DNA break- and phosphorylation-dependent positive feedback loop promotes immunoglobulin class-switch recombination. Nat Immunol. 2013;14(11):1183–1189. doi: 10.1038/ni.2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Masani S, Han L, Yu K. Apurinic/apyrimidinic endonuclease 1 is the essential nuclease during immunoglobulin class switch recombination. Mol Cell Biol. 2013;33(7):1468–1473. doi: 10.1128/MCB.00026-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.