Significance
Autoimmune type 1 diabetes is characterized by the formation of self-reactive antibodies. A prevalent human autoantigen is glutamate decarboxylase (GAD)65, a highly predictive marker that can precede the emergence of disease by up to several years. Intriguingly, the closely related isoform GAD67 is not immunogenic. What are the determinants of the unique self-reactivity of GAD65 vs. GAD67? We show that, unlike GAD67, GAD65 is highly flexible and exists in multiple structural forms. We show that self-antibodies bind differentially to these GAD65 forms. These properties may be an undesirable consequence of conformational flexibility necessary for enzyme function. Our findings, thus, provide insights into how structural flexibility governs protein immunogenicity in autoimmune diabetes and have implications for therapeutic antibody and vaccine design.
Keywords: conformational dynamics, normal mode analysis, GABA biosynthesis, immunogenicity, autoepitopes
Abstract
The human neuroendocrine enzyme glutamate decarboxylase (GAD) catalyses the synthesis of the inhibitory neurotransmitter gamma-aminobutyric acid (GABA) using pyridoxal 5′-phosphate as a cofactor. GAD exists as two isoforms named according to their respective molecular weights: GAD65 and GAD67. Although cytosolic GAD67 is typically saturated with the cofactor (holoGAD67) and constitutively active to produce basal levels of GABA, the membrane-associated GAD65 exists mainly as the inactive apo form. GAD65, but not GAD67, is a prevalent autoantigen, with autoantibodies to GAD65 being detected at high frequency in patients with autoimmune (type 1) diabetes and certain other autoimmune disorders. The significance of GAD65 autoinactivation into the apo form for regulation of neurotransmitter levels and autoantibody reactivity is not understood. We have used computational and experimental approaches to decipher the nature of the holo → apo conversion in GAD65 and thus, its mechanism of autoinactivation. Molecular dynamics simulations of GAD65 reveal coupling between the C-terminal domain, catalytic loop, and pyridoxal 5′-phosphate–binding domain that drives structural rearrangement, dimer opening, and autoinactivation, consistent with limited proteolysis fragmentation patterns. Together with small-angle X-ray scattering and fluorescence spectroscopy data, our findings are consistent with apoGAD65 existing as an ensemble of conformations. Antibody-binding kinetics suggest a mechanism of mutually induced conformational changes, implicating the flexibility of apoGAD65 in its autoantigenicity. Although conformational diversity may provide a mechanism for cofactor-controlled regulation of neurotransmitter biosynthesis, it may also come at a cost of insufficient development of immune self-tolerance that favors the production of GAD65 autoantibodies.
The two mammalian isoforms of glutamate decarboxylase (GAD; EC 4.1.1.15), GAD65 and GAD67, are responsible for the biosynthesis of the inhibitory neurotransmitter gamma-aminobutyric acid (GABA) (1, 2). GAD65 and GAD67 belong to the group II pyridoxal 5′-phosphate (PLP)-dependent enzymes of fold type I, which comprise two other evolutionarily related human enzymes: aromatic l-amino acid decarboxylase (AADC; synonym: l-dopa decarboxylase; EC 4.1.1.28) and histidine decarboxylase (EC 4.1.1.22) (3). Expression of these enzymes in the brain is responsible for the synthesis of the biogenic amines GABA, dopamine, serotonin, and histamine. Because they are implicated in a wide range of biological activities from central homeostatic functions to cognitive phenomena, it is not surprising that their activities need to be finely regulated. In this respect, the binding of the cofactor PLP to the apo forms of these enzymes might represent an important mechanism for regulation (4). The interaction of apoGAD with PLP is a major factor in the short-term regulation of GAD activity (5). Although GAD67 is responsible for basal production of GABA and constantly active (i.e., in the holo form), the major pool of GAD65 (at least 50%) exists as an inactive apoenzyme, which can be activated when extra GABA synthesis is required. The interconversion of holoGAD65 and apoGAD65 occurs by a cycle of reactions that include the primary α-decarboxylation reaction (which is the required step for GABA formation) followed by an alternative transamination reaction, leading to the production of succinic semialdehyde and formation of apoGAD65 as the consequence of pyridoxamine 5′-phosphate (PMP) release (5, 6).
GAD65 is a major autoantigen in patients with type 1 diabetes (T1D) and other autoimmune disorders (7). The series of events responsible for initiation of these autoimmune responses is unknown. GAD65 autoantibodies can be detected up to several years before the clinical onset of disease (8), which usually becomes manifest when >80% of β-cells are destroyed (9). Intriguingly, the other GAD isoform, GAD67, is seldom self-sufficiently autoantigenic (10). Such peculiarities of the GAD isoforms reflect their contrasting autoantigenic potential (11), despite their high sequence and structural similarity (6). Human GAD65 and GAD67 share 76% sequence identity overall, differing significantly only in the first 100 N-terminal amino acids. The major epitopes in T1D have been mapped to the PLP- and C-terminal regions (11, 12), and removal of the first 100 amino acids seems to affect neither enzyme activity nor reactivity with sera from diabetic patients (6). Thus, the highly homologous regions are paradoxically the source of differing antigenicity between the GAD isoforms.
Formation of active holoGAD65 from the apo-enzymic form involves a conformational change and increased stability (13). Although similar conformational changes have been observed for AADC and histidine decarboxylase (4, 14), the mechanisms underlying the conformational changes between holo and apo forms of the other members of group II decarboxylases have not been characterized. The crystal structures of both holoGAD65 and holoGAD67 (6) provided an initial opportunity to examine the contrasting autoantigenicities of the GAD isoforms (15, 16), which have been mostly correlated to the higher mobility and charge in the C-terminal domain (CTD) of GAD65 (residues 464–585) compared with that of GAD67 (residues 473–594). However, comparisons of the crystal structures ultimately failed to provide a satisfying explanation for the autoantigenicity of GAD65. Given the known autoinactivation of holoGAD65 in vivo, understanding the conformational changes that drive the holo → apo transition may provide additional insights into the autoantigenicity of GAD65.
Recently, the crystal structure of AADC was determined in an unexpected open conformation: compared with the AADC holoenzyme, the dimer subunits move up to 20 Å apart, and the two active sites become solvent-exposed (4). Intrigued by the possibility that apoGAD65 adopts a similar open conformation, we embarked on an investigation into the structural and biophysical properties of apoGAD65. Our key findings include an overall description of conformational opening of holoGAD65 and the dynamic communication between domains that drive this process. These results have implications for GABA homeostasis in the brain as well as autoimmune reactivity of GAD65 in T1D.
Results and Discussion
Apo- and HoloGAD65 Are More Dynamic than GAD67.
The active site of GAD is located at the center of the PLP-binding domain (residues 188–463 in GAD65 and 197–473 in GAD67) and covered by a catalytic loop (CL) contributed in trans by the other monomer of the functional dimer (6). The CL also contributes a tyrosine residue (Tyr425 in GAD65 and Tyr434 in GAD67) that is essential for catalytic activity. In the X-ray crystal structure of GAD67, the CL adopts a stable conformation, allowing Tyr434 to participate in the reaction. In contrast, the same loop in the GAD65 structure is too flexible to be built into electron density. Recently, a structure of the chimeric GAD6765loop revealed two conformations of the CL. One conformation is similar to that seen in GAD67 (the “in” conformation), whereas in the other conformation, the CL is out of the catalytic site (17). They were accompanied by alternative conformations in the adjacent CTDs, suggesting that the GAD structure may be a dynamic, isoform-specific equilibrium of conformations.
To investigate further the dynamics of GAD65 and GAD67, a series of molecular dynamics (MD) simulations were performed using holoGAD65, holoGAD67, and apoGAD65. Inspection of the RMSD values of the backbone (Fig. S1A) indicates that all simulated systems reach equilibrium in less than 30 ns of simulations, during which time an initial structural rearrangement is seen. Calculated values of backbone RMSD for the production stage (after the initial 50 ns) of each simulation are 0.34 ± 0.05 nm (apoGAD65), 0.37 ± 0.06 nm (holoGAD65), and 0.24 ± 0.03 nm (holoGAD67) (Fig. S1A). These data indicate that all simulated WT GAD systems are stable throughout the simulations. Apo- and holoGAD65 displayed higher RMSD values compared with holoGAD67, indicating that GAD65 is overall more dynamic than GAD67.
Closer inspection of the MD trajectories shows high flexibility of the CTDs of all simulated GAD isoforms, which are greater in apo- and holoGAD65 compared with holoGAD67 (Fig. 1A, Fig. S1B, and Movie S1). Moreover, whereas the CL of holoGAD67 is stable, the CLs of both apo- and holoGAD65 show high flexibility (Fig. S1C). This finding is in agreement with the crystal structures of GAD65 and GAD67 (6). A major functional aspect of loop flexibility is that the conserved catalytically important residue Tyr425 shows high flexibility in GAD65. The mean distances between the Nε atom of the PLP-Lys Schiff base (internal aldimine) and the Oη atom of the catalytic Tyr for the production stage of holoGAD simulations are 1.25 ± 0.53 nm (holoGAD65) and 0.74 ± 0.20 nm (holoGAD67). This finding indicates that, compared with Tyr434 of GAD67, Tyr425 of GAD65 spends less time in its catalytically competent conformation. Finally, the PLP-binding domain of apoGAD65 has higher flexibility compared with that of holoGAD65 and GAD67 (Fig. 1A, Fig. S1B, and Movie S1). This observation is also consistent with crystallographic data (6, 17), where the covalently bound PLP forms a network of interactions in the PLP domain, stabilizing this region. On PLP release, GAD65 dynamics are, therefore, expected to increase, which was observed in our MD analysis.
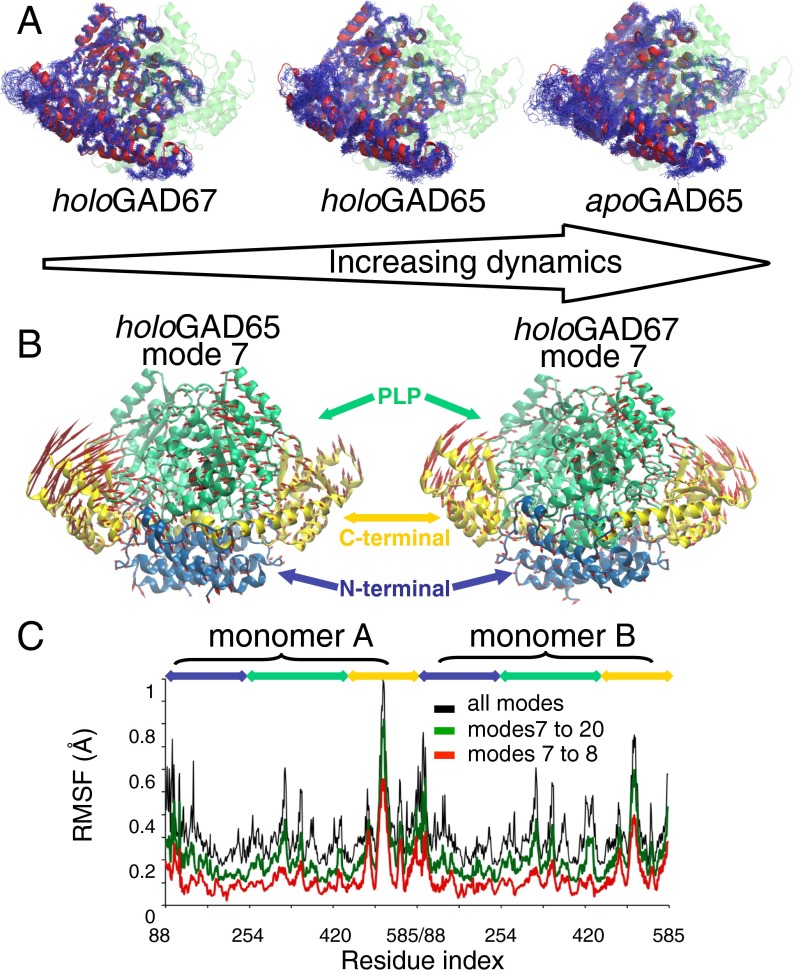
Fig. 1.
MD and NM analyses of GAD proteins. (A) Atomistic MD of GAD65 vs. GAD67 indicating the higher flexibility of apoGAD65 compared with holoGAD65 and holoGAD67. Backbone atoms are shown after a superposition of 25 structures sampled every 10 ns from a 250-ns simulation of (Right) apoGAD65 and (Center) holoGAD65 and (Left) 20 structures sampled every 10 ns from a 200-ns simulation of holoGAD67. (B) Motions described by the lowest frequency NM (mode 7) of holoGAD65 and holoGAD67. The directions and amplitudes of the motions are represented by arrows. (C) Flexibility profile [root mean square fluctuation (RMSF)] provided by distinct sets of holoGAD65 NMs. Blue, N terminus; green, PLP; yellow, C terminus.
Cofactor Release Enhances the Asymmetrical Dynamics of GAD65 CL and Its Interaction with the CTD.
The structure and activity of GAD65 is regulated by the PLP cofactor, with binding (uptake) inducing a conformational change (13) and release causing autoinactivation (6, 13). Our MD analysis illustrates how the relatively high dynamics of the CTD and CL promote PMP formation and autoinactivation of GAD65 (6). Furthermore, after PLP is lost, increased dynamics are seen in the PLP-binding domain, CL, and CTDs. Structural differences between apo/holoGAD65 and holoGAD67 are observed in the dynamics of the CL and its interactions with the C-terminal helix 14 (H14) of the adjacent monomer (Fig. 1A and Fig. S1 B and C). After the first 50 ns, the average number of contacts below 0.6 nm was 60 ± 10 (apoGAD65), 46 ± 5 (holoGAD65), and 37 ± 7 (holoGAD67) (Fig. S1C). Therefore, the absence of PLP not only destabilizes the CL but also, increases its clashes with H14. A range of CL/H14 dynamics is observed, where apoGAD65 and holoGAD67 sit at opposite ends of the mobility spectrum. MD analysis also shows asymmetrical domain movements within the dimer of holoGAD65 but not holoGAD67. This asymmetry is likely caused by the differences in CL conformation in the starting structure of holoGAD65. This observation is in agreement with recently solved crystal structures of the open form of AADC (4) and holoGAD6765loop (17). The contacts between H14 and the CL in both holoGAD67 monomers are similar, which may be a consequence of the nearly identical conformation of the two CLs in the crystal structure (Fig. S1C). The minimum distance and number of interactions below 0.6 nm between the CL and H14 of both monomers are similar throughout the simulations of GAD67. Conversely, in apoGAD65, the number of these interactions differs between monomers A and B, indicating asymmetrical dynamics for this isoform.
Taken together, the MD data indicate that holoGAD65 is intrinsically more flexible compared with holoGAD67, that release of PLP enhances the dynamics of the CL of GAD65 and its interaction with H14, and that apoGAD65 motions are asymmetrical.
CTDs in the Symmetrical Dimer of GAD65 Move Asymmetrically.
To investigate the observed asymmetrical motion, we performed an all-atom normal mode (NM) analysis to probe large-amplitude motions that are often inaccessible to atomistic MD simulations (18–20). NM analysis is insensitive to the presence of small molecules in the context of large-protein systems, such as PLP in GAD. However, a comparison of GAD65/GAD67 showed that the magnitude of the fluctuations was highest in the CTDs and that GAD65 was more flexible than GAD67 (Fig. 1 B and C). Notably, the NM data corroborates the findings from MD and the flexibility inferred from crystallographic B factors (6). Furthermore, holoGAD65, but not holoGAD67, displayed an asymmetric profile of fluctuations at the CTDs (Figs. 1C and 2 A and B). This observation is consistent with the higher interchain correlation coefficients obtained for holoGAD67 (R = 0.85) compared with holoGAD65 (R = 0.67).
Fig. 2.
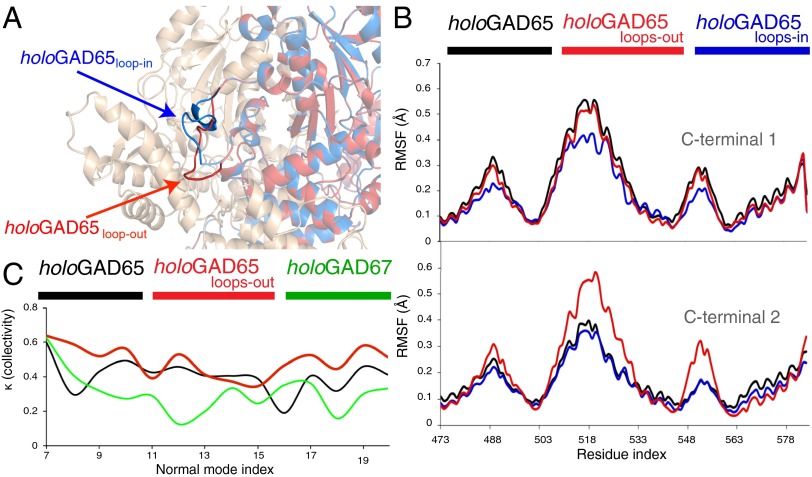
(A) Modeling and dynamics of the distinct CL conformations and effect on the collectivity of the CTD dynamics of GAD65. The in conformation (similar to that in holoGAD67) is represented in blue, and the out conformation is represented in red. (B) RMSFs of the CTDs provided by modes 7–20: holoGAD65 (black), holoGAD65loops-in (blue), and holoGAD65loops-out (red). Residues are numbered as in Fig. 1. (C) Collectivity of motions described by low frequency NMs. The collectivity index κ represents the fraction of atoms participating significantly in a given displacement.
CL Conformation Dictates the Collectivity of CTD Motions.
The dependency of the CTD mobility on the adjacent CL conformation was investigated by NM analysis of the crystal structure of chimeric holoGAD6567loop and holoGAD65 models with in and out loop conformations (Fig. 2 A and B and SI Results and Discussion) (17). The fluctuation amplitude of the CTD is dependent on the CL conformation (i.e., increasing when the loop is in the out conformation) (Fig. 2 and Fig. S2). We noticed an increase in the overall collectivity of holoGAD65 low-frequency modes when the loop is in the out conformation, contrasting with the minor values obtained for GAD67. These findings indicate a higher flexibility encoded in the GAD65 fold, probably related to the out CL conformation not observed in GAD67, suggesting that flexibility makes an important contribution to the differential autoinactivation rates of GAD65 and GAD67.
Contrasting Dynamics of Apo- and HoloGAD Isoforms Revealed by Limited Proteolysis.
Previous biophysical studies indicated a conformational change of GAD65 upon PLP removal (13, 21–23). Our simulation results are consistent with these data, adding atomic detail. We set out to investigate these dynamics further using limited proteolysis, a well-studied approach for probing structural changes (reviewed in ref. 24). Apoenzymes were generated by omitting PLP during all purification steps. This material, referred to as apoGAD65/GAD67, was incubated with trypsin, and a time course of proteolysis was compared with holoGAD65/GAD67, with no protease and protease-only controls (Fig. 3A and SI Results and Discussion).
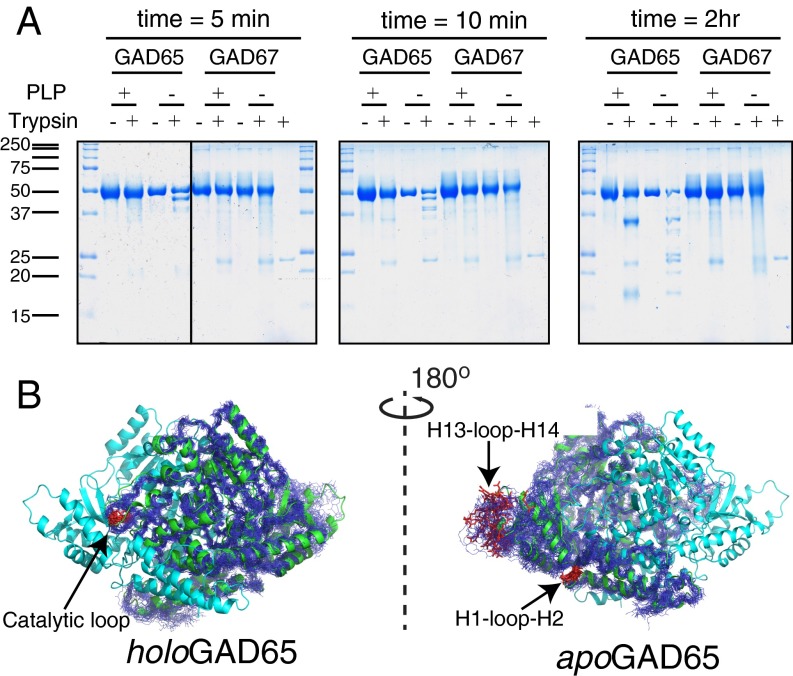
Fig. 3.
Limited proteolysis of GAD65/GAD67. (A) GAD proteins purified in the presence/absence of PLP were exposed to trypsin or buffer only. At the indicated times, an aliquot of the reaction mixture was boiled in SDS sample buffer, and the reactions were resolved by SDS/PAGE. Trypsin (∼23 kDa) was run alone for comparison as indicated. (B) Models (MD snapshots) of GAD65 mobility are shown with the cleavage sites (red) and labeled.
ApoGAD65 underwent more extensive cleavage, with a higher frequency of three cleavage products that migrated ∼5, 10, and 12 kDa lower than the intact precursor at the earliest time point (45-, 40-, and 38-kDa fragments). These fragments were degraded and not detected at the end of the time course. A second set of fragments (35 and 17 kDa) was seen in both apo- and holoGAD65 but was the most dominant species in holoGAD65. Although apo- and holoGAD67 were less cleaved than GAD65 counterparts, differences in the cleavage rate and patterns were apparent. Whereas holoGAD67 remains intact over the time course of the experiment, apoGAD67 is significantly decreased (by 40%) (Fig. 3A and Fig. S3). Identification of proteolytic fragments by N-terminal sequencing is consistent with our dynamics results, notably for the CL and loop regions flanking the C-terminal H14 (Fig. 3B, SI Results and Discussion, and Fig. S3A). These data show that limited availability of PLP renders GAD65 and GAD67 more susceptible to proteolysis and alters their proteolytic profile, thus indicating increased dynamics. Consistent with our MD and NM analysis, this finding indicates that GAD65 is more dynamic than GAD67 and that a conformational change in GAD65 occurs on release of PLP.
Cofactor-Dependent Dynamic Communication Between the CTD and the CL.
Because GAD65 showed the most PLP-dependent movement, indicated by limited proteolysis and computational methods, we examined the fragmentation pattern of this isoform more closely. Limited proteolysis of GAD65 seems to take one of two mutually exclusive pathways that are shifted dependent on the presence of PLP. In the presence of PLP, holoGAD65 is relatively protease-resistant but exposes a cut site in the CL (431K^HYDLS436), producing stably associated 35- and 17-kDa fragments that are resistant to additional cleavage (Fig. 3 and Fig. S3C). Conversely, apoGAD65 is cleaved in two different regions early in the time course. Cleavages are observed in the N-terminal region H1-loop-H2 that contacts the C terminus of H14, the less-exposed adjoining H3-loop-H4, and the C-terminal H13-loop-H14 region. These initial cleavages of apoGAD65 are unstable during the time course and undergo subsequent digestion. The data, therefore, are consistent with a dramatic remodeling of GAD65 structure on loss of PLP and an increase in CTD motions. Furthermore, these data suggest that there is PLP-dependent dynamic communication between the CTD and the CL through direct contacts mediated by H14. Cleavage of the CL of holoGAD65 removes this structural connection and therefore, causes the H1-loop-H2 and the H13-loop-H14 regions to be stable and resistant to proteolysis. The mutually exclusive nature of the cleavage events results in two different end points and indicates the critical role of the PLP-binding domain in the CL and CTD dynamics. Extending a recent crystallographic analysis of a GAD6567loop chimera that revealed interplay between the CL and the CTD (17), these data provide important structural clues for the holo → apo conversion.
ApoGAD65 Is a Conformational Ensemble.
The crystal structures of holoGAD65 and holoAADC are highly similar (Cα RMSD of 0.144 nm). In addition, inspection of the physiochemical properties of the dimer interfaces for GAD65, GAD67, and AADC reveals that GAD65 and AADC share solvation properties that favor dimer opening, which are not observed in GAD67 (SI Results and Discussion and Fig. S4). Therefore, we reasoned that the recently solved crystal structure of an open form of AADC (4) could be used to make a homology model of apoGAD65 (apoGAD65open). Opening of the GAD65 dimer involves a dramatic conformational change (Fig. 4 and Fig. S5). NM analysis of apoGAD65open revealed a symmetric rigid-body motion of the PLP-binding domain, leading to a closed conformation. Remarkably, in both apoGAD65open and apoAADCopen, the CTDs moved in concert with the opening of PLP-binding domains (Fig. 4, SI Results and Discussion, Figs. S4 B–D, S6, and S7, and Movie S2).
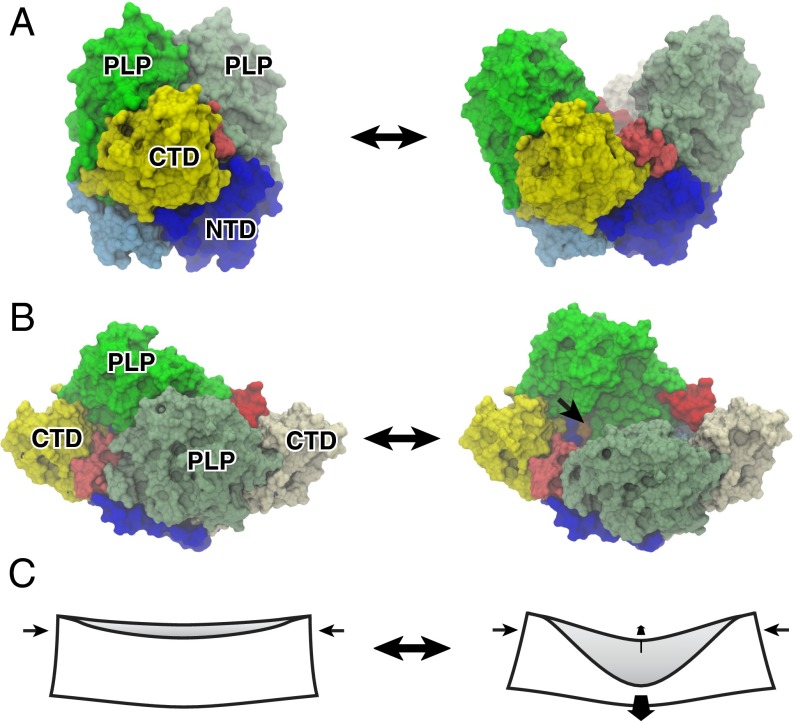
Fig. 4.
Open and closed dimers of GAD65 and the mechanism of dimer opening in holo → apo conversion. (A) Molecular surface representations of (Left) the closed form of holoGAD65 and (Right) model of apoGAD65 in an open conformation. The CL (red/pink) connects the CTD and PLP-binding domain, mediating the dimer opening as supported by our MD and proteolysis data and ref. 17. (B) Orthogonal view facing the dimer interface. The active site lysine (396) that is buried in the closed form but solvent-accessible in the open form is shown in orange and indicated by an arrow (only one of the two sites is visible). (C) Origami analogy illustrating the mechanism of dimer opening in same view as in B. Coupled orthogonal motions of the PLP-binding domains and CTDs that accompany dimer opening are indicated by arrows. Domains are colored and labeled: NTD (blue, monomer A; cyan, monomer B), PLP (green, monomer A; pea green, monomer B), and CTD (yellow). NTD, N-terminal domain.
We next performed small-angle X-ray scattering (SAXS) analysis of both apo- and holoGAD65. The measured SAXS data (Table S1) for holoGAD65 are in good agreement with the crystal structure (Fig. 5A). However, for apoGAD65, the P(r) peak has broadened significantly instead of moving to the predicted longer interatomic distances (Fig. 5B). In the calculated apoGAD65 P(r), the peak of the P(r) function is moved to the right with respect to holoGAD65 (Fig. 5B). This finding is correlated with the increase in average interatomic distance in the open (apo) structure compared with the closed (holo) form. These data are consistent with apoGAD65 existing as a conformational ensemble of open, closed, and intermediate dimeric forms, which were indicated by our MD, rather than a binary model. In support, the conformational differences between holo (closed) and apo (open) forms are consistent with proteolysis data (Fig. 3) as well as emission fluorescence and CD spectra (Fig. 5C and SI Results and Discussion).
Fig. 5.
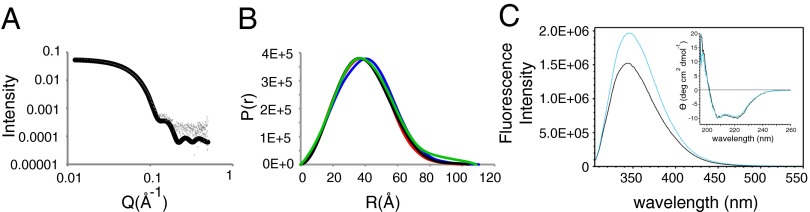
(A and B) SAXS analysis of apo- and holoGAD65. (A) HoloGAD65 scattering data fitted to the crystal structure (χ2 = 3.3). (B) P(r) functions from measured data (black, holoGAD65; green, apoGAD65) and scattering intensities calculated from models (blue, apoGAD65; red, holoGAD65). (C) Fluorescence emission spectra on excitation at 295 nm and CD spectra (Inset) of holoGAD65 (black) and apoGAD65 (cyan).
A Dynamic ApoGAD65 Ensemble May Elicit a Mode of Antibody Production and Binding Different to That Available to the Rigid Closed Holo Form.
We hypothesized that conformational plasticity will influence the way that B cells and antibodies interact with GAD65. We have previously shown that GAD65–antibody binding kinetics can be measured efficiently using Surface Plasmon Resonance Imaging (SPRi) (25). To test our hypothesis, we immobilized, alternatively, apo- and holoGAD65 (purified and analyzed as in the proteolysis experiments) on an SPRi surface before the addition of mouse GAD1 anti-GAD65 mAb. GAD1 recognizes a highly conformational epitope of GAD65 and is sensitive to structural changes in GAD65 (26–29). In the case of holoGAD65, only one Ab-binding site was exposed [even at high (100 nM) concentrations of Ab]. Conversely, for apoGAD65, the best fit to the binding curve corresponded to a single association constant and two dissociation constants (SI Results and Discussion and Fig. S8). The Kd value of 1.3 nM calculated for the holo form corresponds well with the value obtained for binding at low antibody concentration observed previously (25). The Kd values obtained for apoGAD65 are less precise because of the heterogeneity of the interaction, and therefore, absolute values are of comparative interest only. However, the reactive species in the apoGAD65 form interacted more slowly with the antibody (SI Results and Discussion and Fig. S8B) and then formed a majority complex (77%) that dissociated even more slowly than the holo form as well as a minority rapidly dissociating species. These observations suggest that the closed holo form exposes a single epitope, but on conversion to a more open dynamic apo form ensemble, the antibody has more difficulty accessing this site; then, it forms a majority of stable species but with at least 25% of the population shunting into significantly less stable complexes (SI Results and Discussion). These data suggest that the dynamics of the apoGAD65 ensemble, while creating initial frustration for the engaging antibody, exploit mutually induced conformational changes to achieve a mode of binding not available to the relatively rigid closed holo dimer.
Conformationally Controlled Autoinactivation of HoloGAD65—Implications for Neurotransmitter Biosynthesis and Autoantigenicity.
In vivo GAD65 exists predominantly in the inactive apo form. Coupling the binding of PLP to domain motions that we have described seems to drive the closing of the GAD65 dimer and therefore, may facilitate a mechanism for the regulation of GABA homeostasis by PLP availability. The flexibility and decreased stability of the open apo form may also influence proteasomal processing, possibly offering another level of regulation. Intriguingly, the conformational properties and unique autoinactivation of GAD65 that differentiate it from GAD67 may offer an explanation for its autoantigenicity. The structurally diverse apo form ensemble may influence both B and T cell-mediated immunogenicity by conformationally induced antibody binding, epitope spreading, and proteasomal-driven antigen presentation. Additional characterization, however, awaits more detailed epitope mapping by mutagenesis aimed at testing hypotheses generated by our structural modeling. Structural studies of GAD65–Ab complexes may ultimately reveal the most detailed information. Understanding the insufficiency of tolerance that accompanies the production of anti-GAD65 antibodies remains enigmatic. We can speculate that the GAD65 ensemble is poorly represented in the primary lymphoid organs (bone marrow and thymus), where natural immune tolerance is initiated, and additionally, our findings implicate the likely importance of flexibility and dynamics of apoGAD65 in achieving high-affinity antibody–antigen complexes through mutually induced conformational changes. A scheme illustrating how the conformational properties of GAD65 may dictate its function and autoantigenicity is shown in Fig. 6.
Fig. 6.
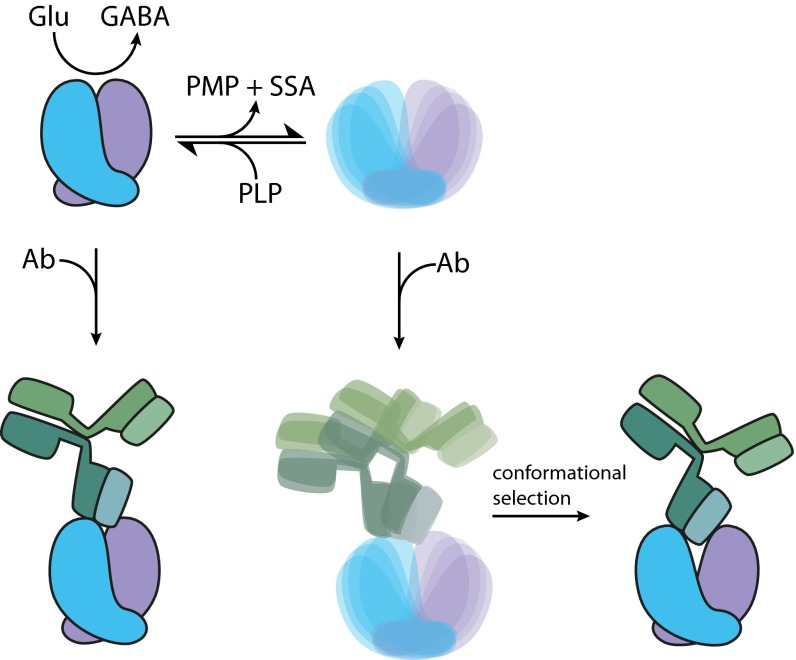
Implications of GAD65 autoinactivation for neurotransmitter biosynthesis and autoantigenicity. HoloGAD65 readily loses its PLP cofactor and autoinactivates through a secondary reaction, yielding a diverse ensemble. Supply of PLP shifts the equilibrium in favor of the primary reaction that catalyzes the conversion of Glu to GABA, thus regulating GABA production. This PLP-dependent autoinactivation may play a role in GAD65 autoantigenicity. The rigid closed holoGAD65 dimer binds an anti-GAD65 mAb with high affinity. The antibody also engages with the diverse open ensemble of apoGAD65, with conformational selection and/or induced fit permitting a mode of binding not available with the relatively rigid closed holo dimer. The PLP-dependent dynamic properties of GAD65 may be implicated in the insufficient development of immune self-tolerance that favors the production of GAD65 autoantibodies. PMP, pyridoxamine 5′-phosphate; SSA, succinic semialdehyde.
Conclusion
The neurotransmitter GABA is important in many biological processes. As a result, its synthesis by GAD is heavily regulated. Previous studies indicate that GABA synthesis is inhibited by increased intracellular GABA levels (30) or ATP (31, 32) binding. The conformational properties of the GAD isoforms may offer another mechanism for GABA regulation. Whereas GAD67 is stable in the cell and responsible for basal levels of GABA, GAD65 readily autoinactivates on PLP release and conformational change. Using an array of experimental and theoretical methods to characterize conformational change, we show that holoGAD65 is intrinsically more dynamic than holoGAD67, with conformational coupling between CTDs, the CL, and PLP-binding domains, allowing PLP loss and autoinactivation. Autoinactivated apoGAD65 likely exists as a dynamic ensemble of flexible open forms. Although this conformational plasticity might enable PLP-controlled regulation of GABA synthesis, the cost may be its involvement as a prevalent autoantigen in autoimmune T1D.
Methods
Protein expression and purification was performed as described previously (6). Small angle X-ray scattering was performed at the SAXS-WAXS beamline at the Australian Synchrotron. SPRi was performed as described previously (25). All experimental and computational methods are described in detail in SI Methods.
Supplementary Material
Acknowledgments
M.G.S.C. was financially supported by a French–Brazilian Coordenação de Aperfeiçoamento de Pessoal de Nível Superior/Comité Français d'Evaluation de la Coopération Scientifique et Universitaire avec le Brésil Collaboration Project. E.P. was the recipient of a bursary from the Istituto Pasteur-Fondazione Cenci Bolognetti. D.C. and A.M.B. hold research fellowships from the National Health and Medical Research Council. D.D.B. thanks Fondazione Roma for partially supporting this work. This research was supported by the Victorian Life Sciences Computation Initiative Life Sciences Computation Centre, a collaboration between Melbourne, Monash, and La Trobe Universities and an initiative of the Victorian Government, Australia.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. B.R. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1403182111/-/DCSupplemental.
References
- 1.Erlander MG, Tillakaratne NJ, Feldblum S, Patel N, Tobin AJ. Two genes encode distinct glutamate decarboxylases. Neuron. 1991;7(1):91–100. doi: 10.1016/0896-6273(91)90077-d. [DOI] [PubMed] [Google Scholar]
- 2.Bu DF, et al. Two human glutamate decarboxylases, 65-kDa GAD and 67-kDa GAD, are each encoded by a single gene. Proc Natl Acad Sci USA. 1992;89(6):2115–2119. doi: 10.1073/pnas.89.6.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sandmeier E, Hale TI, Christen P. Multiple evolutionary origin of pyridoxal-5'-phosphate-dependent amino acid decarboxylases. Eur J Biochem. 1994;221(3):997–1002. doi: 10.1111/j.1432-1033.1994.tb18816.x. [DOI] [PubMed] [Google Scholar]
- 4.Giardina G, et al. Open conformation of human DOPA decarboxylase reveals the mechanism of PLP addition to Group II decarboxylases. Proc Natl Acad Sci USA. 2011;108(51):20514–20519. doi: 10.1073/pnas.1111456108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin DL, Rimvall K. Regulation of gamma-aminobutyric acid synthesis in the brain. J Neurochem. 1993;60(2):395–407. doi: 10.1111/j.1471-4159.1993.tb03165.x. [DOI] [PubMed] [Google Scholar]
- 6.Fenalti G, et al. GABA production by glutamic acid decarboxylase is regulated by a dynamic catalytic loop. Nat Struct Mol Biol. 2007;14(4):280–286. doi: 10.1038/nsmb1228. [DOI] [PubMed] [Google Scholar]
- 7.Baekkeskov S, et al. Identification of the 64K autoantigen in insulin-dependent diabetes as the GABA-synthesizing enzyme glutamic acid decarboxylase. Nature. 1990;347(6289):151–156. doi: 10.1038/347151a0. [DOI] [PubMed] [Google Scholar]
- 8.Baekkeskov S, et al. Antibodies to a 64,000 Mr human islet cell antigen precede the clinical onset of insulin-dependent diabetes. J Clin Invest. 1987;79(3):926–934. doi: 10.1172/JCI112903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin JM, Lacy PE. Prediabetic period in partially pancreatectomized rats. Diabetes. 1963;12(3):238–242. [Google Scholar]
- 10.Jayakrishnan B, Hoke DE, Langendorf CG, Buckle AM, Rowley MJ. An analysis of the cross-reactivity of autoantibodies to GAD65 and GAD67 in diabetes. PLoS ONE. 2011;6(4):e18411. doi: 10.1371/journal.pone.0018411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwartz HL, et al. High-resolution autoreactive epitope mapping and structural modeling of the 65 kDa form of human glutamic acid decarboxylase. J Mol Biol. 1999;287(5):983–999. doi: 10.1006/jmbi.1999.2655. [DOI] [PubMed] [Google Scholar]
- 12.Richter W, et al. Human monoclonal islet cell antibodies from a patient with insulin-dependent diabetes mellitus reveal glutamate decarboxylase as the target antigen. Proc Natl Acad Sci USA. 1992;89(18):8467–8471. doi: 10.1073/pnas.89.18.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen CH, Wu SJ, Martin DL. Structural characteristics of brain glutamate decarboxylase in relation to its interaction and activation. Arch Biochem Biophys. 1998;349(1):175–182. doi: 10.1006/abbi.1997.0457. [DOI] [PubMed] [Google Scholar]
- 14.Rodriguez-Caso C, et al. Local changes in the catalytic site of mammalian histidine decarboxylase can affect its global conformation and stability. Eur J Biochem. 2003;270(21):4376–4387. doi: 10.1046/j.1432-1033.2003.03834.x. [DOI] [PubMed] [Google Scholar]
- 15.Arafat Y, et al. Structural determinants of GAD antigenicity. Mol Immunol. 2009;47(2-3):493–505. doi: 10.1016/j.molimm.2009.08.022. [DOI] [PubMed] [Google Scholar]
- 16.Fenalti G, Buckle AM. Structural biology of the GAD autoantigen. Autoimmun Rev. 2010;9(3):148–152. doi: 10.1016/j.autrev.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 17.Langendorf CG, et al. Structural characterization of the mechanism through which human glutamic acid decarboxylase auto-activates. Biosci Rep. 2013;33(1):137–144. doi: 10.1042/BSR20120111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kass I, Reboul CF, Buckle AM. Computational methods for studying serpin conformational change and structural plasticity. Methods Enzymol. 2011;501:295–323. doi: 10.1016/B978-0-12-385950-1.00014-6. [DOI] [PubMed] [Google Scholar]
- 19.Batista PR, et al. Consensus modes, a robust description of protein collective motions from multiple-minima normal mode analysis—application to the HIV-1 protease. Phys Chem Chem Phys. 2010;12(12):2850–2859. doi: 10.1039/b919148h. [DOI] [PubMed] [Google Scholar]
- 20.Floquet N, et al. Activation of the ghrelin receptor is described by a privileged collective motion: A model for constitutive and agonist-induced activation of a sub-class A G-protein coupled receptor (GPCR) J Mol Biol. 2010;395(4):769–784. doi: 10.1016/j.jmb.2009.09.051. [DOI] [PubMed] [Google Scholar]
- 21.Rust E, Martin DL, Chen CH. Cofactor and tryptophan accessibility and unfolding of brain glutamate decarboxylase. Arch Biochem Biophys. 2001;392(2):333–340. doi: 10.1006/abbi.2001.2466. [DOI] [PubMed] [Google Scholar]
- 22.Chen CH, Colón W, Myer YP, Martin DL. ATP’s impact on the conformation and holoenzyme formation in relation to the regulation of brain glutamate decarboxylase. Arch Biochem Biophys. 2000;380(2):285–293. doi: 10.1006/abbi.2000.1931. [DOI] [PubMed] [Google Scholar]
- 23.Chen CH, Battaglioli G, Martin DL, Hobart SA, Colón W. Distinctive interactions in the holoenzyme formation for two isoforms of glutamate decarboxylase. Biochim Biophys Acta. 2003;1645(1):63–71. doi: 10.1016/s1570-9639(02)00522-8. [DOI] [PubMed] [Google Scholar]
- 24.Fontana A, et al. Probing protein structure by limited proteolysis. Acta Biochim Pol. 2004;51(2):299–321. [PubMed] [Google Scholar]
- 25.Nogues C, et al. Characterisation of peptide microarrays for studying antibody-antigen binding using surface plasmon resonance imagery. PLoS ONE. 2010;5(8):e12152. doi: 10.1371/journal.pone.0012152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richter W, Shi Y, Baekkeskov S. Autoreactive epitopes defined by diabetes-associated human monoclonal antibodies are localized in the middle and C-terminal domains of the smaller form of glutamate decarboxylase. Proc Natl Acad Sci USA. 1993;90(7):2832–2836. doi: 10.1073/pnas.90.7.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Butler MH, Solimena M, Dirkx R, Jr, Hayday A, De Camilli P. Identification of a dominant epitope of glutamic acid decarboxylase (GAD-65) recognized by autoantibodies in stiff-man syndrome. J Exp Med. 1993;178(6):2097–2106. doi: 10.1084/jem.178.6.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gottlieb DI, Chang YC, Schwob JE. Monoclonal antibodies to glutamic acid decarboxylase. Proc Natl Acad Sci USA. 1986;83(22):8808–8812. doi: 10.1073/pnas.83.22.8808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Myers MA, et al. Conformational epitopes on the diabetes autoantigen GAD65 identified by peptide phage display and molecular modeling. J Immunol. 2000;165(7):3830–3838. doi: 10.4049/jimmunol.165.7.3830. [DOI] [PubMed] [Google Scholar]
- 30.Porter TG, Martin DL. Evidence for feedback regulation of glutamate decarboxylase by gamma-aminobutyric acid. J Neurochem. 1984;43(5):1464–1467. doi: 10.1111/j.1471-4159.1984.tb05409.x. [DOI] [PubMed] [Google Scholar]
- 31.Sze PY, Sullivan P, Alderson RF, Towle AC. ATP binding to brain l-glutamate decarboxylase: A study by affinity chromatography. Neurochem Int. 1983;5(1):51–56. doi: 10.1016/0197-0186(83)90008-6. [DOI] [PubMed] [Google Scholar]
- 32.Meeley MP, Martin DL. Inactivation of brain glutamate decarboxylase and the effects of adenosine 5′-triphosphate and inorganic phosphate. Cell Mol Neurobiol. 1983;3(1):39–54. doi: 10.1007/BF00734997. [DOI] [PMC free article] [PubMed] [Google Scholar]