Significance
The toxic composition of snake venom varies between species. Such variation can have major medical implications for the treatment of human snakebite victims. Venom variation is largely attributed to differences in toxin-encoding genes present in the genome or venom gland of snakes. Here, we demonstrate that mechanisms affecting the transcription, translation, and posttranslational modification of toxins also significantly contribute to the diversity of venom protein composition. Venom variation observed between related snake species is therefore the result of a complex interaction between a variety of genetic and postgenomic factors acting on toxin genes. Ultimately, this variation results in significant differences in venom-induced pathology and lethality and can undermine the efficacy of antivenom therapies used to treat human snakebite victims.
Abstract
Variation in venom composition is a ubiquitous phenomenon in snakes and occurs both interspecifically and intraspecifically. Venom variation can have severe outcomes for snakebite victims by rendering the specific antibodies found in antivenoms ineffective against heterologous toxins found in different venoms. The rapid evolutionary expansion of different toxin-encoding gene families in different snake lineages is widely perceived as the main cause of venom variation. However, this view is simplistic and disregards the understudied influence that processes acting on gene transcription and translation may have on the production of the venom proteome. Here, we assess the venom composition of six related viperid snakes and compare interspecific changes in the number of toxin genes, their transcription in the venom gland, and their translation into proteins secreted in venom. Our results reveal that multiple levels of regulation are responsible for generating variation in venom composition between related snake species. We demonstrate that differential levels of toxin transcription, translation, and their posttranslational modification have a substantial impact upon the resulting venom protein mixture. Notably, these processes act to varying extents on different toxin paralogs found in different snakes and are therefore likely to be as important as ancestral gene duplication events for generating compositionally distinct venom proteomes. Our results suggest that these processes may also contribute to altering the toxicity of snake venoms, and we demonstrate how this variability can undermine the treatment of a neglected tropical disease, snakebite.
Venom systems are important adaptations that have evolved independently on many occasions in different animal lineages (1). Of all venomous animals, snakes are the most well-known because of their medical importance: As many as 90,000 people die each year as the result of snakebite, with the majority of those inhabiting rural poor regions of the tropics (2, 3). This substantial mortality burden of snakebite victims is surprising because antivenom treatment (immunoglobulins from venom-immunized horses/sheep) can be highly effective at neutralizing the toxic components present in snake venom (4, 5). However, the efficacy of these therapies is largely restricted to the snake species whose venom was used in manufacture. This limitation arises because variation in venom composition is ubiquitous at every level of snake taxonomy, including interspecifically and intraspecifically and even ontogenetically (6–9). Importantly, the extent of this variation is not simply reflected by taxonomic distance (9–11) and, therefore, cannot be readily predicted. The consequence of venom variation is that antivenoms raised against any particular species of snake are often ineffective in treating snakebite by different, even closely related, species (5, 12, 13).
Snake venoms are used for predation. They primarily consist of proteins and peptides (commonly referred to as toxins) that exert neurotoxic, hemotoxic, and/or cytotoxic pathologies in envenomed prey and humans. Typically, toxins are encoded by relatively few (approximately 5–10) multilocus gene families, with each family capable of producing related isoforms generated by gene duplication events occurring over evolutionary time (1, 14, 15). The birth and death model of gene evolution (16) is frequently invoked as the mechanism giving rise to venom gene paralogs, with evidence that natural selection acting on surface exposed residues of the resulting gene duplicates facilitates subfunctionalization/neofunctionalization of the encoded proteins (15, 17–19). The result of these processes is a complex suite of toxins that act synergistically to cause rapid prey death. Consequently, it has been hypothesized that variation in venom composition is the result of adaptation in response to dietary selection pressures (1, 9, 11, 20).
There is some evidence that genome-level effects, i.e., the presence or absence of key toxin genes in the genome of venomous snakes, can dictate major shifts in venom composition (21). However, the assumption that the presence or absence of toxin gene paralogs is responsible for causing all cases of observed venom variation is overly simplistic. Few studies have attempted to investigate the factors impacting upon the transcription of toxins from genes housed in the genome of the venomous animal to the proteins secreted in venom. Although some studies have detected concordance between the abundance of both toxin gene transcripts and proteins in snake venom systems (22, 23), many others have demonstrated that the toxin genes detected in venom glands do not correlate well with the composition of secreted venom (7, 24), suggesting that some level of regulatory control acts on protein translation. However, most studies typically focused on comparisons at the toxin family level by using a single species, which could be misleading if mechanisms affecting translation act differentially on toxin paralogs and, therefore, result in different outcomes in different species.
Here, we take an integrated multispecies approach, coupling molecular, proteomic, and evolutionary methodologies, to rigorously investigate the mechanisms responsible for generating venom variation in snakes. We demonstrate that there is discordance between the number of genomic loci encoding toxin genes with the number and abundance of genes transcribed in the venom gland and with the number and abundance of toxins translated as functional proteins. We find that transcriptional and translational control mechanisms are likely to play a critical role in the generation of compositionally and functionally distinct venoms found among related snake species. Ultimately, we demonstrate that the action of these postgenomic processes can restrict the snake-species efficacy of therapeutic antivenoms.
Results and Discussion
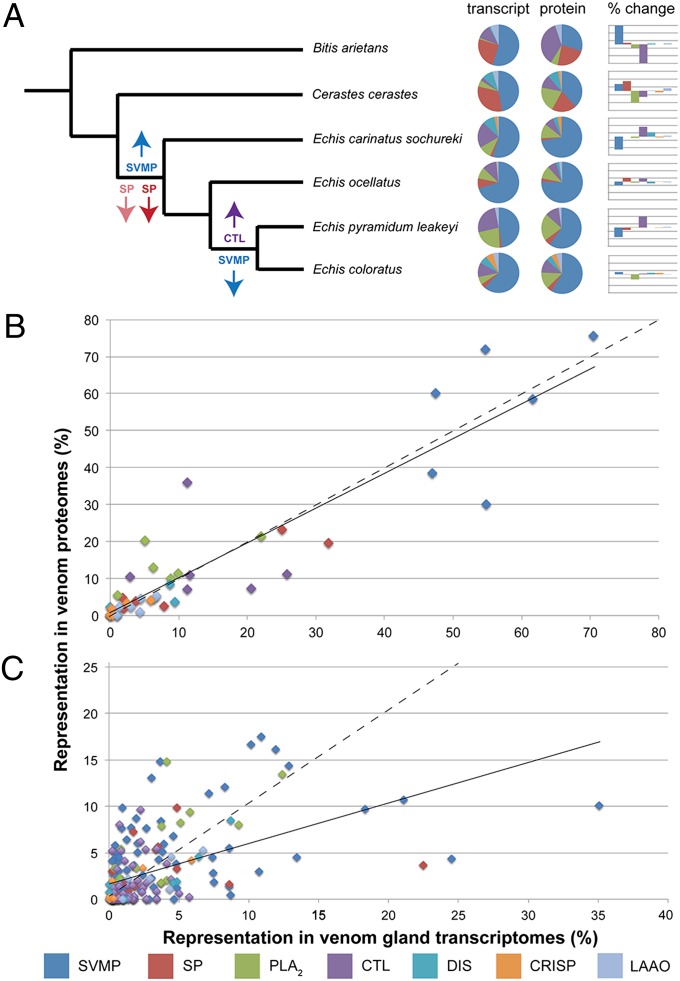
To investigate the mechanisms governing venom variation we selected a related group of medically important viperid snakes: four species of saw-scaled vipers (Echis ocellatus, Echis coloratus, Echis pyramidum leakeyi, and Echis carinatus sochureki) and two related species from different genera, the puff adder (Bitis arietans) and the Saharan horned viper (Cerastes cerastes) (Fig. 1). These species were selected based on their medical importance in Africa and our previous descriptions of interspecific variation in venom composition, dietary preference, and prey lethality (12, 20, 25, 26). We used our previously constructed and assembled venom gland transcriptomes for the four Echis species (26, 27) and prepared venom gland trancriptomes for B. arietans and C. cerastes by using identical protocols. We generated proteomes from venom extracted from each of these species (28) and used translations of the transcriptomic datasets to facilitate protein identification (SI Appendix, Figs. S1–S6 and Tables S1–S6). We next compiled gene and protein abundance profiles for the toxin families identified in the venom proteomes of the majority of the sampled species [snake venom metalloproteinases (SVMPs), serine proteases (SPs), C-type lectins (CTLs), phospholipase A2s (PLA2s), disintegrins (DISs), cysteine rich secretory proteins (CRISPs), and l-amino acid oxidases (LAAOs)]. These profiles were mapped to the species phylogeny to reveal considerable interspecific variation in both gene and protein abundance over evolutionary time (Fig. 1).
Fig. 1.
The complex relationship between snake venom composition at the gene and protein level. (A) Comparisons of gene transcription (transcript) in the venom gland and protein abundance (protein) in secreted venom of the sampled species. Percentage change from transcript to protein is displayed in bar charts for visual purposes (see SI Appendix, Fig. S7 for detail). Arrows indicate significant increases or decreases in transcriptomic (transparent) and proteomic (solid) toxin family abundance over evolutionary time (calculated by paired two-tailed t test of toxin representation between taxa found within and outside each node, P < 0.05). The phylogeny is taken from refs. 20, 29, and 30). (B) Comparisons of total venom gland transcriptome and venom proteome abundance summarized for all gene paralogs encoded by each toxin family for each species. (C) Comparisons of individual gene paralog abundance for each toxin family for each species. Solid lines indicate linear trendlines (R2 values: B, 0.83; C, 0.26) and, for visual purposes, dotted lines indicate equal gene and protein abundance.
The high abundance of SVMPs in the venom proteomes (>50% of all toxins) of the four Echis species correlates with our previous reports that these toxins are transcribed at the highest level in the venom glands (26). The SVMPs, alongside the SPs, represent the majority of toxin genes transcribed in the venom glands of B. arietans and C. cerastes, and these are also the most abundant proteins secreted in their venoms. However, the quantitative proteomic representation of CTLs in B. arietans and PLA2s in C. cerastes was greater than that predicted by the gene transcription data (Fig. 1A). This disparity in transcriptomic and proteomic abundance is notably high in some species (e.g., B. arietans and E. p. leakeyi), yet largely absent from others (e.g., E. ocellatus and E. coloratus) (Fig. 1A and SI Appendix, Fig. S7).
Illustrating this comparison in scatter plots reveals a reasonable correlation between the total transcription of toxin gene paralogs encoded by each gene family and their cumulative proteomic abundance (Fig. 1B). However, this correlation disappears when analyzing each gene paralog for each toxin family individually, demonstrating that a variety of highly transcribed toxin genes exhibit little to no proteomic representation, and vice versa (Fig. 1C). The starkest examples involve the SVMPs: A highly transcribed SVMP (24.49% of all toxins) in the venom gland of E. ocellatus has relatively low abundance in venom (4.33%), which is a pattern repeated with a paralogous SVMP found in C. cerastes (8.73% transcriptome; 0.46% proteome). Contrastingly, other toxins encoded by the same gene family show the opposite pattern: Individual SVMPs from E. p. leakeyi and E. c. sochureki exhibit considerably greater proteomic than transcriptomic abundance (14.83% and 9.83% proteome; 3.69% and 0.97% transcriptome, respectively). Notably, none of these toxins found in the different species are homologous. Our results here demonstrate that intratoxin family variation between the transcriptome and proteome of snake species can greatly affect the composition of toxins found in snake venom. Mechanisms affecting protein translation therefore appear capable of acting differentially on toxin paralogs within a single gene family, rather than supressing or enhancing the translation of all toxin genes encoded by the same gene family.
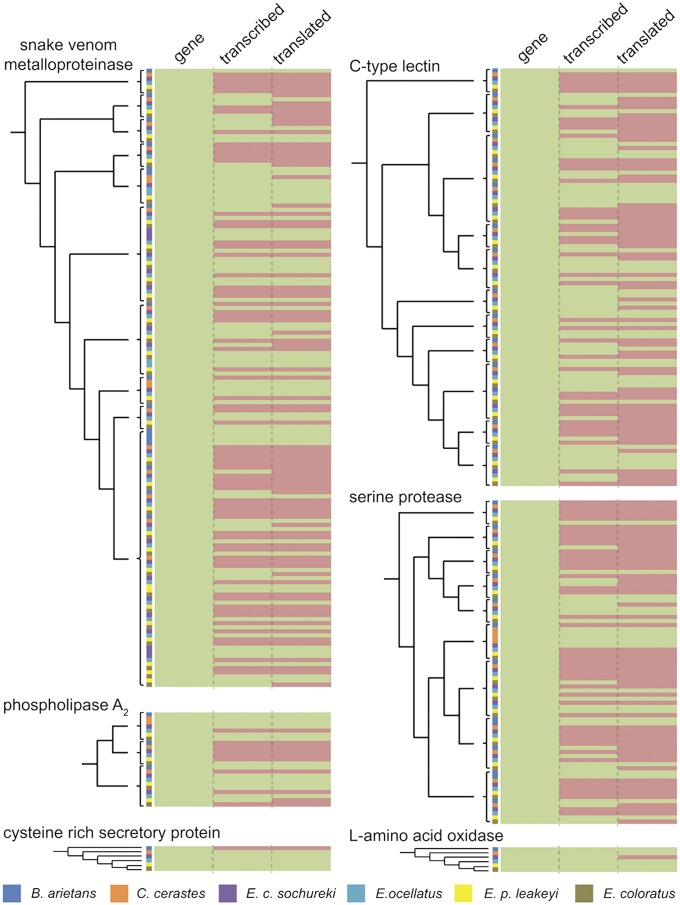
To better understand the evolutionary steps underpinning this complex interaction between gene transcription and protein secretion of snake venom, we reconstructed gene phylogenies for each toxin family identified across the venom proteomes (SVMP, SP, CTL, PLA2, CRISP, and LAAO), by coupling venom gland transcriptome data to phylogenetic analyses. Because relevant genome information was lacking, we reconciled the resulting gene trees to the previously described species tree (20, 29, 30) to reconstruct the timing of gene duplication events and to predict the number of untranscribed (or pseudogenic) gene loci. Such reconciliation approaches have successfully been used elsewhere, including with venom toxins, to predict the number of genes housed in a genome in the absence of genomic information (31–33). To determine which loci are translated into venom proteins, we mapped the proteomic representation and abundance of each toxin to the reconciled trees (SI Appendix, Figs. S8–S14).
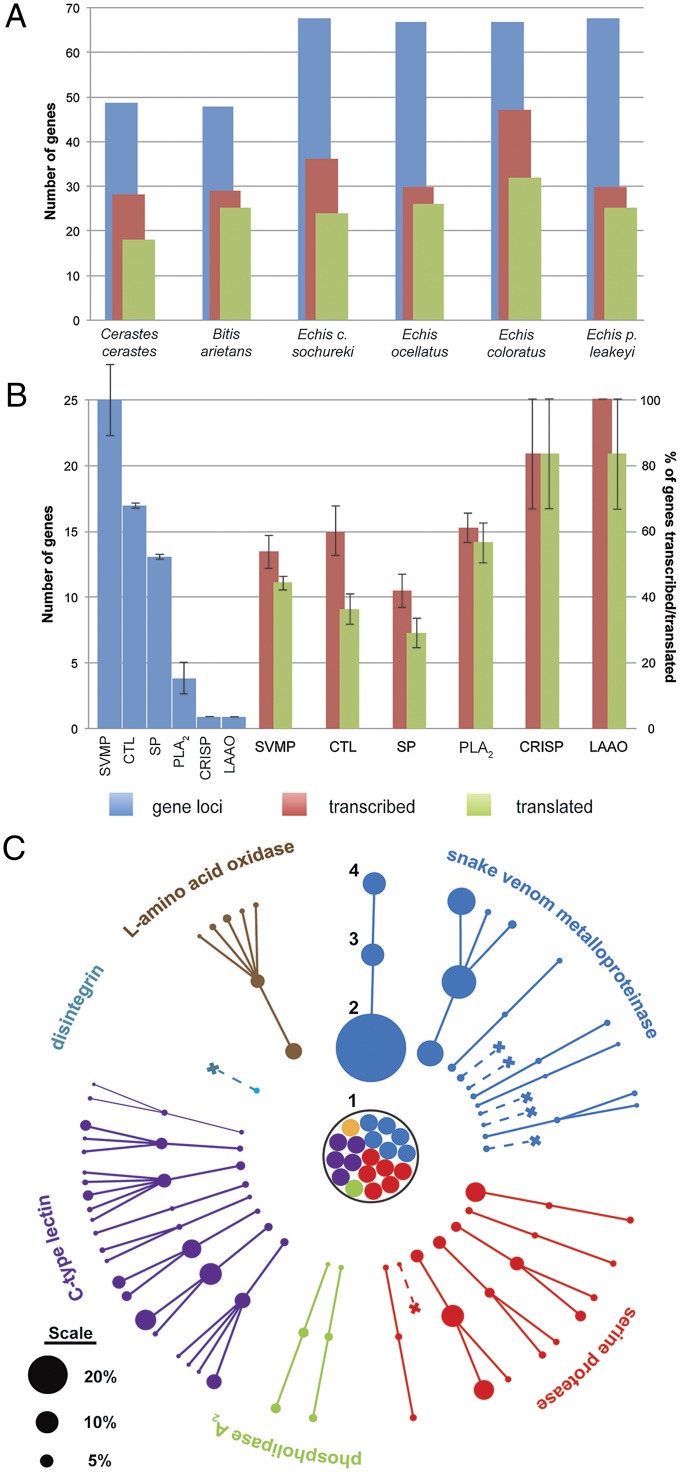
Comparing the number of genomic loci inferred by our reconciliation analyses with the number of transcribed genes (identified in the venom gland transcriptomes) and with the number of translated proteins (identified in the venom proteomes) revealed a surprisingly high pattern of gene redundancy (Figs. 2 and 3A). The percentage of genomic loci transcribed as toxin genes in the venom gland ranges from 44.12 to 70.15% across the six species, whereas the percentage of those genomic loci being translated into secreted venom toxins ranges from 35.29 to 52.08% (Fig. 3A). Whereas these results highlight the likely influence of many genes becoming pseudogenic, as predicted by the birth and death model of gene evolution (16), they also strongly indicate that transcriptional and translational regulatory control mechanisms have a major influence on the production of venom secretions. Venom gland microRNAs have recently been demonstrated to influence the translation of venom proteins from genes transcribed in the venom gland (8), and their action here may also be partially responsible for generating the venom variation observed. However, our data suggests that factors acting on the transcription of genomic loci in the venom gland may also play an important role in influencing venom composition.
Fig. 2.
Heat map visualization of venom toxins demonstrating variable patterns of transcription and translation for gene homologs and paralogs. The heat map is colored for genes predicted by reconciliation analysis, those transcribed in the venom gland transcriptome, and those translated as secreted proteins detected in venom. Green represents presence, and red represents absence. To the left of each heat map is the tree topology for: (i) the major clades found in the multilocus gene families (SVMP, CTL, SP, and PLA2) and (ii) the single locus gene families (CRISP and LAAO). Genes are ordered by phylogenetic clades (SI Appendix, Figs. S8–S14) with the species that each gene was recovered from indicated by different colors displayed in the key.
Fig. 3.
Variation in gene loci, gene transcription, and gene translation in the snake venom system. (A) Comparisons between species. Data are displayed as the total number of gene loci, genes transcribed, and genes translated for all venom toxin families in each species. (B) Comparisons between toxin families. Data are displayed as the mean number of gene loci per species (Left) and the mean percentage of those genes transcribed and translated summarized for all species (Right). Error bars represent SEM. Also see SI Appendix, Fig. S15. (C) Variation in the composition of toxins found in the venom system of B. arietans. (1) Predicted toxin genes that are untranscribed. (2) Venom gland transcribed toxin genes. (3) Corresponding toxins detected in the venom proteome. (4) Proteolysed protein products derived from each protein. Toxins are represented by nodes and their percentage abundance by node size (scale does not apply to part 1). Crosses indicate transcribed genes not translated. The orange node represents a predicted CRISP gene that was untranscribed.
Our analyses also reveal an extremely variable pattern of differential transcription, translation, and relative abundance of toxin paralogs at the gene family level (Fig. 2). Notably, the most extensive variation in genes encoding toxic proteins was found in the larger multilocus gene families, with the single loci gene families approaching a typically 1:1:1 ratio of genomic loci to transcribed gene to secreted venom protein (Fig. 3B and SI Appendix, Fig. S15). These results are perhaps unsurprising, but they further reflect the apparent distinction between pathogenically important toxin families (e.g., SVMP and SP) and so-called “ancillary” toxin families (e.g., LAAO and CRISP) described (15). Here, we demonstrate that this dichotomy appears to extend from the mode and tempo of gene family evolution noted (15) to the mechanisms governing toxin transcription and translation.
Venom complexity is further influenced by the effect of posttranslational protein modifications (34, 35). Across the sampled species, we detected multiple instances of the same toxin being identified in distinct proteomic fractions (SI Appendix, Tables S1–S6), signifying the presence of multiple protein products as the result of proteolysis (SI Appendix, Fig. S16). These observations likely reflect (i) the proteolytic cleavage of single gene proteins forming multiple products (e.g., SVMPs; ref. 34) and (ii) the cleavage of multimeric structures, which can be encoded by the same gene or different genes from either the same or distinct toxin families (35). Considerable interspecific variation in the number of proteolysed toxins was observed, ranging from three genes in E. p. leakeyi producing six venom protein products, to 13 forming 36 in B. arietans venom (SI Appendix, Fig. S16A). Therefore, the extensive interspecies variation observed for gene transcription and protein translation also extends to the proteolytic cleavage of venom toxins. We also detected extensive variation across the different toxin families, with the vast majority detected in the CTLs (43%) and SVMPs (38%). Each species contained multiple proteolysed toxins from these gene families in their venom, with the exception of CTLs in C. cerastes (SI Appendix, Fig. S16B). In contrast, no PLA2 or CRISP toxins showed evidence of proteolytic cleavage in any venom. Because evidence of proteolysis appears to be more commonplace in some toxin families than others, the importance of their respective contribution to the venom mixture is likely to be intrinsically associated to the evolution of each gene family and the representation of such genes in the genome and venom gland of each species.
The multiple layers of regulation acting on the production of venom generate surprising diversity in toxin composition, even between closely related species (Figs. 1 and 3). Variation in genomic loci, through the duplication or loss of toxin encoding genes over evolutionary time, is likely to have a major influence on the generation of venom variation, particularly between taxonomically distinct groups of snakes, as seen here when comparing Echis with B. arietans and C. cerastes (e.g., SI Appendix, Figs. S8, S9, and S12). However, factors influencing the transcription of these genes, and then their translation, clearly influence the mixture of toxins present in these secretions to varying extents (Fig. 3C), although the action of these mechanisms appears indiscriminate. Nonetheless, the result of variation in gene transcription, protein translation, and proteolytic processing results in secreted venom containing a drastically different toxin composition than predicted by our genome analyses and the venom gland transcriptome (Fig. 3C), thereby emphasizing the relative importance of these processes to the generation of snake venom variation.
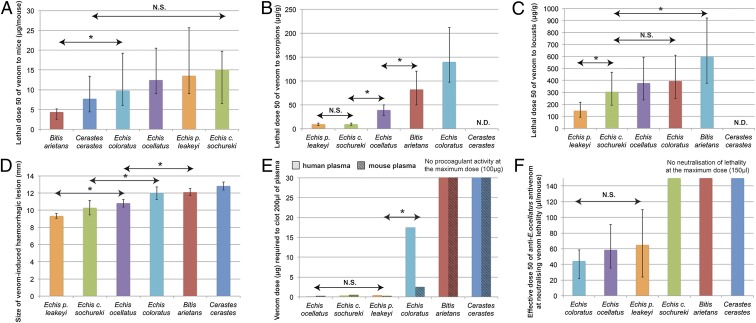
Importantly, the generation of venom variation may have serious medical consequences by causing significant differences in the toxicity and pathogenicity of different venoms (Fig. 4 and SI Appendix, Table S7). We previously found no significant difference between the lethality of the four Echis venoms to laboratory mice (12), and here we demonstrate that all exhibit comparable lethality to the venom of C. cerastes (Fig. 4A). However, we find that the venom of B. arietans is significantly more potent to mice that any of the Echis venoms (Fig. 4A). Contrastingly, the Echis venoms show significant interspecific variation in their lethality to (i) scorpions, where all, apart from E. coloratus, are more potent than the venom of B. arietans to this prey species (20) (Fig. 4B) and (ii) locusts, to which E. p. leakeyi venom is significantly more toxic than venoms from the other Echis species and from B. arietans (25) (Fig. 4C). Here, we also describe significant variation in specific venom-induced pathologies that characterize bites by viperid snakes: hemorrhage and coagulopathy. The Echis venoms exhibit significant interspecific variation in their ability to cause hemorrhage in a murine in vivo model, although the venoms of B. arietans and C. cerastes are significantly more hemorrhagic than Echis, with the exception of E. coloratus (Fig. 4D). In contrast, the venoms of the Echis species are highly procoagulant and capable of clotting 200 μL of human and mouse plasma with less than 0.6 μg of venom (Fig. 4E). The exception to this observation is the venom of E. coloratus, which is significantly less procoagulant, although this disparity is noticeably higher with human plasma than mouse, further highlighting the apparent unpredictability of taxon-specific venom activities. Contrastingly, the venoms of B. arietans and C. cerastes were incapable of inducing clotting, even at 1,000 times the dose required by E. ocellatus venom (Fig. 4E).
Fig. 4.
The effect of venom variation on venom-induced lethality and pathogenicity and the neutralizing efficacy of an antivenom. Comparisons of the in vivo lethality (LD50) of each venom to mice (A), scorpions (B), and locusts (C), their in vivo hemorrhagic (D) and in vitro procoagulant activities (E), and their neutralization by an E. ocellatus-monospecific antivenom, EchiTAbG (F). Asterisks indicate significant differences (P < 0.05) detected between venoms; N.D. indicates not determined; and N.S. indicates no significant difference. Error bars in A–C and F represent 95% confidence intervals and in D represent SEM. Note that B. arietans and C. cerastes venoms did not cause procoagulant effects at the highest dose tested (100 μg) and that EchiTAbG was ineffective at preventing lethality caused by E. c. sochureki, B. arietans, and C. cerastes venoms at the highest dose permitted by the ED50 assay (150 μL). Parts of the data shown in A–C and F are reproduced from refs. 12, 20, and 25).
The variations in venom-induced lethality and pathology described here have medical implications relating to antivenom therapy. Thus, the antivenom EchiTAbG (manufactured from the IgG of sheep hyperimmunized with E. ocellatus venom; ref. 4) is equally effective at neutralizing the lethal effects of venom from E. ocellatus, E. coloratus, and E. p. leakeyi in vivo (12). These results emphasize that a certain degree of venom variation does not render a monospecific antivenom ineffective, so long as there are sufficient species-common toxin epitopes present to ensure the cross-reactive efficacy of antivenom antibodies. However, we find that EchiTAbG antivenom is ineffective at neutralizing venom from E. c. sochureki, B. arietans, and C. cerastes in a murine preclinical model (Fig. 4F). In all cases, the maximum dose of antivenom permitted in the assay failed to prevent venom-induced lethality. The result for E. c. sochureki is particularly notable because the gross venom composition of this species appears similar to its congeners (Fig. 1), thereby highlighting that subtle variation within toxin families is sufficient to undermine antivenom efficacy. Therefore, antivenom failure may be the result of a combination of variation in both genomic and postgenomic processes. Importantly, the highly variable nature of postgenomic processes acting differentially on gene paralogs highlights why predicting the cross-reactive efficacy of an antivenom can be exceedingly problematic.
The multiple layers of regulation acting on venom toxins results in substantial species differences in venom protein composition and may also be at least partially responsible for differences in venom toxicity. Ultimately, the combination of genomic and postgenomic processes causes substantial variation in venom composition that makes the design of a universal snakebite therapy extremely problematic. We therefore hope that this research will stimulate additional investigations to elucidate the specific mechanisms controlling venom proteome complexity and, thereby, improve our understanding of how to better treat an important neglected tropical disease, snakebite.
Methods
SI Appendix, SI Materials and Methods has additional information relating to the methodologies described below.
Venom Gland Transcriptomes.
Venom gland transcriptomes for B. arietans (Nigeria) and C. cerastes (Egypt) were constructed using the protocols previously described for E. ocellatus (Nigeria), E. p. leakeyi (Kenya), E. coloratus (Egypt), and E. c. sochureki (United Arab Emirates) (26, 27). Briefly, libraries were constructed by using mRNA extracted from venom glands pooled from 10 individuals of each species by using the CloneMiner method, with ∼1,000 clones sequenced by using Sanger sequencing for each library. Resulting ESTs were assembled into contigs (putative gene products) and annotated, and their transcription level was quantified as previously described (26, 27). Each dataset was then subjected to six frame translations and used as a reference database to facilitate proteomic identification.
Venom Proteomes.
Proteins from each crude, lyophilized venom (2 mg extracted from the same individuals used for venom gland transcriptomics) were separated by reverse-phase HPLC with isolated fractions subjected to N-terminal sequencing and molecular mass determination as previously described (28). The relative abundances (expressed as percentage of the total venom proteins) of the different protein families were calculated from the relation of the sum of the areas of the reverse-phase chromatographic peaks (containing proteins from the same family), to the total area of venom protein peaks in the reverse-phase chromatogram. The relative contributions of different proteins eluting in the same chromatographic fraction was estimated by densitometry of Coomassie brilliant blue-stained SDS-PAGE gels, as previously outlined (28).
Toxin Evolution.
Toxin gene sequences annotated as SVMP, PLA2, CTL, SP, LAAO, or CRISP in the transcriptomes were extracted and analyzed to reconstruct their evolutionary history. These gene families were selected based on the results of the proteomic analyses, which identified the presence of these toxin types in the venom of the majority of the sampled species. The remaining toxin family identified and analyzed, the short-coding disintegrins (DIS), were discarded from phylogenetic analysis due to their apparent convergent evolution from SVMPs (18, 36). For each toxin family, nonredundant nucleotide sequences from each of the six transcriptomes were aligned with published gene homologs isolated from the venom systems of other viperid snakes, using the MUSCLE algorithm (37). Phylogenetic analyses for each toxin family were performed by incorporating optimized models of sequence evolution (SI Appendix, Table S8) into MrBayes v3.2 (38). Nucleotide gene trees were generated in duplicate using four chains for 1 × 107 generations, sampling every 500th cycle from the chain and using default settings in regards to priors. Tracer v1.4 (39) was used to estimate effective sample sizes for all parameters and to verify the point of convergence (burnin), with trees generated prior to this point discarded. The resulting sequence alignments have been submitted to the Dryad Digital Repository, http://datadryad.org/ (doi:10.5061/dryad.1j292).
Gene and Protein Comparisons.
Proteomic matches to transcriptome gene products were overlaid on to the generated gene trees, alongside calculations of protein abundance. We applied the following rules to the data: (i) We conservatively retained the longest nucleotide sequence for each transcriptome contig and discarded all other sequence variants. These sequences are unlikely to represent distinct genes, but rather are the result of allelic variation (as 10 specimens were used for library construction). We removed these sequences to prevent artificial inflation of gene numbers generated by the reconciliation analyses described below. (ii) Where different contigs from the same species exhibited monophyly and did not result in distinct proteome matches, we merged the contigs and discarded nonmatching sequences because of sequence redundancy. (iii) Sequences from contigs that were nonmonophyletic with other contigs from the same species and had no proteomic match were retained and annotated with “no proteomic hit”, indicative of nontranslation of the transcript. (iv) Posttranslational modifications (i.e., proteolytic cleavage) were assigned where multiple proteomic matches to the same transcriptome cluster were found in distinct nonoverlapping proteomic fractions.
Gene/Species Tree Reconciliation.
We reconciled the resulting gene trees with the species tree presented in Fig. 1 by using NOTUNG.v2.6 (31). Each Echis/Bitis/Cerastes toxin clade was analyzed separately by using the reconciliation option, which produces a reconciled tree displaying the timing of gene duplication and loss events, and the predicted number of gene loci. Because NOTUNG permits the reconciliation of genes trees to nonbinary species trees (31), we used this option when clade support in the gene tree exhibited a Bayesian posterior probability of <0.95. This approach allowed nodes that were not robustly supported in the gene tree to be reconciled with a polytomous species tree. We used this conservative approach to mitigate the generation of spurious gene duplication events that would otherwise be generated as artifacts of uncertainty in the gene tree.
Functional Studies.
The same venom samples used for proteomic analyses were used for functional assessments. All animal experimentation was conducted using standard protocols approved by the Liverpool School of Tropical Medicine Animal Welfare and Ethical Review Board and the UK Home Office (license no. 40/3216, 40/3151, 40/3718). Murine in vivo lethality studies (12) were conducted to calculate the lethal dose 50 (LD50; the amount of venom that kills 50% of injected mice) for B. arietans and C. cerastes venom. We used the same murine model to test the efficacy of the E. ocellatus monospecific antivenom, EchiTAbG, at neutralizing five times the venom LD50 for B. arietans and C. cerastes in the effective dose 50 (ED50) assay (12). Both sets of experiments were undertaken as previously described using venom from the four Echis species (12), with comparisons undertaken using the 95% confidence limits generated by probit analysis (40). LD50 values for venom lethality to scorpions and locusts were reproduced from our earlier work (20, 25). Modified minimum hemorrhagic dose (MHD) experiments (41) were undertaken to compare the hemorrhagic activity of each venom. Ten-microgram doses of venom were injected intradermally into the dorsal skin of groups of anesthetized mice and the skin lesion size measured after 24 h. The mean diameter of each lesion was compared using paired two-tailed t tests with a P value threshold of 0.05. The procoagulant activity of each venom was determined using the minimum coagulant dose (MCD-P) assay (41). Varying doses of venom were incubated with 200 μL of human or mouse plasma (Sigma) at 37°C, and the clotting time was recorded. The 60-s clotting time was calculated by plotting clotting time against venom dose, and statistical comparison of different venoms were undertaken using regression analysis of the resulting lines with a P value threshold of 0.05.
Supplementary Material
Acknowledgments
We thank Paul Rowley (Liverpool School of Tropical Medicine) and Tim Booth [Natural Environmental Research Council (NERC) Environmental Bioinformatics Centre] for their help with this study. This work was funded by NERC UK Research Studentship NER/S/A/2006/14086 and Postdoctoral Research Fellowship NE/J018678/1 (to N.R.C.), access to the NERC Molecular Genetics Facility at the University of Edinburgh from Grant MGF150 (to W.W.), a Leverhulme Trust Grant F/00174/I (to W.W. and R.A.H.), a National Centre for the Replacement, Refinement and Reduction of Animals in Research Studentship NC/K500288/1 (to R.A.H.), Ministerio de Ciencia é Innovación (currently Ministerio de Economía y Competitividad), Madrid Grant BFU2010-17373 and the Generalitat Valenciana Grant PROMETEO/2010/005 (to J.J.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence alignments reported in this paper have been deposited in the Dryad Digital Respository, http://datadryad.org (doi:10.5061/dryad.1j292).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1405484111/-/DCSupplemental.
References
- 1.Casewell NR, Wüster W, Vonk FJ, Harrison RA, Fry BG. Complex cocktails: The evolutionary novelty of venoms. Trends Ecol Evol. 2013;28(4):219–229. doi: 10.1016/j.tree.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 2.Kasturiratne A, et al. The global burden of snakebite: A literature analysis and modelling based on regional estimates of envenoming and deaths. PLoS Med. 2008;5(11):e218. doi: 10.1371/journal.pmed.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrison RA, Hargreaves A, Wagstaff SC, Faragher B, Lalloo DG. Snake envenoming: A disease of poverty. PLoS Negl Trop Dis. 2009;3(12):e569. doi: 10.1371/journal.pntd.0000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abubakar IS, et al. Nigeria-UK EchiTab Study Group Randomised controlled double-blind non-inferiority trial of two antivenoms for saw-scaled or carpet viper (Echis ocellatus) envenoming in Nigeria. PLoS Negl Trop Dis. 2010;4(7):e767. doi: 10.1371/journal.pntd.0000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams DJ, et al. Ending the drought: New strategies for improving the flow of affordable, effective antivenoms in Asia and Africa. J Proteomics. 2011;74(9):1735–1767. doi: 10.1016/j.jprot.2011.05.027. [DOI] [PubMed] [Google Scholar]
- 6.Chippaux JP, Williams V, White J. Snake venom variability: Methods of study, results and interpretation. Toxicon. 1991;29(11):1279–1303. doi: 10.1016/0041-0101(91)90116-9. [DOI] [PubMed] [Google Scholar]
- 7.Durban J, et al. Profiling the venom gland transcriptomes of Costa Rican snakes by 454 pyrosequencing. BMC Genomics. 2011;12:259. doi: 10.1186/1471-2164-12-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durban J, et al. Integrated “omics” profiling indicates that miRNAs are modulators of the ontogenetic venom composition shift in the Central American rattlesnake, Crotalus simus simus. BMC Genomics. 2013;14:234. doi: 10.1186/1471-2164-14-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibbs HL, Sanz L, Sovic MG, Calvete JJ. Phylogeny-based comparative analysis of venom proteome variation in a clade of rattlesnakes (Sistrurus sp.) PLoS ONE. 2013;8(6):e67220. doi: 10.1371/journal.pone.0067220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casewell NR, Wagstaff SC, Harrison RA, Wüster W. Gene tree parsimony of multilocus snake venom protein families reveals species tree conflict as a result of multiple parallel gene loss. Mol Biol Evol. 2011;28(3):1157–1172. doi: 10.1093/molbev/msq302. [DOI] [PubMed] [Google Scholar]
- 11.Daltry JC, Wüster W, Thorpe RS. Diet and snake venom evolution. Nature. 1996;379(6565):537–540. doi: 10.1038/379537a0. [DOI] [PubMed] [Google Scholar]
- 12.Casewell NR, et al. Pre-clinical assays predict pan-African Echis viper efficacy for a species-specific antivenom. PLoS Negl Trop Dis. 2010;4(10):e851. doi: 10.1371/journal.pntd.0000851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petras D, et al. Snake venomics of African spitting cobras: Toxin composition and assessment of congeneric cross-reactivity of the pan-African EchiTAb-Plus-ICP antivenom by antivenomics and neutralization approaches. J Proteome Res. 2011;10(3):1266–1280. doi: 10.1021/pr101040f. [DOI] [PubMed] [Google Scholar]
- 14.Fry BG. From genome to “venome”: Molecular origin and evolution of the snake venom proteome inferred from phylogenetic analysis of toxin sequences and related body proteins. Genome Res. 2005;15(3):403–420. doi: 10.1101/gr.3228405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vonk FJ, et al. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc Natl Acad Sci USA. 2013;110(51):20651–20656. doi: 10.1073/pnas.1314702110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nei M, Gu X, Sitnikova T. Evolution by the birth-and-death process in multigene families of the vertebrate immune system. Proc Natl Acad Sci USA. 1997;94(15):7799–7806. doi: 10.1073/pnas.94.15.7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kini RM, Chan YM. Accelerated evolution and molecular surface of venom phospholipase A2 enzymes. J Mol Evol. 1999;48(2):125–132. doi: 10.1007/pl00006450. [DOI] [PubMed] [Google Scholar]
- 18.Casewell NR, Wagstaff SC, Harrison RA, Renjifo C, Wüster W. Domain loss facilitates accelerated evolution and neofunctionalization of duplicate snake venom metalloproteinase toxin genes. Mol Biol Evol. 2011;28(9):2637–2649. doi: 10.1093/molbev/msr091. [DOI] [PubMed] [Google Scholar]
- 19.Casewell NR, Huttley GA, Wüster W. Dynamic evolution of venom proteins in squamate reptiles. Nat Commun. 2012;3:1066. doi: 10.1038/ncomms2065. [DOI] [PubMed] [Google Scholar]
- 20.Barlow A, Pook CE, Harrison RA, Wüster W. Coevolution of diet and prey-specific venom activity supports the role of selection in snake venom evolution. Proc Biol Sci. 2009;276(1666):2443–2449. doi: 10.1098/rspb.2009.0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wooldridge BJ, et al. Mojave rattlesnakes (Crotalus scutulatus scutulatus) lacking the acidic subunit DNA sequence lack Mojave toxin in their venom. Comp Biochem Physiol B Biochem Mol Biol. 2001;130(2):169–179. doi: 10.1016/s1096-4959(01)00422-5. [DOI] [PubMed] [Google Scholar]
- 22.Wagstaff SC, Sanz L, Juárez P, Harrison RA, Calvete JJ. Combined snake venomics and venom gland transcriptomic analysis of the ocellated carpet viper, Echis ocellatus. J Proteomics. 2009;71(6):609–623. doi: 10.1016/j.jprot.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 23.Aird SD, et al. Quantitative high-throughput profiling of snake venom gland transcriptomes and proteomes (Ovophis okinavensis and Protobothrops flavoviridis) BMC Genomics. 2013;14:790. doi: 10.1186/1471-2164-14-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Margres MJ, et al. Linking the transcriptome and proteome to characterize the venom of the eastern diamondback rattlesnake (Crotalus adamanteus) J Proteomics. 2014;96:145–158. doi: 10.1016/j.jprot.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 25.Richards DP, Barlow A, Wüster W. Venom lethality and diet: Differential responses of natural prey and model organisms to the venom of the saw-scaled vipers (Echis) Toxicon. 2012;59(1):110–116. doi: 10.1016/j.toxicon.2011.10.015. [DOI] [PubMed] [Google Scholar]
- 26.Casewell NR, Harrison RA, Wüster W, Wagstaff SC. Comparative venom gland transcriptome surveys of the saw-scaled vipers (Viperidae: Echis) reveal substantial intra-family gene diversity and novel venom transcripts. BMC Genomics. 2009;10:564. doi: 10.1186/1471-2164-10-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagstaff SC, Harrison RA. Venom gland EST analysis of the saw-scaled viper, Echis ocellatus, reveals novel α9β1 integrin-binding motifs in venom metalloproteinases and a new group of putative toxins, renin-like aspartic proteases. Gene. 2006;377:21–32. doi: 10.1016/j.gene.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 28.Calvete JJ. Proteomic tools against the neglected pathology of snake bite envenoming. Expert Rev Proteomics. 2011;8(6):739–758. doi: 10.1586/epr.11.61. [DOI] [PubMed] [Google Scholar]
- 29.Pook CE, Joger U, Stümpel N, Wüster W. When continents collide: Phylogeny, historical biogeography and systematics of the medically important viper genus Echis (Squamata: Serpentes: Viperidae) Mol Phylogenet Evol. 2009;53(3):792–807. doi: 10.1016/j.ympev.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 30.Wüster W, Peppin L, Pook CE, Walker DE. A nesting of vipers: Phylogeny and historical biogeography of the Viperidae (Squamata: Serpentes) Mol Phylogenet Evol. 2008;49(2):445–459. doi: 10.1016/j.ympev.2008.08.019. [DOI] [PubMed] [Google Scholar]
- 31.Vernot B, Stolzer M, Goldman A, Durand D. Reconciliation with non-binary species trees. J Comput Biol. 2008;15(8):981–1006. doi: 10.1089/cmb.2008.0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vandepoele K, De Vos W, Taylor JS, Meyer A, Van de Peer Y. Major events in the genome evolution of vertebrates: Paranome age and size differ considerably between ray-finned fishes and land vertebrates. Proc Natl Acad Sci USA. 2004;101(6):1638–1643. doi: 10.1073/pnas.0307968100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang D, Duda TF., Jr Extensive and continuous duplication facilitates rapid evolution and diversification of gene families. Mol Biol Evol. 2012;29(8):2019–2029. doi: 10.1093/molbev/mss068. [DOI] [PubMed] [Google Scholar]
- 34.Fox JW, Serrano SMT. Insights into and speculations about snake venom metalloproteinase (SVMP) synthesis, folding and disulfide bond formation and their contribution to venom complexity. FEBS J. 2008;275(12):3016–3030. doi: 10.1111/j.1742-4658.2008.06466.x. [DOI] [PubMed] [Google Scholar]
- 35.Doley R, Kini RM. Protein complexes in snake venom. Cell Mol Life Sci. 2009;66(17):2851–2871. doi: 10.1007/s00018-009-0050-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanz-Soler R, et al. Recombinant expression of mutants of the Frankenstein disintegrin, RTS-ocellatusin. Evidence for the independent origin of RGD and KTS/RTS disintegrins. Toxicon. 2012;60(4):665–675. doi: 10.1016/j.toxicon.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 37.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ronquist F, et al. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Finney DJ. Probit Analysis; A Statistical Treatment of the Sigmoid Response Curve. Oxford: Macmillan; 1947. [Google Scholar]
- 41.Theakston RD, Reid HA. Development of simple standard assay procedures for the characterization of snake venom. Bull World Health Organ. 1983;61(6):949–956. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.