Significance
Hemagglutinin (HA), the major influenza virus envelope glycoprotein, is the principal target of neutralizing antibodies. Wide diversity and variation of HA entails annual vaccination, as current vaccines typically fail to elicit/boost cross-reactive, broadly neutralizing antibodies (bnAbs). Although several bnAbs bind at the conserved stem of HA making it an attractive universal vaccine candidate, the metastable conformation of this domain imposes challenges in designing a stable, independently folding HA stem immunogen. We rationally designed a stem-fragment immunogen, mimicking the native HA stem that binds conformation-specific bnAbs with high affinity. The immunogen elicited bnAbs and conferred robust protection against lethal, heterologous virus challenge in vivo. Additionally, soluble bacterial expression of such a thermotolerant, disulfide-free immunogen allows for rapid scale-up during pandemic outbreak.
Keywords: hemagglutinin stalk, stabilization, cross-protection, pandemic preparedness, Escherichia coli
Abstract
Influenza hemagglutinin (HA) is the primary target of the humoral response during infection/vaccination. Current influenza vaccines typically fail to elicit/boost broadly neutralizing antibodies (bnAbs), thereby limiting their efficacy. Although several bnAbs bind to the conserved stem domain of HA, focusing the immune response to this conserved stem in the presence of the immunodominant, variable head domain of HA is challenging. We report the design of a thermotolerant, disulfide-free, and trimeric HA stem-fragment immunogen which mimics the native, prefusion conformation of HA and binds conformation specific bnAbs with high affinity. The immunogen elicited bnAbs that neutralized highly divergent group 1 (H1 and H5 subtypes) and 2 (H3 subtype) influenza virus strains in vitro. Stem immunogens designed from unmatched, highly drifted influenza strains conferred robust protection against a lethal heterologous A/Puerto Rico/8/34 virus challenge in vivo. Soluble, bacterial expression of such designed immunogens allows for rapid scale-up during pandemic outbreaks.
Seasonal influenza outbreaks across the globe cause an estimated 250,000–500,000 deaths annually (1). Current influenza vaccines need to be updated every few years because of antigenic drift (2). Despite intensive monitoring, strain mismatch between vaccine formulation and influenza viruses circulating within the population has occurred in the past (2). Public health is further compromised when an unpredictable mixing event among influenza virus genomes leads to antigenic shift facilitating a potential pandemic outbreak. These concerns have expedited efforts toward developing a universal influenza vaccine.
Neutralizing antibodies (nAbs) against hemagglutinin (HA) are the primary correlate for protection in humans and hence HA is an attractive target for vaccine development (3). The precursor polypeptide, HA0, is assembled into a trimer along the secretory pathway and transported to the cell surface. Cleavage of HA0 generates the disulfide-linked HA1 and HA2 subunits. Mature HA has a globular head domain which mediates receptor binding and is primarily composed of the HA1 subunit, whereas the stem domain predominantly comprises the HA2 subunit. The HA stem is trapped in a metastable state and undergoes an extensive low-pH-induced conformational rearrangement in the host-cell endosomes to adopt the virus–host membrane fusion-competent state (4, 5).
The antigenic sites on the globular head of HA are subjected to heightened immune pressure resulting in escape variants, thereby limiting the breadth of head-directed nAbs (6). However, extensive efforts have resulted in the isolation of monoclonal antibodies (mAbs) that bind within the globular head and inhibit receptor attachment, which neutralize drifted variants of an HA subtype or heterosubtypic HA (7–16). The HA stem is targeted by several broadly neutralizing antibodies (bnAbs) with neutralizing activity against diverse influenza A virus subtypes (17). The epitopes of these bnAbs in the HA stem are more conserved across different influenza HA subtypes compared with the antigenic sites in the HA globular head (18).
During a primary infection, the immunodominant globular head domain suppresses the response toward the conserved stem. Several efforts have been made to circumvent this problem. Repeated immunizations with full-length, chimeric HAs (cHAs) in a protracted vaccination regimen have been shown to boost stem-directed responses in mice (19). Alternatively, full-length HA presented on nanoparticles (np) has been shown to elicit stem-directed nAbs (20). Attempts have also been made to steer the immune response toward the conserved HA stem by hyperglycosylating the head domain (21). Although the aforementioned strategies need to be further evaluated and provide novel alternatives, detrimental interference from the highly variable immunodominant head domain in eliciting a broad functional response cannot be completely evaded. A “headless” stem domain immunogen offers an attractive solution. However, early attempts at expressing the HA2 subunit independently in a native, prefusion conformation were unsuccessful. In the absence of the head domain, the HA2 subunit expressed in Escherichia coli spontaneously adopted the low-pH conformation (22) in which the functional epitopes of stem-directed bnAbs are disrupted. More recently, the entire HA stem region has been expressed in a prefusion, native-like conformation in both prokaryotic and eukaryotic systems adopting multiple strategies (23–26).
Design of independently folding HA stem fragments which adopt the prefusion HA conformation presents another approach to elicit bnAbs against influenza (27, 28). The A helix of the HA2 subunit contributes substantial contact surface to the epitope of stem-directed bnAbs such as CR6261, F10, and others. Although multivalent display of A helix on the flock house virus as a virus-like particle platform elicited cross-reactive antibodies, it conferred only minimal protection (20%) against virus challenge in mice (29).
We report the design and characterization of engineered headless HA stem immunogens based on the influenza A/Puerto Rico/8/34 (H1N1) subtype. H1HA10-Foldon, a trimeric derivative of our parent construct (H1HA10), bound conformation-sensitive, stem-directed bnAbs such as CR6261 (30), F10 (31), and FI6v3 (32) with a high-affinity [equilibrium dissociation constant (KD) of 10–50 nM]. The designed immunogens elicited broadly cross-reactive antiviral antibodies which neutralized highly drifted influenza virus strains belonging to both group 1 (H1 and H5 subtypes) and 2 (H3 subtype) in vitro. Significantly, stem immunogens designed from unmatched, highly drifted influenza strains conferred protection against a lethal (2LD90) heterologous A/Puerto Rico/8/34 virus challenge in mice. Our immunogens confer robust subtype-specific and modest heterosubtypic protection in vivo. In contrast to previous stem domain immunogens (23–25), the designed immunogens were purified from the soluble fraction in E. coli. The HA stem-fragment immunogens do not aggregate even at high concentrations and are cysteine-free, which eliminates the complications arising from incorrect disulfide-linked, misfolded conformations. The aforementioned properties of the HA stem-fragment immunogens make it amenable for scalability at short notice which is vital during pandemic outbreaks.
Results
Immunogen Design.
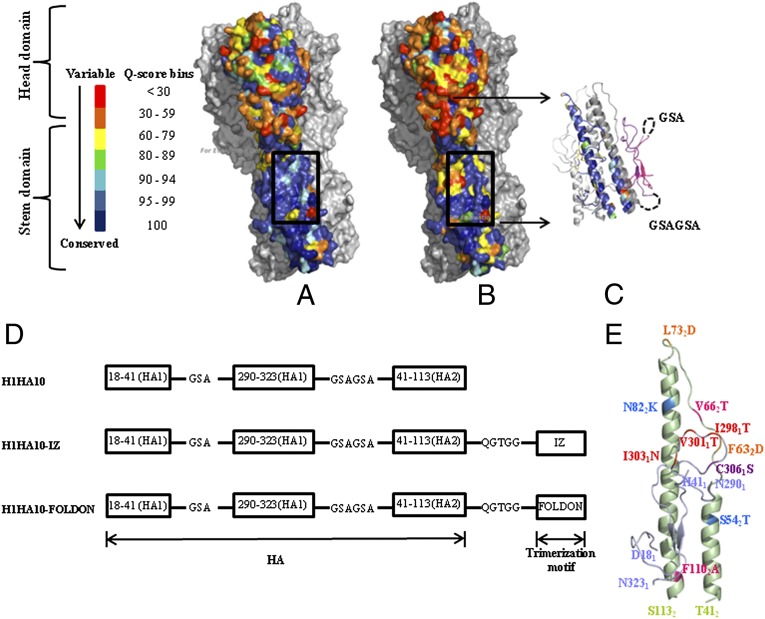
Antigenic differences between the HA surface glycoprotein of various influenza A viruses provide the basis for classification into 18 subtypes (H1–H18) (33). Wide diversity and rapid antigenic variation of HA remains the principal challenge to the development of potent vaccines. Targeting conserved regions of HA offers a promising strategy to combat viral evolution and escape. We analyzed a large dataset (4,241 sequences for H1N1 and 182 sequences for H5N1) of group 1 influenza virus sequences to identify conserved targets on HA. Consistent with previous results (18), the HA stem is more conserved as opposed to the highly variable, globular head domain. The residue conservation across all full-length, human isolates of H1 HA (Fig. 1A) and group 1 HA (Fig. 1B) was mapped onto the crystal structure of H1N1 A/Puerto Rico/8/34 HA [Protein Data Bank (PDB) ID code 1RU7] (34). The HA stem comprising the epitope of bnAbs is therefore a plausible target for developing a broadly protective vaccine (35).
Fig. 1.
The HA stem is conserved. (A) Residue conservation among H1 HA isolates mapped onto the surface representation of H1N1 A/Puerto Rico/8/34 HA trimer (PDB ID code 1RU7) (34). All residues in a monomer are colored according to the quality score (Q score) at that position obtained from a multiple-sequence alignment of H1N1 HA sequences using ClustalX (56). The head domain is highly variable as opposed to the conserved stem domain. The stem-directed bnAb CR6261 epitope (box) is highly conserved. Rest of HA (gray). (B) Residue conservation across group 1 HA (H1N1 and H5N1 subtype HA sequences were analyzed) mapped onto a H1N1 A/Puerto Rico/8/34 HA trimer (PDB ID code 1RU7). Despite the increased variation in HA across group 1 influenza A subtypes as opposed to within a subtype (H1), the epitope of the stem-directed bnAb CR6261 (box) is well conserved. Rest of HA (gray). (C) Cartoon representation of H1HA10. One monomer is colored according to the conservation scheme shown in A. In a second monomer, linker connectivity is shown (dashed black line). The first fragment 181–411 (pink) was connected to 2901–3231 (purple) by a three-residue GSA linker. The HA1 (purple) and HA2 fragments (gray) were connected by a six-residue linker (GSAGSA). The HA2 fragment (412–1132) included in H1HA10 incorporates elements of A helix and LAH of the HA2 subunit. Rest of HA (light gray). (D) Fragments of the HA stem defined by stable breakpoints were included in our preliminary construct H1HA10. The fragments were connected by flexible, soluble linkers. Derivatives of H1HA10 with C-terminal trimerization motifs, IZ or Foldon, were made (41, 42). (E) Mutations introduced in H1HA10 are shown in a monomer model. The HA1 fragments are colored light blue, whereas the HA2 fragment is in light green. Mutations were introduced to mask the hydrophobic patch [I2981T, V3011T, I3031N in the HA1 fragments (red); and V662T, F1102A in the HA2 fragment (pink)]. An additional mutation [C3061S (purple)] was made to prevent intermolecular disulfide bond formation. Mutations were also introduced to destabilize the low-pH conformation of HA [F632D and L732D (orange)]. Residues S542 and N822 present in the A/Puerto Rico/8/34 HA crystal structure (PDB ID code 1RU7) were mutated to the most commonly occurring residues T542 and K582 (sky blue) in a multiple-sequence alignment of all available full-length H1N1 A/Puerto Rico/8/34 HA sequences. The figures were rendered using PyMOL.
Mimicking the epitope of these stem-directed bnAbs in a native, prefusion conformation in a headless stem immunogen is challenging because of the metastable conformation of HA. The HA2 subunit, when expressed independently, adopts the low-pH conformation spontaneously (22). Extensive rearrangement at low pH displaces the A helix by over 100 Ǻ (5), disrupting the conformation-specific epitope of these bnAbs.
To enhance the immune response to the epitope of the HA stem-directed bnAbs like CR6261, F10, and FI6v3 (30–32), we adopted a protein-minimization approach to refine the previously reported HA stem immunogens, H1HA6 and H1HA0HA6. These HA stem immunogens consisted of the entire stem region of HA and were 258 (H1HA6) and 243 (H1HA0HA6) residues in length (24). Smaller, headless HA stem immunogens that contained the above epitope were designed from the influenza A (H1N1) A/Puerto Rico/8/34 subtype. We analyzed the interaction network of residues in the antibody footprint of these bnAbs with the rest of HA using the in-house software PREDBURASA as described previously (36). Briefly, the accessible surface area (ASA) of every residue in HA was calculated in the absence and presence of at least three-residue-long stretches of the CR6261 antibody footprint (321–361, 2921–2941, 182–212, 362–562), covering ∼95% of the CR6261 epitope. Residues belonging to either HA1 or HA2 subunits are distinguished by subscripts 1 and 2, respectively. All residues of HA, which had a total side-chain ASA difference of ≥5 Ǻ2 in the aforementioned calculations, were identified as interacting “network” residues. We repeated the PREDBURASA calculations now including the network residues to identify HA stem fragments defined by stable breakpoints with optimal termini distances, having minimalistic interactions with the rest of HA. Residue fragments 181–411, 2901–3231, and 412–1132 were included in H1HA10 (Fig. 1 C and D). H1HA10 includes ∼80% of the CR6261 antibody footprint. H1HA10 is 139 residues in length and is therefore ∼46% smaller than the full-length HA stem immunogen described previously (24). The newly generated exposed hydrophobic patches in H1HA10 due to interactions lost with the rest of HA were mutated using the software ROSETTA DESIGN (Version 3.0) (37) to minimize potential protein aggregation. A similar approach has been previously used by us to design stable influenza and HIV immunogens and inhibitors (24, 38, 39). The following mutations were incorporated to mask the hydrophobic patch: I2981T, V3011T, I3031N, V662T, and F1102A (Fig. 1E; Fig. S1). Cys2811 and Cys3061 form an intramolecular disulfide bond in full-length HA. Because Cys2811 was not incorporated in our design, we mutated Cys3061 to Ser to prevent incorrect, intermolecular disulfide bond formation. Low-pH conformation destabilizing mutations F632D and L732D, which were previously characterized (24) by us, were also incorporated in the design. Residues S542 and N822 present in the A/Puerto Rico/8/34 HA crystal structure (PDB ID code 1RU7) were replaced in H1HA10 with the most commonly occurring residue (T542 and K822) at that position among all of the available H1N1 A/Puerto Rico/8/34 sequences deposited with the National Center for Biotechnology Information (NCBI) Influenza Virus Database (www.ncbi.nlm.nih.gov/genomes/FLU/FLU.html). None of the introduced mutations were in the epitopes of the previously reported stem-directed bnAbs CR6261, F10, or FI6v3. These independent HA fragments were connected by flexible, soluble linkers of appropriate length as described previously (40) (Fig. 1D; Fig. S1). Also to promote the formation of a trimer, we made derivatives of H1HA10 with C-terminal trimerization motifs connected by flexible linkers. The parallel, coiled-coil trimerization motif isoleucine zipper (IZ) (41) was used in H1HA10-IZ. H1HA10-Foldon had the globular, β-rich trimerization motif “Foldon” (42) (Fig. 1D).
To assess the ability of our stem immunogens to confer cross-protection, constructs similar to H1HA10-Foldon were designed from unmatched, highly drifted influenza strains (Table S1) and tested against heterologous A/Puerto Rico/8/34 virus challenge in mice. The alternative approach of immunizing with our headless stem constructs designed from H1N1 A/Puerto Rico/8/34 and carrying out heterologous virus challenge could not be tested because of the unavailability of appropriate mouse-adapted virus strains other than H1N1 A/Puerto Rico/8/34. Constructs from other strains [H1N1 A/New Caledonia/20/99 (NCH1HA10-Foldon), H1N1 A/California/04/2009 (pH1HA10-Foldon), and H5N1 A/Viet Nam/1203/2004 (H5HA10-Foldon)] were designed using a facile strategy. A simplistic, pair-wise sequence alignment, which can guide immunogen design, emphasizes the utility of our design (Fig. S2). The hydrophobic residues mutated in H1HA10 to mask the newly generated hydrophobic patches are identical/similar within a subtype, therefore, analogous mutations can be included in H1HA10-like designs from other strains. Wide applicability of the mutations destabilizing the low-pH conformation of HA has been previously discussed (24). The sequences of all of the designed constructs are listed in Fig. S1.
Protein Purification and Biophysical Characterization of HA Stem Immunogens.
Protein solubility is a coarse indicator of proper folding and remains a crucial problem in heterologous expression systems (43). HA fragments expressed previously in E. coli formed inclusion body aggregates and required refolding (24, 44). In contrast, all our designed immunogens expressed in E. coli BL21(DE3) cells were purified from the soluble fraction of the cell culture lysate (Fig. S3), suggesting proper folding and validating our rational design. The protein yields were about 10–15 mg/L culture using unoptimized shake flask cultures and were purified using a single, affinity purification step.
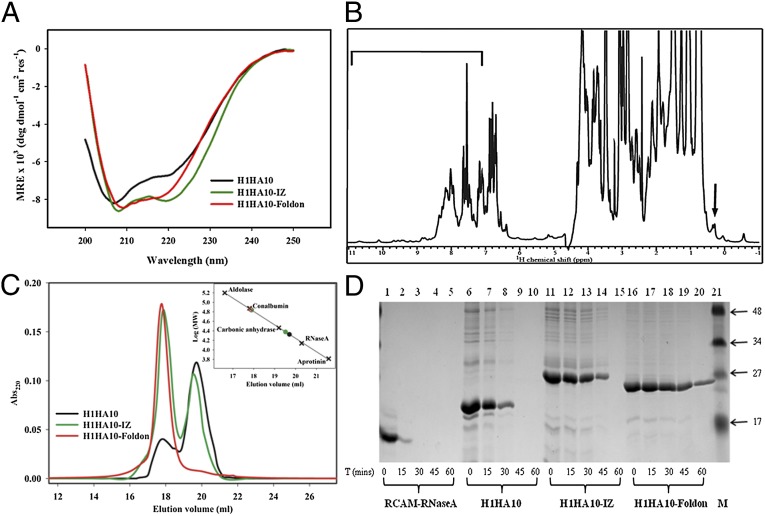
Circular dichroism (CD) spectra indicated that all of the proteins were folded and predominantly α-helical as expected. The trimerization motifs assist in the folding of H1HA10. H1HA10-IZ and H1HA10-Foldon are more helical than the parent construct, H1HA10, as observed from the double minima at 208 and 222 nm (Fig. 2A). The thermal stability of H1HA10-Foldon was monitored using CD. H1HA10-Foldon showed a reversible and cooperative unfolding thermal melt profile with an apparent transition midpoint (Tm) of ∼323 K (50 °C) at a protein concentration (in monomer units) of ∼15 µM. Because the folded protein is a trimer and the unfolded protein is likely to be a monomer, the Tm is expected to be concentration dependent (41). Consecutive scans recorded after cooling the sample back to 15 °C overlapped well with each other (Fig. S4A). In contrast, the full-length H1N1 A/Puerto Rico/8/34 rHA showed a broad transition without clear baselines and it was therefore not possible to estimate a Tm. The rescan showed no transition, indicating that thermal denaturation for rHA is irreversible (Fig. S4B). Intrinsic tryptophan fluorescence measurements also affirmed a folded, native structure for H1HA10 and its derivatives. All proteins showed a significant red-shift in the emission maxima upon denaturation with guanidine hydrochloride (GdnCl) (Fig. S5).
Fig. 2.
Biophysical and biochemical characterization of headless HA stem immunogens. (A) Secondary structure of the designed immunogens were monitored by CD spectroscopy. H1HA10 was folded and predominantly α-helical. The trimerization motifs, IZ and Foldon, assist in the folding of H1HA10. The double minima at 208 and 222 nm indicate that H1HA10-IZ and H1HA10-Foldon are more helical than H1HA10. The samples were in PBS (pH 7.4). (B) One-dimensional 1H NMR spectra of H1HA10-Foldon. The chemical shift dispersion indicates the protein is well folded. The upfield-shifted lines in the methyl group region starting at 0.6 ppm are indicated by an arrow. The chemical shift dispersion in the amide region is highlighted by a bar. (C) Oligomeric state of the designed immunogens was determined by analytical gel-filtration chromatography under nondenaturing conditions on a Superdex-200 column in PBS (pH 7.4) at room temperature. H1HA10 eluted predominantly as a monomer, H1HA10-IZ as a mixture of trimer and monomer, and H1HA10-Foldon as a homogenous trimer in solution. The column was calibrated using a broad range of molecular weight markers (×). Trimer peaks of the HA stem immunogens are represented by open circles (○) and the monomer peaks are represented by closed circles (●) (Inset). (D) Proteolysis of HA stem immunogens. Aliquots of the test proteins at the indicated time points (0, 15, 30, 45, and 60 min) were quenched with 0.1% formic acid. The samples were analyzed by SDS/PAGE. Lanes: 1–5, RCAM-RNaseA; 6–10, H1HA10; 11–15, H1HA10-IZ; 16–20, H1HA10-Foldon, and 21, prestained broad range marker (Biorad). RCAM-RNaseA was completely digested within 15 min. Proteolytic resistance increased as follows: H1HA10 < H1HA10-IZ < H1HA10-Foldon, confirming that H1HA10-Foldon adopts a compact, folded conformation.
Furthermore, the 1D 1H-NMR spectrum of H1HA10-foldon exhibits solution properties characteristic of a well-folded protein molecule. The presence of resolved resonance lines that appear in the downfield (9–11 ppm) and the upfield (0.5 to –1 ppm) regions of the spectrum are clear indicators that the molecule adopts a stable tertiary structure (Fig. 2B). The upfield-shifted signals are those of methyl protons that are spatially proximal to aromatic rings in the interior of the protein (hydrophobic core). The conformational stability of the folded state of H1HA10-Foldon was further probed by hydrogen exchange studies. The slow exchange of the amide protons in the downfield (9–11 ppm) region of 1D 1H-NMR spectrum is suggestive of a well-packed molecule (Fig. S6).
H1HA10-Foldon Is a Homogenous Trimer in Solution and Resistant to Proteolysis.
The long central α-helices (LAHs) located in the HA stem assemble together into a parallel, trimeric coiled-coil promoting oligomerization. The oligomeric state of the designed immunogens was probed by analytical gel-filtration chromatography under native conditions. H1HA10 eluted predominantly as a monomer and a minor trimeric peak. This was probably because in the absence of the transmembrane (TM) domain of HA, the trimer is not stable (45). It has previously been demonstrated that trimerization motifs facilitate oligomerization in the absence of the HA TM domain (46). Both IZ and Foldon trimerization sequences aided in trimerization of H1HA10. H1HA10-IZ formed a mixture of stable trimeric (∼65%) and monomeric (∼35%) conformers in solution which did not reequilibrate when partitioned. H1HA10-Foldon eluted exclusively as a trimer in solution (Fig. 2C). In contrast to previously designed immunogens H1HA6 and H1HA0HA6, which contain the entire stem domain (24), none of the proteins characterized in this study were aggregation prone. Gel-filtration studies indicate that we have effectively resurfaced exposed hydrophobic patches in our designed constructs.
Misfolded proteins having disordered sectors are subjected to increased proteolysis. H1HA10 showed limited resistance to proteolysis (trypsin digestion) compared with a control unfolded protein [reduced and carboxamidated ribonuclease A (RCAM-RNaseA)]. Proteolytic stability increased in the order H1HA10 < H1HA10-IZ < H1HA10-Foldon. Proteolysis confirmed a compact, folded conformation for H1HA10-Foldon (Fig. 2D).
Headless Stem Immunogens Bind Conformation-Specific bnAbs.
Stem-directed bnAbs like CR6261, F10, and FI6v3 bind the native, neutral-pH conformation of HA with high affinity (30–32). Epitopes of these bnAbs are disrupted in the low-pH, fusion-competent conformation of HA. Therefore, the ability of stem-derived immunogens to bind these bnAbs offers a robust validation of their conformation.
Binding of the designed headless stem immunogens to the bnAbs was determined by surface plasmon resonance (SPR). H1HA10 bound IgG CR6261 with submicromolar affinity (315.4 ± 14.5 nM) (Table 1; Fig. S7). As previously mentioned, H1HA10 has ∼80% of the antibody-footprint. H1HA10 although folded, is monomeric and not particularly compact as implied from proteolysis. These factors may contribute to the high koff, which would decrease the binding affinity (Table 1). Derivatives of H1HA10, which had improved biophysical and biochemical properties had considerably tighter binding to CR6261. H1HA10-IZ had a KD of 73.9 ± 2.3 nM. The conformational heterogeneity of H1HA10-IZ in solution may possibly lead to the observed biphasic binding to CR6261 at higher concentrations (trace 1 in Fig. S7B) as observed previously with the HA stem immunogen H1HA6 (24). H1HA10-Foldon, which assembles into a homogenous trimer in solution, bound CR6261 with the highest affinity among the designed stem immunogens (52.4 ± 1.8nM). Although H1HA10-Foldon binds CR6261 with about a sixfold weaker affinity than full-length A/Puerto Rico/8/34 recombinant HA (rHA) (8.9 ± 0.3 nM) (Table 1), it is a significant improvement over the previously reported stem immunogen H1HA6 (∼260 nM) (24), which comprised the entire HA stem with the complete CR6261 epitope.
Table 1.
HA stem immunogens bind bnAbs with high affinity
| Immunogen | Ligand* | kon, M−1⋅s−1 | koff, s−1 | KD, nM |
| H1HA10 | CR6261-IgG | 2.49 ± 0.08 × 104 | 7.85 ± 0.09 × 10−3 | 315.4 ± 14.5 |
| H1HA10-IZ | CR6261-IgG | 6.55 ± 0.51 × 104 | 4.84 ± 0.11 × 10−3 | 73.9 ± 2.3 |
| CR6261-IgG | 3.72 ± 0.22 × 104 | 1.95 ± 0.19 × 10−3 | 52.4 ± 1.8† | |
| H1HA10-Foldon | F10-scFv | 1.54 ± 0.14 × 105 | 1.50 ± 0.20 × 10−3 | 9.8 ± 2.1 |
| FI6v3-scFv | 1.69 ± 0.53 × 105 | 2.05 ± 0.12 × 10−3 | 12.1 ± 3.4 | |
| CR6261-IgG | 2.89 ± 0.02 × 105 | 2.58 ± 0.08 × 10−3 | 8.9 ± 0.3 | |
| H1 A/Puerto Rico/8/34 rHA | F10-scFv | 4.15 ± 0.61 × 105 | 1.21 ± 0.21 × 10−3 | 2.9 ± 0.9 |
| FI6v3-scFv | 2.45 ± 0.06 × 105 | 2.16 ± 0.27 × 10−3 | 8.8 ± 1.1 |
Kinetic parameters for binding of conformation-specific bnAbs to HA stem immunogens and full-length rHA by SPR.
Five hundred RU of CR6261-IgG were immobilized on the surface of a CM5-chip. F10-scFv and FI6v3-scFv were immobilized to a density of 750 RU.
Binding of H1HA10-Foldon to CR6261 was monitored after heat stress. The protein was incubated at 40 °C, 60 °C, and 80 °C for 1 h and the KD to CR6261 at 25 °C was determined. H1HA10-Foldon retained its ability to bind CR6261 even after heating at 80 °C. The KD after various heat stresses; 40 °C (55.7 ± 0.9 nM), 60 °C (61.4 ± 1.3 nM), and 80 °C (71.6 ± 0.5 nM).
Binding of H1HA10-Foldon to the single-chain variable fragment (scFv) derivatives of other stem-directed bnAbs (F10 and FI6v3) was also determined to confirm the native, prefusion HA-like conformation of H1HA10-Foldon. The slower off-rates for both H1HA10-Foldon and A/Puerto Rico/8/34 rHA result in a higher affinity to F10-scFV in comparison with IgG CR6261 (Table 1; Fig. S8 A and C). The KD of H1HA10-Foldon binding to F10-scFv was 9.8 ± 2.1nM, about threefold weaker than full-length A/Puerto Rico/8/34 rHA (2.9 ± 0.9 nM). H1HA10-Foldon also bound FI6v3 (the pan-influenza binding antibody) with a low KD of 12.1 ± 3.4 nM (Table 1; Fig. S8 B and D).
Thermal tolerance, a pharmaceutically relevant parameter, was assessed by determining the KD of H1HA10-Foldon binding to CR6261 after prolonged heat stress. H1HA10-Foldon bound CR6261 with a KD of 71.6 ± 0.5 nM even after incubating the protein at 80 °C for 1 h (see the second footnote to Table 1).
The specificity of H1HA10-Foldon binding to IgG CR6261 was also confirmed in a pull-down assay. Protein G beads (GE HealthCare) specific for human IgG CR6261 were used to pull down the antibody–antigen complex (Fig. S9).
HA Stem Immunogens Elicit Broadly Cross-Reactive, Neutralizing Antibodies.
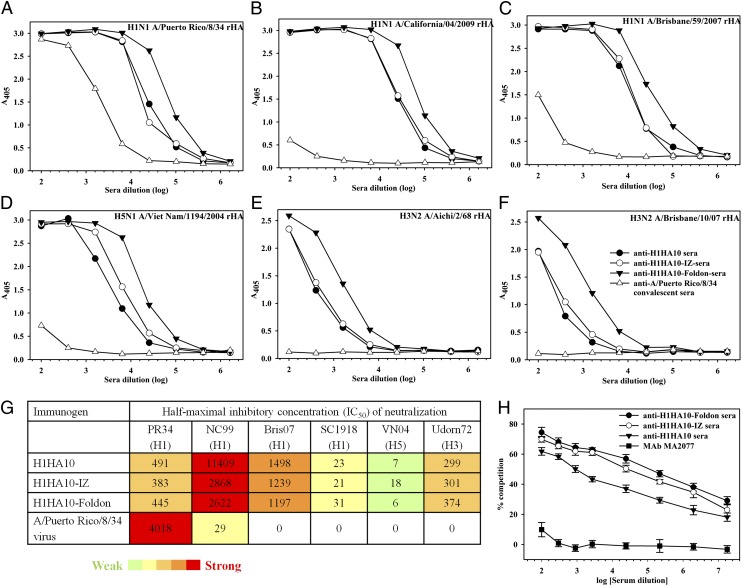
During natural infection, the immunodominant head domain steers the immune response away from the conserved stem (47). This is probably the reason why there are low/undetectable cross-reactive antibodies in the anti-A/Puerto Rico/8/34 convalescent sera. In contrast, high titers of cross-reactive antibodies are elicited by our stem domain immunogens (Fig. 3; Table S2) confirming that the immunogens have adopted a native-like, neutral-pH conformation. H1HA10-Foldon elicited the highest cross-reactive antibody titers (in the range of 409,600 to 1,638,400) with group 1 HAs (Fig. 3 A–D). H1HA10-Foldon bound the pan-influenza nAb FI6v3 with high affinity, which prompted us to examine the ability of anti–H1HA10-Foldon sera to bind group 2 HAs. Interestingly, H1HA10-Foldon elicited moderate titers (25,600) of cross-reactive antibodies against (group 2) H3 HAs (Fig. 3 E and F). However, additional design optimization incorporating the sequence variation information among the stem of all HA subtypes will be essential to formulate an immunogen that can elicit a truly universal response.
Fig. 3.
Headless HA stem immunogens elicit broadly cross-reactive, nAbs in mice. Breadth of the antibody response elicited by the designed immunogens was determined by ELISA of the pooled antisera (n = 10 mice per group) against purified rHA proteins (A) H1N1 A/Puerto Rico/8/34, (B) H1N1 A/California/04/2009, (C) H1N1 A/Brisbane/59/2007, (D) H5N1 A/Viet Nam/1194/2004, (E) H3N2 A/Aichi/2/68, and (F) H3N2 A/Brisbane/10/07. All of the stem immunogens elicited higher cross-reactive antibody titers compared with mice immunized with A/Puerto Rico/8/34 virus. H1HA10-Foldon elicited the highest cross-reactive antibody titers. (G) Neutralization of heterologous influenza virus strains by the H1N1 A/Puerto Rico/8/34 sequence-derived HA stem-immunized mice sera. IC50 titer is the reciprocal of sera dilution at which half-maximal neutralization was observed. The neutralization strength is color coded from green (weak) to red (strong). An IC50 value of zero indicates no detectable neutralization. The neutralization assay was performed against a panel of influenza HA strains: A/Puerto Rico/8/34 [PR34 (H1)], A/New Caledonia/20/1999 [NC99 (H1)], A/Brisbane/59/2007 [Bris07 (H1)], A/South Carolina/1/1918 [SC1918 (H1)], A/Viet Nam/1194/2004 [VN04 (H5)], and A/Udorn/307/1972 [Udorn72 (H3)]. (H) HA stem-immunized mice sera compete with the bnAb CR6261. Antisera from mice immunized with the designed headless stem immunogens were tested for competition binding to H1N1 A/California/04/2009 rHA in the presence of 300 ng/mL CR6261 (24). H1HA10-Foldon antisera showed maximum competition with CR6261. Although all of the stem-immunized mice sera competed with CR6261, the control head-specific mAb MA2077 (binds at the Sa antigenic site on HA) (50) did not compete with CR6261.
Neutralization titers for the various antisera were measured using a pseudotype neutralization assay (48). Current vaccination strategies with whole inactivated viruses typically fail to elicit/boost cross-reactive nAbs (49). Consistent with this, the anti-A/Puerto Rico/8/34 convalescent sera showed low or undetectable neutralization of drifted influenza strains. In contrast, sera from mice immunized with the HA stem immunogens showed robust neutralization of the highly divergent H1 A/New Caledonia/20/1999 and H1 A/Brisbane/59/2007 influenza viruses. The sera also showed measurable neutralization of the highly pathogenic H1 A/South Carolina/1/1918 and avian H5 A/Viet Nam/1194/2004 influenza viruses. Impressively, the sera also showed neutralization activity against a group 2 virus (H3 A/Udorn/307/1972) (Fig. 3G).
Antibodies elicited by the HA stem immunogens probably mediate virus neutralization by inhibiting virus–host cell membrane fusion as inferred from their ability to compete with the stem-directed bnAb IgG-CR6261 for binding to H1N1 A/California/04/2009 rHA (Fig. 3H). The sera elicited against H1HA10-Foldon showed maximum competition with CR6261, in accordance with the improved biophysical/biochemical properties of the immunogen. The competition assay demonstrates the presence of CR6261-like antibodies following immunization with headless stem immunogens. As a control, the nAb MA2077 which binds at the Sa antigenic site on H1N1 A/California/04/2009 HA (50) failed to compete with CR6261.
The sera were not tested in a HA inhibition assay because the stem-directed bnAbs mediate neutralization by inhibiting membrane-fusion and not by blocking the virus from binding to receptors on the host cells (51).
H1HA10-Foldon Completely Protects Against a Lethal Homologous Virus Challenge.
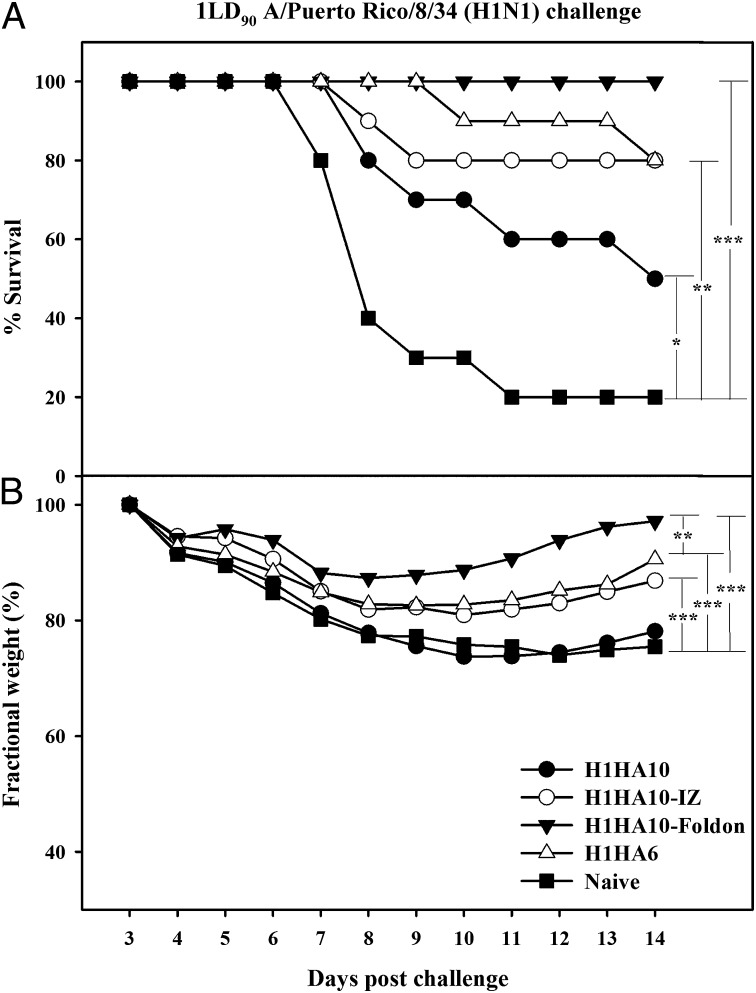
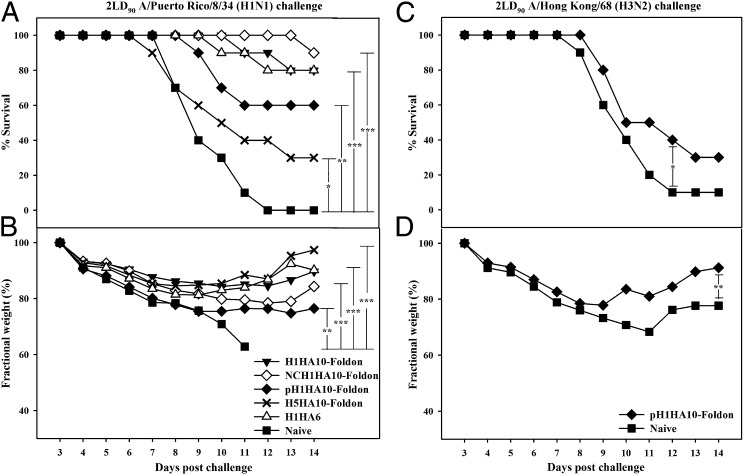
All of the headless constructs elicited a robust immune response in mice with high serum antibody self-titers (>1,638,400). Twenty-one days postsecondary immunization, mice were challenged intranasally with a lethal dose (1LD90) of homologous A/Puerto Rico/8/34 virus. The challenged mice showed significant weight recovery by the end of the observation period after initial weight loss (Fig. 4B). The monomeric immunogen H1HA10 conferred 50% protection, whereas its compact, trimeric derivative H1HA10-Foldon protected mice completely from a lethal homologous virus challenge (Fig. 4A). Notably, immunization with H1HA10-Foldon gave improved protection in comparison with a previously reported stem immunogen H1HA6 (24). The extent of weight loss was also slightly lower for mice immunized with H1HA10-Foldon relative to H1HA6. Immunization with H1HA10-Foldon also provided significant protection against a higher challenge dose (2LD90) of the virus (Fig. 5A).
Fig. 4.
Immunization with HA stem immunogens protects mice against lethal homologous challenge. Mice (n = 10 per group) were primed (day 0) and boosted (day 28) with 20 μg of the indicated immunogens and challenged intranasally 21 d after the boost with 1LD90 of mouse-adapted homologous A/Puerto Rico/8/34 virus. Survival (A) and average weight changes (only for surviving mice) (B) were monitored for 14 d postchallenge. Naïve and adjuvant-treated mice were used as controls and showed similar responses. H1HA10-Foldon conferred complete protection and minimum weight loss in vivo. H1HA6, a previously characterized stem immunogen (24) conferred 80% protection. Survival curves were evaluated by Kaplan–Meier survival analysis with the log-rank significance test (***P ≤ 0.0005, **P ≤ 0.05, and *P < 0.2). Each data point in B represents the percent change in mean body weight post-virus challenge. The fractional SE in measurement was ≤0.82%. The differences in fractional body weight (at day 14) between the immunized and naïve groups were analyzed by Student’s t test (***P ≤ 0.0005 and **P ≤ 0.05).
Fig. 5.
Headless HA stem immunogens confer robust subtype-specific and limited cross-group protection in vivo. Mice (n = 10 per group) immunized with the indicated immunogens were challenged (21 d postsecondary immunization) with an increased lethal challenge dose (2LD90) of mouse-adapted A/Puerto Rico/8/34 or A/Hong Kong/68 virus. Survival (A and C) and average weight changes (only for surviving mice) (B and D) were monitored for 14 d postchallenge. All of the HA stem immunogens delayed infection. H1HA10-Foldon conferred robust protection against an increased dose of homologous A/Puerto Rico/8/34 virus challenge. Significantly, NCH1HA10-Foldon protected mice against a heterologous A/Puerto Rico/8/34 virus challenge. H5HA10-Foldon, designed from an H5 subtype also provided partial protection against heterologous A/Puerto Rico/8/34 virus challenge and protected mice recovered completely. H1HA6 (designed from H1N1 A/Puerto Rico/8/34), a previously characterized stem immunogen (24) conferred 80% protection. pH1HA10-Foldon also conferred limited protection against a lethal H3N2 A/Hong Kong/68 virus challenge. The surviving mice showed significant weight recovery. Survival curves were evaluated by Kaplan–Meier survival analysis with the log-rank significance test (***P ≤ 0.0005, **P ≤ 0.05, and *P < 0.2). Each data point in B and D represents the percent change in mean body weight post-virus challenge. The fractional SE in measurement was ≤0.82%. The differences in fractional body weight (at day 14) between the immunized and naïve groups were analyzed by Student’s t test (***P ≤ 0.0005 and **P ≤ 0.05). In B the fractional body weight of the naïve group at day 11 was used for analysis as all of the mice in this group had succumbed to virus challenge by the end of the study period.
HA Stem Immunogens Confer Robust Subtype-Specific Protection.
Immunogens designed from unmatched, highly drifted influenza strains (Fig. S2) also elicited a robust immune response in mice with high serum antibody self-titers (≥1,638,400). The ability of stem immunogens to provide cross-protection was tested against a heightened challenge dose (2LD90) of heterologous A/Puerto Rico/8/34 virus in mice. All of the immunogens significantly delayed viral infection (Fig. 5 A and C). NCH1HA10-Foldon conferred robust protection (Fig. 5A), emphasizing the protective ability of stem immunogens across decades of genetic drift. Impressively, the stem fragment NCH1HA10-Foldon designed from a drifted strain (H1N1 A/New Caledonia/20/1999) had greater efficacy relative to the full-length stem domain H1HA6 (designed from H1N1 A/Puerto Rico/8/34) against A/Puerto Rico/8/34 virus challenge. H5HA10-Foldon, designed from an H5 subtype influenza strain (H5N1 A/Viet Nam/1203/2004) also provided partial protection and the surviving mice showed significant weight recovery (Fig. 5B). H1HA10-Foldon elicited significant titers of cross-group HA-specific antibodies which neutralized an H3 virus (Fig. 3 E–G). Hence, the protective ability of pH1HA10-Foldon against a group 2 H3N2 A/Hong Kong/68 virus challenge (2LD90) was also tested. The immunogen delayed infection and conferred weak protection (Fig. 5C). Surviving mice showed significant weight recovery (Fig. 5D). Our designed headless stem-fragment immunogens confer robust subtype-specific and weak cross-group protection in vivo.
Discussion
We have successfully engineered stem-domain fragments of HA to adopt the native, neutral-pH-like conformation in the absence of the globular head. Our design can readily be extended to other strains as discussed, emphasizing its wide applicability. The headless immunogens provide an efficient strategy to evade interference from the highly variable immunodominant head domain in the hope of broadening a vaccine response, which is validated by the broad spectrum neutralization profile of the antibodies elicited by our HA stem-fragment immunogens.
In a series of earlier studies, immunization with full-length cHA with a globular head from heterologous influenza strains (H9, H6, and H5, all belonging to group 1) and stem from H1N1 A/Puerto Rico/8/34 (group 1) was shown to protect against homologous and heterologous group 1 virus challenge. The animals were immunologically naïve to the head domain of the challenge virus (19). A prolonged immunization regimen was adopted in the cHA study with a primary DNA vaccination followed by two boosts with cHA recombinant protein administered both intranasally and intramuscularly. More recently, intramuscular immunizations (prime-boost) with different combinations of cHA proteins with diverse heads and a A/Puerto Rico/8/34 stem, were shown to protect mice against A/Puerto Rico/8/34 and pandemic H1N1 challenge. A precise comparison with the present work is difficult because of the differences in challenge viruses and doses used in the two studies (52). Although this is a promising approach and immunizations with cHAs elicit stem-directed Abs, the contribution of the head-directed immune response to protection is difficult to evaluate in these studies. Antibody responses to the head domains used in the cHAs may also interfere or depress titer of stem directed antibodies. We have circumvented this issue altogether by designing stable headless stem immunogens. Our study highlights the applicability of the reported HA stem-fragment immunogens to elicit stem-directed nAbs in a curtailed prime-boost immunization regimen.
In another study, repetitive arrays of full-length HA were successfully engineered on ferritin np to induce a strong immune response (20). Immunization with HA-np (H1N1 A/New Caledonia/20/1999) induced a robust antibody response in mice which was tested against a panel of H1 pseudotyped viruses in a neutralization assay. Although the antibodies mediated strong neutralization of related seasonal strains, they only weakly neutralized the unmatched, highly divergent H1N1 A/Puerto Rico/8/34 influenza strain [serum half-maximal inhibitory concentrations (IC50) titer = 210]. In contrast, antibodies elicited by our HA stem immunogens designed from H1N1 A/Puerto Rico/8/34 showed strong neutralization of the highly drifted H1 A/New Caledonia/20/1999 pseudotyped virus (serum IC50 titer ≥ 2622). In addition, the antibodies also mediated weak neutralization of the highly pathogenic avian H5 A/Viet Nam/1194/2004 influenza virus. Examining the immune response of our HA stem immunogens on an np platform is an interesting future prospect.
The present study demonstrates that the designed HA stem immunogens elicit nAbs which compete with the bnAb CR6261 for binding to HA. These nAbs probably inhibit the critical step of virus–host membrane fusion during influenza virus entry. Additionally, as previously shown in several studies (23, 53, 54), antibody-dependent effector functions such as antibody-dependent cell-mediated cytotoxicity may also contribute to the observed protection. The detailed mechanism(s) by which our headless stem immunogens provide protection need further investigation. HA stem-fragment immunogens induce heterosubtypic immunity. They also confer limited cross-group protection, something that has been difficult to achieve so far. These immunogens can also potentially be used as a vaccine strategy to specifically boost low levels of preexisting, stem-directed, cross-reactive Abs in the human population. Headless, folded, HA stem-fragment immunogens add to the repertoire of reagents to unambiguously detect influenza HA-stalk specific antibodies in immunological assays. Most existing influenza vaccine strategies involve expression in eukaryotic systems and require a cold chain. In contrast, the immunogens described in this work are bacterially expressed in soluble form and thermotolerant, allowing for rapid scale-up during a pandemic outbreak, including in low resource settings.
Materials and Methods
Sequence Analysis.
All nonidentical, full-length influenza sequences (4,241 sequences for H1N1 and 182 sequences for H5N1, derived from human hosts) were obtained from the NCBI Influenza Virus Database. H1N1 sequences were clustered at 99% homology using Cluster Database at High Identity with Tolerance to filter out 465 unique, representative H1N1 sequences, which were used for further analysis (55). These H1N1 sequences were multiply aligned using ClustalX (56). H1N1 (n = 465) and H5N1 (n = 182) sequences were also simultaneously aligned using ClustalX. The quality score for each column in the alignment file is a measure of residue conservation at that position. The quality scores were binned and mapped onto the crystal structure of H1N1 A/Puerto Rico/8/34 HA (PDB ID code 1RU7) (34).
Cloning, Expression, and Protein Purification.
The gene sequence of our designed immunogen H1HA10 was synthesized (GenScript) and cloned in the expression vector pET-28a (+) (Novagen) between NdeI and BamHI restriction sites. Gene sequences corresponding to the trimerization motifs IZ and Foldon were synthesized (Abexome) with flanking KpnI and HindIII restriction sites. H1HA10-IZ and H1HA10-Foldon derivatives of our preliminary construct were generated by cloning the trimerization motifs at the C terminus of H1HA10. The stop codon in H1HA10 was mutated to generate a unique KpnI restriction site using site-directed mutagenesis to facilitate the cloning of the trimerization motifs. Cloning was confirmed by sequencing (Macrogen). Gene sequences corresponding to NCH1HA10-Foldon, pH1HA10-Foldon and H5HA10-Foldon were synthesized (GenScript) and cloned in the expression vector pET-28a (+) between NdeI and HindIII restriction sites. All constructs were codon-optimized for expression in E. coli.
The proteins were overexpressed in E. coli BL21(DE3) cells and purified from the soluble fraction of the cell culture lysate. All proteins in this study were purified using a similar protocol. Briefly, a single colony of E. coli BL21(DE3) transformed with the plasmid of interest was inoculated into 50 mL Tartoff-Hobbs HiVeg media (HiMedia). The primary culture was grown overnight at 37 °C. Two liters of Tartoff-Hobbs HiVeg media (500 mL ×4) (HiMedia) was inoculated with 1% of the primary inoculum and grown at 37 °C until an OD600 of ∼0.6–0.8 was reached. Cells were then induced with 1 mM isopropyl-β-thiogalactopyranoside and grown for another 12–16 h at 20 °C. Cells were harvested at 5,000 × g and resuspended in 100 mL PBS (pH 7.4). The cell suspension was lysed by sonication on ice and subsequently centrifuged at 14,000 × g. The supernatant was incubated with buffer-equilibrated Ni-NTA resin (GE HealthCare) for 2 h at 4 °C under mild mixing conditions to facilitate binding. The protein was eluted using an imidazole gradient (in PBS, pH 7.4) under gravity flow. Fractions containing the protein of interest were pooled and dialyzed against PBS (pH 7.4) containing 1 mM EDTA. The dialyzed protein was concentrated in an Amicon (Millipore) stirred cell apparatus to a final concentration of ∼1 mg/mL. Protein purity was assessed by SDS/PAGE and its identity confirmed by electrospray ionization MS.
CD.
CD spectra for all proteins were recorded on a Jasco J-715C spectropolarimeter flushed with nitrogen gas. The concentration for all proteins was ∼5–10 µM. Measurements were done at 25 °C in a 1-mm-path-length quartz cuvette with a scan rate of 50 nm/min, response time of 4 s, and a bandwidth of 2 nm. Each spectrum was an average of five scans. Mean residue ellipticity was calculated as described previously (57). The protein spectrum was corrected for buffer (PBS, pH 7.4) signals.
For thermal unfolding experiments, the CD signal was monitored as a function of temperature. Thermal denaturation of H1HA10-Foldon was monitored at 208 nm, and for H1N1 A/Puerto Rico/8/34 rHA at 215 nm. The buffer used was PBS (pH 7.4). The data were collected between 15 °C and 90 °C with a 1-°C/min gradient and a data pitch of 0.2 °C. The bandwidth was 2 nm and the response time 4 s. A quartz cuvette of 1-mm-path length was used. The scan was repeated after cooling the samples back to 15 °C to measure the reversibility of thermal unfolding.
Fluorescence Spectroscopy.
All fluorescence spectra were recorded at 25 °C on a Jasco FP-6300 spectrofluorimeter. For intrinsic fluorescence measurements, the protein concentration used was in the range of 1 to 3 µM. The protein samples were excited at 280 nm, and emission was monitored from 300 to 400 nm. The excitation and emission slit widths were 3 and 5 nm, respectively. Each spectrum was an average of five scans. The data were analyzed after buffer correction. Fluorescence measurements were carried out under native conditions in PBS (pH 7.4) or under denaturing conditions of 7 M GdnCl in PBS (pH 7.4).
NMR Spectroscopy.
One-dimensional 1H NMR spectra were recorded on an Agilent 600 MHz NMR spectrometer using a triple resonance probe fitted with a z-axis-only pulsed field gradient accessory. Spectra were recorded at 25 °C. Chemical shifts were referenced to external 2,2-dimethyl-2-silapentane-5-sulfonic acid. A spectral width of 9,615.4 Hz was sampled. Solvent suppression was achieved using the excitation sculpting pulse scheme (58). H1HA10-Foldon protein samples for NMR were prepared in PBS (pH 7.4) [90% H2O:10% D2O (vol/vol)]. A total of 2,048 scans were recorded with a 1-s relaxation delay. Protein sample (H1HA10-Foldon) was prepared in PBS (pH 7.4) [80% D2O:20% H2O (vol/vol)] for the hydrogen exchange studies. One-dimensional 1H NMR spectra of H1HA10-Foldon were recorded as a function of time over 24 h at every 2-h interval.
Gel-Filtration Chromatography.
The oligomeric status of purified proteins was analyzed under nondenaturing conditions by gel-filtration chromatography at room temperature on a Superdex-200 analytical gel-filtration column (GE HealthCare). The column was equilibrated with PBS (pH 7.4) and calibrated using broad range molecular weight markers (GE HealthCare).
Proteolysis.
Proteolytic digestion of the designed immunogens was carried out using a protease (trypsin)/substrate molar ratio of 1:50. RCAM-RNase A was used as a positive control for trypsin activity. Proteolysis was carried out in the presence of 50 mM Hepes (pH 8.0) and 2 mM CaCl2 on ice. Aliquots were collected at various time points and quenched for trypsin activity with 0.1% formic acid. Samples collected at different time points were analyzed on 12% SDS/PAGE followed by staining with Coomassie Brilliant Blue R250 (Sigma).
Binding Affinity Studies Using SPR.
Binding affinity of the designed immunogens (H1HA10, H1HA10-IZ, and H1HA10-Foldon) and full-length H1N1 A/Puerto Rico/8/34 rHA (Sino Biological Inc.) to the stem-directed bnAb (CR6261 IgG) or their scFv derivatives (F10-scFv and FI6v3-scFv) was determined by SPR experiments performed with a Biacore 3000 optical biosensor (Biacore) at 25 °C. Recombinant CR6261 IgG was produced in 293T cells as described previously (24). Plasmids encoding F10-scFv and FI6v3-scFv were synthesized (GenScript) based on the published sequence (31, 32) and expressed in E. coli. Five hundred to seven hundred response units (RU) of the ligand (CR6261 IgG, F10-scFv, or FI6v3-scFv) were immobilized by standard amine coupling to the surface of a research-grade CM5 chip (GE HealthCare). An ovalbumin-immobilized sensor channel served as a negative control for each binding interaction. Multiple concentrations of the analyte were passed over each channel in a running buffer of PBS (pH 7.4) with 0.05% P20 surfactant. Both binding and dissociation events were measured at a flow rate of 30 μL/min. The sensor surface was regenerated after every binding event by repeated washes with 4 M MgCl2. Each binding curve was analyzed after correcting for nonspecific binding by subtraction of signal obtained from the negative control flow channel. The concentration of the monomeric fraction of H1HA10 was used for obtaining the kinetic paramenters, whereas for H1HA10-IZ, H1HA10-Foldon, and H1N1 A/Puerto Rico/8/34 rHA, concentrations of the trimeric fraction were used. The kinetic parameters were obtained by globally fitting the data to a simple 1:1 Langmuir interaction model using BIA EVALUATION 3.1 software as described previously (38). Trace 1 (Fig. S7B) was omitted from global fitting while obtaining the kinetic parameters for H1HA10-IZ because it could not be fitted to a 1:1 interaction model.
Thermal tolerance of H1HA10-Foldon was assessed by its ability to bind CR6261 after heat stress. The protein sample was incubated at 40 °C, 60 °C, and 80 °C for 1 h in a PCR cycler (BioRad) with a heated lid to prevent evaporation. The samples were cooled to 25 °C and binding affinity to CR6261 was determined by SPR experiments as described above.
Pull-Down Assay.
The ability of H1HA10-Foldon to form a stable complex with the bnAb CR6261 IgG was further confirmed in a pull-down assay. CR6261 and H1HA10-Foldon were mixed together at different molar ratios and incubated for 2 h at 4 °C. Buffer (PBS, pH 7.4)-equilibrated Protein G (GE HealthCare) beads were added to the complex and incubated for 1 h to specifically pull down CR6261 IgG. The beads were spun down at 3,000 × g for 15 min at 4 °C. The unbound supernatant was separated and the beads were washed with PBS (pH 7.4). The antibody bound to the beads was eluted with 100 mM glycine⋅HCl (pH 3.0). The eluted fractions were neutralized with 1 M Tris⋅HCl (pH 9.0). The unbound and eluted fractions were subsequently analyzed by SDS/PAGE.
Immunization and Challenge Studies.
Female BALB/c mice (4–5 wk old) (Taconic Farms, Inc.) were maintained at the animal facilities of Merck Research Laboratories. The study design was approved by the Merck Research Laboratories Institutional Animal Care and Use Committee. Mice (10 mice per group) were immunized intramuscularly with 20 µg test immunogen along with 100 µg CpG7909 adjuvant (TriLink BioTechnologies) at days 0 (prime) and 28 (boost). Naïve (buffer-only) mice and adjuvant-treated mice were used as controls. Serum samples obtained from tail vein venipuncture were collected in Microtainer serum separator tubes (BD Biosciences) 21 d after the prime- and 14 d postboost from all of the mice. Twenty-one days after the secondary immunization, mice were anesthetized with ketamine/xylazine and challenged intranasally with 1LD90 of mouse-adapted H1N1 A/Puerto Rico/8/34 virus in 20 µL PBS. To test for protection against a higher dose of the virus, one group of mice primed and boosted with H1HA10-Foldon was challenged with 2LD90 of homologous A/Puerto Rico/8/34 virus. The ability of NCH1HA10-Foldon, pH1HA10-Foldon and H5HA10-Foldon to confer cross-protection was evaluated against a stringent 2LD90 heterologous H1N1 A/Puerto Rico/8/34 virus challenge. Another group of mice (n = 10) immunized with pH1HA10-Foldon were challenged with 2LD90 heterologous, mice-adapted H3N2 A/Hong Kong/68 virus. Survival and weight change of the challenged mice were monitored daily for 14 d postchallenge. At each time point, all surviving mice of a group were weighed together and the mean weight calculated. Errors in the mean weight were estimated from three repeated measurements of the mean weight of the same number of healthy mice.
Determination of Serum Antibody Titers.
Antibody titers against test immunogens were determined by ELISA. Briefly, test immunogens were coated on 96-well Nunc plates (Thermo Fisher Scientific) at 4 μg/mL in 50 μL PBS at 4 °C overnight. Plates were then washed with PBS containing 0.05%Tween-20 (PBST) and blocked with 3% skim milk in PBST for 1 h. One hundred microliters of the antisera raised against the test immunogens was diluted in a fourfold series in milk–PBST and added to each well. Plates were incubated for 2 h at room temperature followed by washes with PBST. Fifty microliters of HRP-conjugated goat anti-mouse IgG (H+L) secondary antibody (Invitrogen) in milk–PBST was added to each well at a predetermined dilution (1:5,000) and incubated at room temperature for 1 h. Plates were washed with PBST followed by development with 100 µL per well of the substrate 3,3′,5,5′-tetramethylbenzidine (TMB) solution (Virolabs) and stopped after 3–5 min of development with 100 μL per well of the stop solution for TMB (Virolabs). OD at 450 nm was measured and the antibody titer was defined as the reciprocal of the highest dilution that gave an OD value above the mean plus 2 SDs of control wells.
Binding of Antisera to Recombinant HAs.
Binding of antisera raised against the test immunogens to several rHA proteins was determined by ELISA. Briefly, mammalian-expressed rHA proteins (H1N1 A/Puerto Rico/8/34, H1N1 A/California/04/2009, H1N1 A/Brisbane/59/2007, H5N1 A/Viet Nam/1203/2004, H3N2 A/Aichi/2/68, and H3N2 A/Brisbane/10/07; Sino Biological Inc.) were coated on 96-well Nunc plates (Thermo Fisher Scientific) at 2.5 μg/mL in 50 μL PBS at 4 °C overnight. Ovalbumin (125 ng per well)-coated wells were used as a negative control. Plates were washed with PBST and blocked with 1% BSA in PBST (PBSB). Antisera were then added to each well at a starting dilution of 1:100 followed by a fourfold dilution series and incubated for 2 h. Plates were washed with PBST. Alkaline phosphatase (ALP)-conjugated goat anti-mouse secondary antibody (Sigma) in PBSB was added to each well at a predetermined dilution (1:10,000) and incubated at room temperature for 2 h. Plates were washed and developed using the chromogenic substrate p-nitrophenyl phosphate (Sigma). Plates were read at 405 nm (SPECTRAmax Plus 384; Molecular Devices). Antibody titer was defined as the reciprocal of the highest dilution that gave an OD value above the mean plus 2 SDs of control wells.
Neutralization Assay.
HA-pseudotyped lentiviral particles were produced as described previously (48, 59). Briefly, HIV gag-pol plasmid p8.91, firefly luciferase expressing plasmid pCSFLW, HA (H1N1 A/Puerto Rico/8/1934, H1N1 A/New Caledonia/20/1999, H1N1 A/Brisbane/59/2007, H1N1 A/South Carolina/1/1918, H5N1 A/Viet Nam/1194/2004, and H3N2 A/Udorn/307/1972) expressing plasmid, and pCAGGS-HAT or pCAGGS-TMPRSS2 (60) (for H1 and H3 pseudotypes) expressing plasmid were cotransfected into human embryonic kidney (HEK 293T/17) cells using Fugene-6 transfection reagent (Promega). Twenty-four hours postincubation, recombinant neuraminidase (Sigma) was added to facilitate the release of the pseudotype virus. Forty-eight hours posttransfection, supernatant containing the released pseudotype virus was harvested, filtered through 0.45-µm filter, and stored at −80 °C.
Pseudotype virus [1 × 106 relative luminescence units per well] was incubated with serial dilutions of immunized mice sera in 96-well flat-bottomed white plates (Nunc) in a final volume of 100 µL and incubated at 37 °C. After 1 h, 1 × 104 HEK293T/17 cells were added to each well and the plates were incubated for another 48 h at 37 °C. Luminescence was evaluated using the Bright-Glo assay system (Promega). IC50 of neutralization were calculated with Prism Version 6.0e (GraphPad Software).
Competition ELISA.
Competition ELISA between the antisera raised against the test immunogens and the bnAb CR6261 IgG was carried out as described elsewhere (50). Briefly, 96-well half area plates (Corning Incorporated) were coated with 3 µg pandemic H1N1 A/California/04/2009 rHA (Sino Biological Inc.) in 50 µL PBS and kept overnight at 4 °C. Ovalbumin (3 µg)-coated wells were used as negative control for antisera binding. Plates were washed with PBST and blocked for 1 h with PBSB. Twenty-five microliters of antisera were added to each well starting at a 1:100 dilution followed by a threefold serial dilution in PBSB. As a control for nonspecific competition, a previously characterized head-specific neutralizing mAb IgG MA2077 was used (50). Twenty-five microliters of mAb MA2077 was added to each well starting at a concentration of 2 mg/mL followed by threefold serial dilutions. After 2 h of incubation, the plates were washed and blocked with PBSB for 15 min. CR6261 was then added to each well at a fixed concentration (300 ng/mL) as determined from the titration curve of CR6261 with pandemic H1N1 rHA. After 2 h of incubation with CR6261, the plates were washed with PBST. The wells were then probed with 25 μL of ALP-conjugated goat anti-human antibody (Sigma) at a predetermined dilution (1:10,000) to detect the bound CR6261. The plates were washed and developed using the chromogenic substrate p-nitrophenyl phosphate (Sigma). The optical density was measured at 405 nm (SPECTRAmax Plus 384L; Molecular Devices). The percent competition was calculated as follows: % competition = [(A − P)/A] × 100, where A is the signal of CR6261 binding to rHA in the absence of antiserum and P is the binding signal of CR6261 to rHA in the presence of antiserum (24).
Statistical Analysis.
Differences in antibody/neutralization titers and mean fractional body weights of surviving mice between different groups were analyzed by analysis of variance and Student’s t test. The fractional body weight of mice is calculated relative to their starting body weight. Differences in survival were calculated by Kaplan–Meier survival analysis with the log-rank significance test.
Supplementary Material
Acknowledgments
V.V.A.M. is a recipient of a fellowship from the Council of Scientific and Industrial Research, Government of India. This work was supported in part by a grant from the Department of Biotechnology, Government of India (to R.V.).
Footnotes
Conflict of interest statement: V.V.A.M. and R.V. are authors in a patent application on the designed immunogen sequences described in this work.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1402766111/-/DCSupplemental.
References
- 1.World Health Organization Influenza fact sheet. 2014. Available at www.who.int/mediacentre/factsheets/fs211/en. Accessed January 1, 2014.
- 2.Pica N, Palese P. Toward a universal influenza virus vaccine: Prospects and challenges. Annu Rev Med. 2013;64:189–202. doi: 10.1146/annurev-med-120611-145115. [DOI] [PubMed] [Google Scholar]
- 3.Gerhard W. The role of the antibody response in influenza virus infection. Curr Top Microbiol Immunol. 2001;260:171–190. doi: 10.1007/978-3-662-05783-4_9. [DOI] [PubMed] [Google Scholar]
- 4.Carr CM, Kim PS. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993;73(4):823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- 5.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: The influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 6.Knossow M, Skehel JJ. Variation and infectivity neutralization in influenza. Immunology. 2006;119(1):1–7. doi: 10.1111/j.1365-2567.2006.02421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ekiert DC, et al. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature. 2012;489(7417):526–532. doi: 10.1038/nature11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hong M, et al. Antibody recognition of the pandemic H1N1 Influenza virus hemagglutinin receptor binding site. J Virol. 2013;87(22):12471–12480. doi: 10.1128/JVI.01388-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krause JC, et al. A broadly neutralizing human monoclonal antibody that recognizes a conserved, novel epitope on the globular head of the influenza H1N1 virus hemagglutinin. J Virol. 2011;85(20):10905–10908. doi: 10.1128/JVI.00700-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee PS, et al. Heterosubtypic antibody recognition of the influenza virus hemagglutinin receptor binding site enhanced by avidity. Proc Natl Acad Sci USA. 2012;109(42):17040–17045. doi: 10.1073/pnas.1212371109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohshima N, et al. Naturally occurring antibodies in humans can neutralize a variety of influenza virus strains, including H3, H1, H2, and H5. J Virol. 2011;85(21):11048–11057. doi: 10.1128/JVI.05397-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt AG, et al. Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody. Proc Natl Acad Sci USA. 2013;110(1):264–269. doi: 10.1073/pnas.1218256109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsibane T, et al. Influenza human monoclonal antibody 1F1 interacts with three major antigenic sites and residues mediating human receptor specificity in H1N1 viruses. PLoS Pathog. 2012;8(12):e1003067. doi: 10.1371/journal.ppat.1003067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whittle JR, et al. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci USA. 2011;108(34):14216–14221. doi: 10.1073/pnas.1111497108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wrammert J, et al. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med. 2011;208(1):181–193. doi: 10.1084/jem.20101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu R, et al. A recurring motif for antibody recognition of the receptor-binding site of influenza hemagglutinin. Nat Struct Mol Biol. 2013;20(3):363–370. doi: 10.1038/nsmb.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Julien JP, Lee PS, Wilson IA. Structural insights into key sites of vulnerability on HIV-1 Env and influenza HA. Immunol Rev. 2012;250(1):180–198. doi: 10.1111/imr.12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellebedy AH, Ahmed R. Re-engaging cross-reactive memory B cells: The influenza puzzle. Front Immunol. 2012;3:53. doi: 10.3389/fimmu.2012.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krammer F, Pica N, Hai R, Margine I, Palese P. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J Virol. 2013;87(12):6542–6550. doi: 10.1128/JVI.00641-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanekiyo M, et al. Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature. 2013;499(7456):102–106. doi: 10.1038/nature12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eggink D, Goff PH, Palese P. Guiding the immune response against influenza virus hemagglutinin toward the conserved stalk domain by hyperglycosylation of the globular head domain. J Virol. 2014;88(1):699–704. doi: 10.1128/JVI.02608-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen J, et al. A soluble domain of the membrane-anchoring chain of influenza virus hemagglutinin (HA2) folds in Escherichia coli into the low-pH-induced conformation. Proc Natl Acad Sci USA. 1995;92(26):12205–12209. doi: 10.1073/pnas.92.26.12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bommakanti G, et al. Design of an HA2-based Escherichia coli expressed influenza immunogen that protects mice from pathogenic challenge. Proc Natl Acad Sci USA. 2010;107(31):13701–13706. doi: 10.1073/pnas.1007465107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bommakanti G, et al. Design of Escherichia coli-expressed stalk domain immunogens of H1N1 hemagglutinin that protect mice from lethal challenge. J Virol. 2012;86(24):13434–13444. doi: 10.1128/JVI.01429-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu Y, Welsh JP, Swartz JR. Production and stabilization of the trimeric influenza hemagglutinin stem domain for potentially broadly protective influenza vaccines. Proc Natl Acad Sci USA. 2014;111(1):125–130. doi: 10.1073/pnas.1308701110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steel J, et al. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. MBio. 2010;1(1):e00018–10. doi: 10.1128/mBio.00018-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mallajosyula VV, et al. In vitro and in vivo characterization of designed immunogens derived from the CD-helix of the stem of influenza hemagglutinin. Proteins. 2013;81(10):1759–1775. doi: 10.1002/prot.24317. [DOI] [PubMed] [Google Scholar]
- 28.Wang TT, et al. Vaccination with a synthetic peptide from the influenza virus hemagglutinin provides protection against distinct viral subtypes. Proc Natl Acad Sci USA. 2010;107(44):18979–18984. doi: 10.1073/pnas.1013387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schneemann A, et al. A virus-like particle that elicits cross-reactive antibodies to the conserved stem of influenza virus hemagglutinin. J Virol. 2012;86(21):11686–11697. doi: 10.1128/JVI.01694-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ekiert DC, et al. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324(5924):246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sui J, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol. 2009;16(3):265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corti D, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333(6044):850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 33.Tong S, et al. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013;9(10):e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gamblin SJ, et al. The structure and receptor binding properties of the 1918 influenza hemagglutinin. Science. 2004;303(5665):1838–1842. doi: 10.1126/science.1093155. [DOI] [PubMed] [Google Scholar]
- 35.Harris AK, et al. Structure and accessibility of HA trimers on intact 2009 H1N1 pandemic influenza virus to stem region-specific neutralizing antibodies. Proc Natl Acad Sci USA. 2013;110(12):4592–4597. doi: 10.1073/pnas.1214913110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharma D, et al. Protein minimization of the gp120 binding region of human CD4. Biochemistry. 2005;44(49):16192–16202. doi: 10.1021/bi051120s. [DOI] [PubMed] [Google Scholar]
- 37.Kuhlman B, et al. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302(5649):1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- 38.Bhattacharyya S, et al. Design of an Escherichia coli expressed HIV-1 gp120 fragment immunogen that binds to b12 and induces broad and potent neutralizing antibodies. J Biol Chem. 2013;288(14):9815–9825. doi: 10.1074/jbc.M112.425959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saha P, et al. Design and characterization of stabilized derivatives of human CD4D12 and CD4D1. Biochemistry. 2011;50(37):7891–7900. doi: 10.1021/bi200870r. [DOI] [PubMed] [Google Scholar]
- 40.Varadarajan R, et al. Characterization of gp120 and its single-chain derivatives, gp120-CD4D12 and gp120-M9: Implications for targeting the CD4i epitope in human immunodeficiency virus vaccine design. J Virol. 2005;79(3):1713–1723. doi: 10.1128/JVI.79.3.1713-1723.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suzuki K, Hiroaki H, Kohda D, Tanaka T. An isoleucine zipper peptide forms a native-like triple stranded coiled coil in solution. Protein Eng. 1998;11(11):1051–1055. doi: 10.1093/protein/11.11.1051. [DOI] [PubMed] [Google Scholar]
- 42.Güthe S, et al. Very fast folding and association of a trimerization domain from bacteriophage T4 fibritin. J Mol Biol. 2004;337(4):905–915. doi: 10.1016/j.jmb.2004.02.020. [DOI] [PubMed] [Google Scholar]
- 43.Schein CH. Optimizing protein folding to the native state in bacteria. Curr Opin Biotechnol. 1991;2(5):746–750. doi: 10.1016/0958-1669(91)90046-8. [DOI] [PubMed] [Google Scholar]
- 44.Song L, et al. Efficacious recombinant influenza vaccines produced by high yield bacterial expression: A solution to global pandemic and seasonal needs. PLoS ONE. 2008;3(5):e2257. doi: 10.1371/journal.pone.0002257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Copeland CS, Doms RW, Bolzau EM, Webster RG, Helenius A. Assembly of influenza hemagglutinin trimers and its role in intracellular transport. J Cell Biol. 1986;103(4):1179–1191. doi: 10.1083/jcb.103.4.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stevens J, et al. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science. 2006;312(5772):404–410. doi: 10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]
- 47.Kwong PD, Wilson IA. HIV-1 and influenza antibodies: Seeing antigens in new ways. Nat Immunol. 2009;10(6):573–578. doi: 10.1038/ni.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Temperton NJ, et al. A sensitive retroviral pseudotype assay for influenza H5N1-neutralizing antibodies. Influenza Other Respi Viruses. 2007;1(3):105–112. doi: 10.1111/j.1750-2659.2007.00016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wrammert J, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453(7195):667–671. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shembekar N, et al. Isolation of a high affinity neutralizing monoclonal antibody against 2009 pandemic H1N1 virus that binds at the ‘Sa’ antigenic site. PLoS ONE. 2013;8(1):e55516. doi: 10.1371/journal.pone.0055516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okuno Y, Matsumoto K, Isegawa Y, Ueda S. Protection against the mouse-adapted A/FM/1/47 strain of influenza A virus in mice by a monoclonal antibody with cross-neutralizing activity among H1 and H2 strains. J Virol. 1994;68(1):517–520. doi: 10.1128/jvi.68.1.517-520.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goff PH, et al. Adjuvants and immunization strategies to induce influenza virus hemagglutinin stalk antibodies. PLoS ONE. 2013;8(11):e79194. doi: 10.1371/journal.pone.0079194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goins CL, Chappell CP, Shashidharamurthy R, Selvaraj P, Jacob J. Immune complex-mediated enhancement of secondary antibody responses. J Immunol. 2010;184(11):6293–6298. doi: 10.4049/jimmunol.0902530. [DOI] [PubMed] [Google Scholar]
- 54.Jegaskanda S, Weinfurter JT, Friedrich TC, Kent SJ. Antibody-dependent cellular cytotoxicity is associated with control of pandemic H1N1 influenza virus infection of macaques. J Virol. 2013;87(10):5512–5522. doi: 10.1128/JVI.03030-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang Y, Niu B, Gao Y, Fu L, Li W. CD-HIT Suite: A web server for clustering and comparing biological sequences. Bioinformatics. 2010;26(5):680–682. doi: 10.1093/bioinformatics/btq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Higgins DG, Sharp PM. CLUSTAL: A package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73(1):237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 57.Ganesh C, Shah AN, Swaminathan CP, Surolia A, Varadarajan R. Thermodynamic characterization of the reversible, two-state unfolding of maltose binding protein, a large two-domain protein. Biochemistry. 1997;36(16):5020–5028. doi: 10.1021/bi961967b. [DOI] [PubMed] [Google Scholar]
- 58.Hwang TL, Shaka AJ. Water suppression that works. Excitation sculpting using arbitary waveforms and pulsed field gradients. J Magn Reson A. 1995;112:275–279. [Google Scholar]
- 59.Ferrara F, et al. The human Transmembrane Protease Serine 2 is necessary for the production of Group 2 influenza A virus pseudotypes. J Mol Genet Med. 2012;7:309–314. [PMC free article] [PubMed] [Google Scholar]
- 60.Böttcher E, et al. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J Virol. 2006;80(19):9896–9898. doi: 10.1128/JVI.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.