Significance
Increased kinase activity and suppressed phosphatase activity are hallmarks of oncogenic signaling. The transcription factor c-MYC, a master driver of human cancer, is stabilized and activated by persistent serine 62 phosphorylation. The tumor suppressor protein phosphatase 2A (PP2A) targets this site and negatively regulates c-MYC. Here, we show that two cellular inhibitors of PP2A, the SET oncoprotein and cancerous inhibitor of PP2A (CIP2A), are overexpressed in breast cancer, and depletion or inhibition of SET or CIP2A reduces c-MYC expression and activity and decreases the tumorigenic potential of breast cancer cells. These findings strongly suggest that inhibiting SET or CIP2A to reactivate PP2A may be an effective therapeutic strategy for targeting c-MYC in breast cancer.
Keywords: breast cancer therapy, phosphatase activator
Abstract
The transcription factor c-MYC is stabilized and activated by phosphorylation at serine 62 (S62) in breast cancer. Protein phosphatase 2A (PP2A) is a critical negative regulator of c-MYC through its ability to dephosphorylate S62. By inactivating c-MYC and other key signaling pathways, PP2A plays an important tumor suppressor function. Two endogenous inhibitors of PP2A, I2PP2A, Inhibitor-2 of PP2A (SET oncoprotein) and cancerous inhibitor of PP2A (CIP2A), inactivate PP2A and are overexpressed in several tumor types. Here we show that SET is overexpressed in about 50–60% and CIP2A in about 90% of breast cancers. Knockdown of SET or CIP2A reduces the tumorigenic potential of breast cancer cell lines both in vitro and in vivo. Treatment of breast cancer cells in vitro or in vivo with OP449, a novel SET antagonist, also decreases the tumorigenic potential of breast cancer cells and induces apoptosis. We show that this is, at least in part, due to decreased S62 phosphorylation of c-MYC and reduced c-MYC activity and target gene expression. Because of the ubiquitous expression and tumor suppressor activity of PP2A in cells, as well as the critical role of c-MYC in human cancer, we propose that activation of PP2A (here accomplished through antagonizing endogenous inhibitors) could be a novel antitumor strategy to posttranslationally target c-MYC in breast cancer.
The c-MYC (MYC) oncoprotein is overexpressed in human breast cancer and this is associated with poor clinical outcome (1, 2). Expression of MYC is regulated at multiple levels, including protein stability, which is increased in several cancer types (1, 3, 4). MYC stability is regulated in part by sequential and interdependent phosphorylation at two conserved residues, threonine 58 (T58) and serine 62 (S62) (5). MYC is phosphorylated at S62 (pS62) through the mitogen-activated protein kinase (MAPK) pathway or cyclin-dependent kinase (CDK) activation in response to growth signals and this modification increases its stability and oncogenic activity (5–8). When growth signals cease, GSK3, in a manner dependent upon prior phosphorylation at S62, phosphorylates T58 (pT58) (5, 6). T58 phosphorylation facilitates protein phosphatase 2A (PP2A)-mediated dephosphorylation of pS62 and recruitment of the E3 ubiquitin ligase SCFFbw7 to initiate proteasomal destruction of MYC (9, 10). This process is facilitated by AXIN1, which helps nucleate a destruction complex for MYC at target gene promoters (11, 12). Our previous work has shown that MYC stability is increased in breast cancers and that this correlates with high pS62- and low pT58-MYC (4).
PP2A is a ubiquitously expressed, heterotrimeric serine–threonine (S/T) phosphatase that mediates 30–50% of cellular S/T phosphatase activity (13). Target specificity of PP2A is directed by a variable regulatory (B) subunit, and we have shown that B56α is the isoform that directs PP2A to MYC (9, 13). Human cell transformation requires inhibition of PP2A activity and, in an siRNA screen, B56α, B56γ, and PR72/PR130 were the only B subunits shown to be critical for regulating human cell transformation (14). PP2A complexes containing these B subunits regulate MYC, Wnt, and PI3K/Akt signaling, respectively (9, 14).
Whereas loss of PP2A activity is critical for tumor growth, mutations in PP2A subunits are very rare in breast cancers (15, 16). Alterations in the A subunit that impair integration of the C and/or B subunits have only been observed in breast cancers at a low frequency (15–18), suggesting that other mechanisms can affect PP2A activity and, subsequently, MYC protein levels. Indeed, endogenous inhibitors of PP2A such as SET and CIP2A have been shown to be up-regulated in a variety of cancers (13, 19). CIP2A is overexpressed in head and neck squamous cell carcinoma, colon cancer, and 39% of breast tumors (13, 19). CIP2A interacts directly with the N terminus of pS62-MYC and impairs its degradation by inhibiting PP2A-B56α activity (20). SET binds to at least the C subunit of PP2A and inhibits its activity (21). SET is overexpressed in malignant brain tumors, tumors of the head and neck region, testicular cancers, and different types of hematological malignancies (13, 22, 23), but whether its expression is altered in breast cancer has not been reported.
Here we show that SET and CIP2A are up-regulated in breast tumors at both the mRNA and protein levels, and that knockdown of either SET or CIP2A decreases the tumorigenic potential of breast cancer cells in vitro and in vivo. SET inhibition with OP449, a SET antagonist (22), also reduces growth and tumorigenic potential of these cells in vitro and in vivo and induces apoptosis. SET inhibition, either by RNA interference or OP449, decreases pS62-MYC levels and MYC transcriptional activity. Overall, our study suggests that activating PP2A by inhibiting SET and/or CIP2A may be an important therapeutic strategy that posttranslationally targets MYC in breast cancer.
Results
SET and CIP2A Are Frequently Overexpressed in Human Breast Cancer.
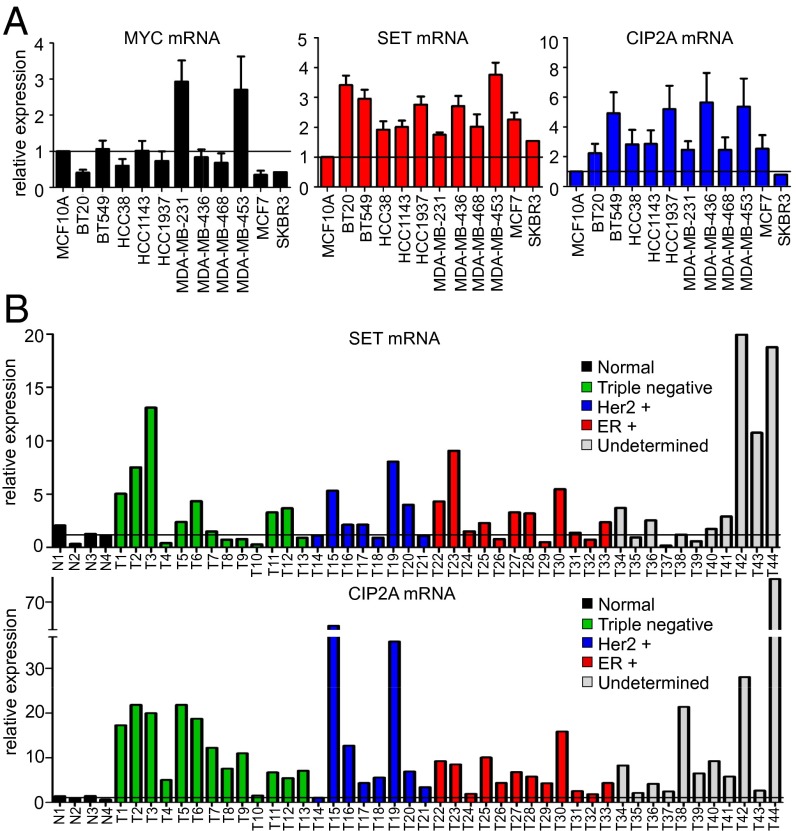
Previously, we have shown that pS62-MYC, a more stable and active form of MYC, is highly expressed in breast cancers (4, 7). In addition, we found that expression of AXIN1, which nucleates the MYC degradation complex, is decreased in some breast tumors (4). To investigate additional mechanisms that lead to MYC overexpression, we focused on SET and CIP2A, two oncogenic cellular inhibitors of PP2A, the phosphatase that removes S62 phosphorylation to destabilize MYC. First, we interrogated the expression of SET, CIP2A, and MYC in multiple breast cancer cell lines by quantitative RT-PCR (qRT-PCR). We found that SET and CIP2A mRNA levels were high in breast cancer cells compared with the immortalized but nontransformed MCF10A cell line (Fig. 1A). To confirm this in primary tumor samples, we performed qPCR for SET and CIP2A levels in a cDNA array of 44 breast tumors and four normal samples. We found that SET was overexpressed in about 60% and CIP2A in about 90% of these tumors (Fig. 1B). Similar results were observed in RNA-sequencing (RNA-seq) data from breast cancer cell lines where SET and CIP2A expression was elevated in about 50% of cell lines (Fig. S1A). In both of these experiments, when tumors were grouped by tumor subtype, we found that high SET expression occurred across all tumor types, whereas high CIP2A expression was enriched in triple negative tumors on the cDNA array (Fig. 1B) and in basal and claudin-low subtypes in the cell lines, molecularly classified using PAM50 (a breast cancer intrinsic classifier using the RT-qPCR assay for 50 genes) (Fig. S1A) (24). Finally, we measured SET mRNA expression by qPCR in a limited set of primary human breast tumor samples with patient-matched adjacent normal tissue and found that 60% of samples showed higher expression of SET in tumor compared with the matched normal (Fig. S1B). Together these data demonstrate that SET is overexpressed in about 50–60% and CIP2A in about 90% of breast cancers, and CIP2A overexpression associates with triple negative, basal, and claudin-low tumor subtypes.
Fig. 1.
SET and CIP2A are frequently overexpressed in human breast cancer. (A) qRT-PCR analysis of MYC, SET, and CIP2A mRNA expression in 12 breast cell lines grown in 0.1% serum. Relative expression is calculated by ∆CT normalized to MCF10A. (B) qPCR analysis of SET and CIP2A expression in 44 breast tumors and four normal samples grouped by histologic subtypes, obtained from TissueScan Breast Cancer and Normal Tissue cDNA array (array 4).
Increased SET, CIP2A, and pS62-MYC Protein Levels Occur in Human Breast Cancer.
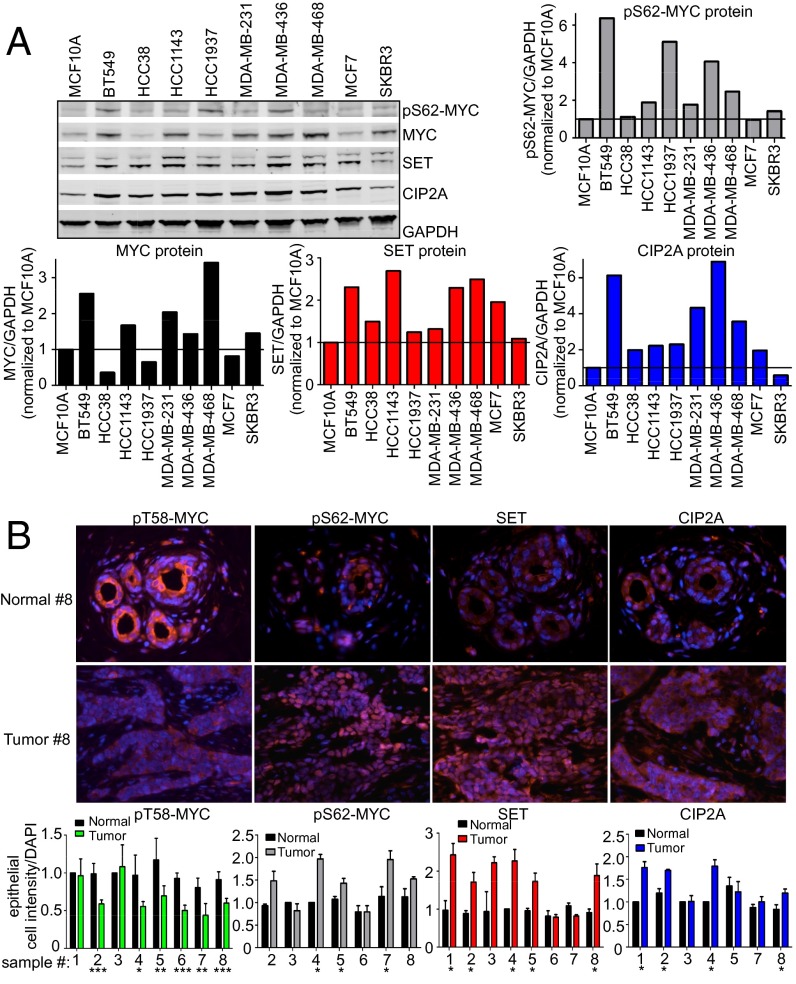
We next measured SET, CIP2A, MYC, and pS62-MYC protein levels in a panel of breast cancer cell lines. As shown in Fig. 2A, compared with immortalized nontransformed MCF10A cells, many of the breast cancer cell lines showed higher expression of these proteins (quantified in Fig. 2A). In addition, increased pS62-MYC positively correlated with increased SET and CIP2A expression (Fig. S2, Left). To extend this observation to human breast tumors, we performed immunofluorescence (IF) to assess SET, CIP2A, pS62-MYC, and pT58-MYC levels in breast tumors and compared them with patient-matched adjacent normal tissue. We found that SET, CIP2A, and pS62-MYC levels were higher in tumors, whereas pT58-MYC, which would be associated with increased PP2A activity, was lower (Fig. 2B). Together, these data show that SET, CIP2A, and pS62-MYC proteins are commonly overexpressed in human breast cancers.
Fig. 2.
Increased SET, CIP2A, and pS62-MYC protein levels occur in human breast cancer. (A) Representative Western blots of SET, CIP2A, pS62-MYC, and MYC protein expression in 10 breast cell lines grown in 0.1% serum. Quantification of SET, CIP2A, pS62-MYC, and MYC protein expression over GAPDH was done using a LICOR scanner and software. Quantification is graphed relative to expression in MCF10A. (B) Immunofluorescence of serial formalin-fixed, paraffin-embedded sections of breast tumors stained for pT58-MYC, pS62-MYC, SET, and CIP2A compared with their adjacent normal breast tissue. The graphs represent quantification of the mean staining intensity per epithelial cell for each protein over DAPI across multiple regions of interest and then normalized to the adjacent normal. Error bars represent SD. *P < 0.05, **P < 0.01, and ***P < 0.001.
SET or CIP2A Knockdown Decreases the Tumorigenic Potential of Breast Cancer Cells.
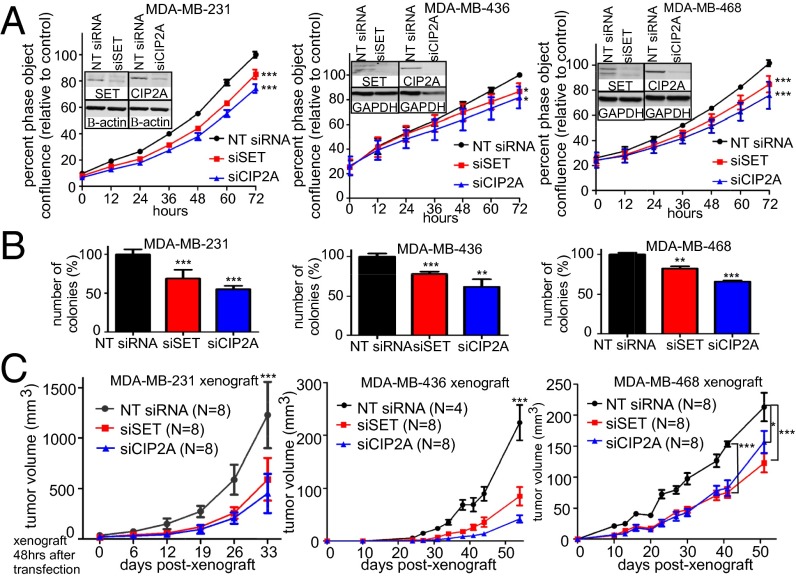
To better understand the impact of SET and CIP2A overexpression in breast cancer, we performed transient knockdown experiments in multiple breast cancer cell lines. We transfected MDA-MB-231, MDA-MB-436, and MDA-MB-468 cells with SET, CIP2A, or nontargeting (NT) siRNAs and measured population expansion capacity over 3 d (Fig. 3A). We observed a modest, but significant decrease in population expansion upon SET or CIP2A knockdown when cells were grown in two-dimensional (2D) culture compared with NT siRNA transfected cells (Fig. 3A). To determine whether decreased expression of SET or CIP2A affected anchorage-independent growth of breast cancer cells, siRNA transfected cells were plated for soft agar colony-forming assays. Whereas only a modest decrease in growth in 2D was observed (Fig. 3A), there was a substantial decrease in colony number in soft agar following knockdown of either protein (Fig. 3B). Because we were able to observe long-term knockdown (12 d) in 2D (Fig. S3A) and reduced soft agar growth (Fig. 3B), we examined the effects of SET or CIP2A loss on tumorigenic potential in vivo by xenografting cells into the mammary gland of nonobese diabetic (NOD)/SCID/γ-chain null (NSG) mice after transfection with siRNA. We found a significant decrease in tumor growth with knockdown of either SET or CIP2A in all three cell lines (Fig. 3C). Interestingly, MDA-MB-436 cells, which maintained longer-term knockdown in comparison with the other cells (Fig. S3A), displayed the most dramatic decrease in tumor growth.
Fig. 3.
SET and CIP2A knockdown decreases tumorigenic potential of breast cancer cell lines. (A) Population expansion analysis of the indicated cell lines over 72 h after transfection with SET or CIP2A siRNA compared with the control NT siRNA from three independent experiments using live cell imaging and IncuCyte analysis software. Representative Western blots show knockdown. (B and C) Soft agar colony assay and xenograft of these cells into the fourth mammary glands of NSG mice. Experimental details and statistics are described in Materials and Methods.
To extend this analysis, we developed stable clones with shRNA-mediated knockdown of SET in MDA-MB-231 cells. We used clones with significant knockdown (Fig. S3B) to test cell population expansion as well as ability to form colonies in soft agar. Similar to the experiments with transient SET knockdown, stable knockdown of SET decreased the rate of cell expansion in 2D (Fig. S3C), but more significantly inhibited the ability of these cells to form colonies in soft agar (Fig. S3D). Together, these results show that SET or CIP2A knockdown in breast cancer cells reduces their oncogenic potential in vitro and in vivo.
The SET Antagonist OP449 Decreases the Growth of Breast Cancer Cells and Induces Apoptosis.
Given that depletion of two endogenous inhibitors of PP2A, SET, and CIP2A, could reduce the oncogenic potential of breast cancer cells, development of a targeted therapeutic that inhibits either of these proteins could be clinically important. Because inhibitors of CIP2A are not currently available, we have collaborated with Oncotide Pharmaceuticals to test their novel SET antagonist, OP449. OP449 (previously referred to as COG449) is a dimer of a chimeric peptide composed of an ApoE mimetic domain that binds to SET, which is fused to antennapedia, a protein transduction domain (25). OP449 interacts with SET in cells, causing the release of SET from PP2A and an increase in PP2A activity as demonstrated in both leukemic cells as well as in some solid tumor cell lines, including MDA-MB-231 (22, 25).
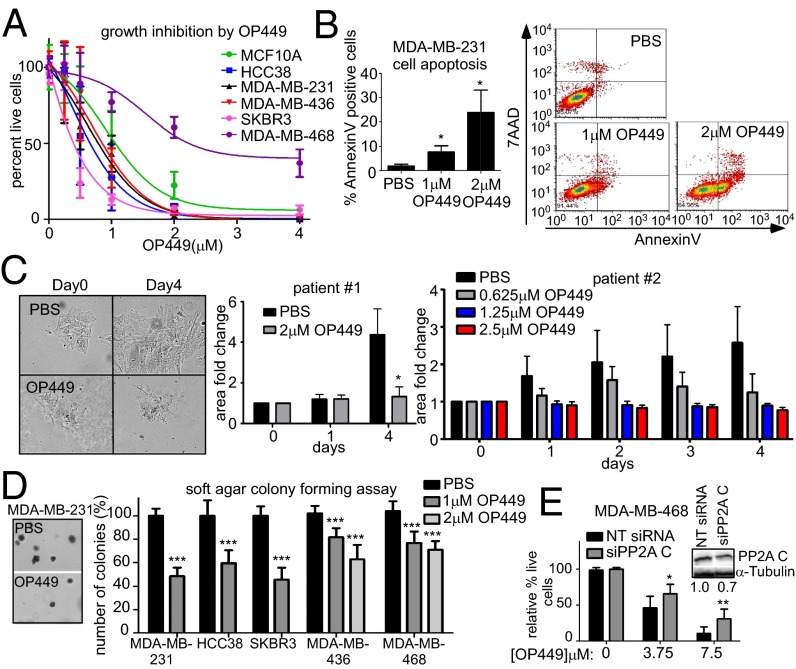
To begin to address the therapeutic potential of SET inhibition using OP449 as a pharmacological antagonist, we treated several breast cancer cell lines as well as immortalized MCF10A cells with OP449 for 24 h and analyzed cell viability by Trypan blue exclusion. OP449 treatment was cytotoxic in a dose-dependent manner (Fig. 4A). Flow cytometry for Annexin V in MDA-MB-231 cells treated with OP449 revealed that OP449 induced apoptosis in these cells as early as 6 h (Fig. 4B). We next isolated cells from two fresh invasive breast carcinoma samples and examined the effect of OP449 on these primary cells. Cells were treated with OP449 for 4 d, and cell colony expansion was measured over time. Whereas vehicle-treated samples for both patients showed significant proliferation, colony expansion was completely inhibited with OP449 treatment at concentrations above 1.25 µM (Fig. 4C). To determine whether OP449 reduced anchorage-independent growth of breast cancer cells, MDA-MB-231, HCC38, SKBR3, MDA-MB-436, and MDA-MB-468 cells were grown in soft agar and treated with OP449. OP449 significantly reduced the anchorage-independent growth of these cells (Fig. 4D). Because OP449 directly inhibits SET, and SET has targets in addition to PP2A, we examined the specificity of OP449 function for PP2A activity. We used siRNA to knock down the PP2A catalytic C subunit in MDA-MB-468 cells and treated them with OP449. We found that with 30% knockdown of PP2A C levels, these cells became partially insensitive to OP449 (Fig. 4E).
Fig. 4.
The SET antagonist OP449 decreases the growth of breast cancer cells and induces apoptosis. (A) Cytotoxicity of multiple breast cancer cell lines after 24 h of OP449 treatment. Cell viability was assessed by Trypan blue exclusion. (B) Apoptosis assay for MDA-MB-231 cells using AnnexinV-7-aminoactinomycin D after 6 h treatment with OP449 or PBS control. (C) Cell colony expansion of two primary invasive breast carcinoma samples treated with OP449 or PBS. Fold colony area ± SD is shown. The images are representative cell colonies (patient 1) taken at day 0 and 4 d with and without OP449 treatment. (D) Soft agar colony assay of breast cancer cell lines after treatment with OP449 or PBS. Representative images of colonies are shown. (E) The 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt (MTS) assay for MDA-MB-468 cells transfected with control NT siRNA or siRNA against PP2A C subunit for 48 h. Cells were then treated with OP449 for another 48 h. A representative Western blot of knockdown is shown. Error bars represent SD from three independent experiments except in C, as indicated, and E, which is from two experiments performed in triplicate. *P < 0.05, **P < 0.01, and ***P < 0.001.
OP449 Decreases S62-Phosphorylated MYC and MYC Transcriptional Activity Contributing to Cytotoxicity in Breast Cancer Cells.
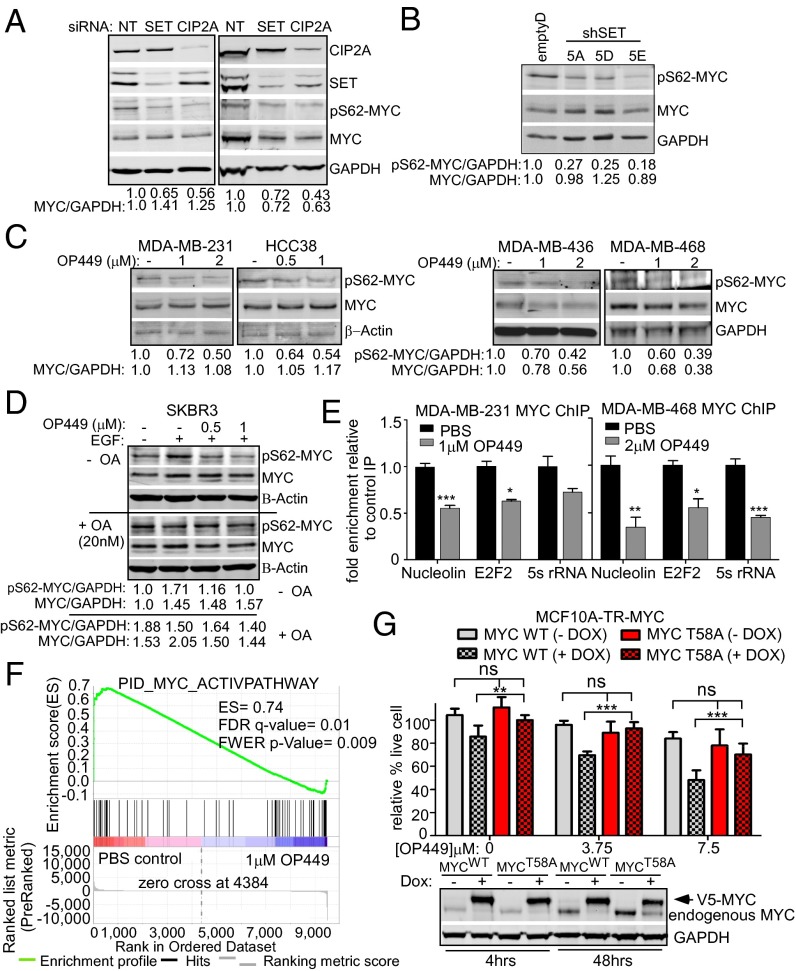
OP449 has been shown to down-regulate PP2A-regulated pathways including NFκB, Rac1, nm23-H1, STAT5, and AKT (23, 25). Because MYC is negatively regulated by PP2A and previous reports showed reduced expression of pS62-MYC and MYC upon CIP2A inhibition (13, 19, 20, 26), we wanted to know whether SET inhibition could also decrease pS62-MYC levels. We therefore measured pS62-MYC and total MYC levels after knocking down SET or CIP2A in MDA-MB-231 and MDA-MB-436 cells. We observed a decrease in pS62-MYC levels in both cell lines and total MYC levels in MDA-MB-436 cells (Fig. 5A). Total MYC levels did not change in some of the cell lines likely due to these cells having lost Fbw7-regulated MYC degradation (27). We also observed decreased pS62-MYC levels in the stable SET knockdown clones from MDA-MB-231 cells (Fig. 5B). To test whether OP449 also affects pS62-MYC levels in breast cancer cells, MDA-MB-231, HCC38, MDA-MB-436, and MDA-MB-468 cells were exposed to OP449 for 4 h and lysates were immunoblotted. We observed decreased pS62-MYC after OP449 treatment in these cells (Fig. 5C). Because OP449 functions at least in part through affecting PP2A activity (Fig. 4E) (22, 25), and PP2A has targets in addition to MYC, we examined the specificity of PP2A activation after OP449 treatment on MYC S62 phosphorylation using okadaic acid (OA), which specifically inhibits PP2A at low doses. SKBR3 cells were starved for 24 h and then treated for 2 h with OP449 in the presence or absence of OA. We then examined the effects on pS62-MYC following stimulation with EGF for 10 min. We found that, as expected, OP449 treatment inhibited the induction of pS62-MYC by EGF (Fig. 5D, Upper). However, in the presence OA, pS62-MYC levels were elevated in control cells and did not change with OP449 treatment (Fig. 5D, +OA, Lower).
Fig. 5.
OP449 decreases S62-phosphorylated MYC and MYC transcriptional activity contributing to cytotoxicity in breast cancer cells. (A) Western blot analysis of pS62-MYC and MYC proteins in MDA-MB-231 and MDA-MB-436 cells after siRNA knockdown of SET or CIP2A. The levels of pS62-MYC and total MYC were quantified on a LICOR scanner and calculated over GAPDH. (B) Western blot analysis of pS62-MYC and MYC protein in MDA-MB-231 stable clones with control or SET knockdown (shown in Fig. S3B). (C) Western blot analysis of pS62-MYC and MYC after treatment of the indicated cells with OP449 for 4 h. (D) Western blot analysis of pS62-MYC and MYC from SKBR3 cells starved for 24 h and then treated with OP449 without or with 20 nM okadaic acid (OA) for 2 h, followed by 10 min EGF (100 ng/mL) treatment. (E) qChIP for MYC at the Nucleolin, E2F2, and 5s rRNA promoters after 24 h of treatment with OP449. The fold enrichment of bound DNA was graphed as the fold enrichment in MYC IP relative to the fold enrichment in IgG control IP. (F) Gene Set Enrichment Analysis of a MYC gene signature (30) in RNA-seq data from MDA-MB-231 cells treated with 1 µM OP449 or PBS for 12 h. The positive enrichment score (ES) and statistical values are listed. (G) MTS assay for MCF10A-TR-MYC cells treated with 1 µg/mL doxycycline (Dox) for 4 h to induce ectopic expression of wild-type (MYCWT) or mutant MYC (MYCT58A). Cells were then treated with different concentrations of OP449 as indicated for 48 h. Representative Western blot of ectopic MYC in these cells with 4 or 48 h of Dox treatment. Error bars represent SD. *P < 0.05, **P < 0.01, and ***P < 0.001.
To examine the effect of OP449 treatment on MYC transcriptional activity, a chromatin immunoprecipitation (ChIP) assay was performed in MDA-MB-231 and MDA-MB-468 cells after OP449 treatment. Consistent with decreased expression of the transcriptionally active pS62-MYC (12, 28) (Fig. 5C), quantitative ChIP (qChIP) analysis showed that OP449 treatment decreased MYC binding to the promoters of its target genes, Nucleolin, E2F2, and 5s rRNA (Fig. 5E). To gain a more comprehensive understanding of the effect of OP449 treatment, or SET or CIP2A knockdown on MYC transcriptional activity, we performed RNA-seq to compare gene expression in MDA-MB-231 cells in control vs. treated or knockdown cells. We then used Gene Set Enrichment Analysis (GSEA) (29) to test whether a curated gene set of validated targets of c-MYC transcriptional activation (30) was affected by treatment or knockdown. We found that this MYC gene signature was enriched in control cells versus OP449-treated cells and in control siRNA versus SET or CIP2A knockdown cells (Fig. 5F and Fig. S4), indicating suppression of MYC target gene expression with OP449 treatment, or SET or CIP2A knockdown. Together, these data show that treatment with OP449, or SET or CIP2A knockdown, causes a decrease in pS62-MYC protein and this leads to a global decrease in MYC’s transcriptional activity.
PP2A has many targets that likely contribute to tumor growth. To examine how much of OP449’s activity is through its effect on pS62-MYC, we took advantage of our MCF10A-tetracycline responsive (TR)-MYC inducible cell lines, in which either wild-type (WT) or a mutant form of MYC (T58A) can be induced with doxycycline (Dox). MYCT58A cannot be phosphorylated at T58 and is resistant to PP2A-mediated S62 dephosphorylation, maintaining constitutive pS62 (5, 6, 9). Ectopic MYC was induced for 4 h before treatment with OP449 for 48 h. Although expression of MYCWT on its own was mildly toxic in MCF10A cells, OP449 still induced cell death, whereas expression of the PP2A-resistant MYCT58A mostly rescued this effect (Fig. 5G). Together, these results demonstrate that MYC activity is suppressed by OP449 treatment through its effects on PP2A and this in part underlies its cytotoxic activity.
OP449 Treatment Decreases Tumor Growth in Vivo and Increases PP2A Activity in Tumors.
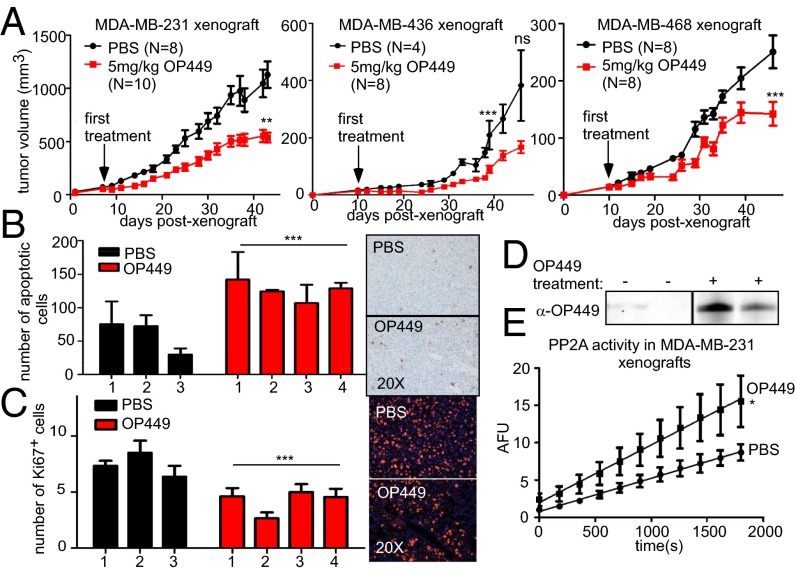
Because we observed that OP449 treatment could decrease the growth of breast cancer cell lines and induce apoptosis in vitro, we next wanted to test whether OP449 has antitumorigenic properties in vivo. MDA-MB-231, MDA-MB-436, and MDA-MB-468 cells were xenografted into the mammary glands of NSG mice. Once tumors were palpable, mice were randomized into two groups and treated intraperitoneally with 5 mg/kg of OP449 or PBS control, three times per week. Tumor size was determined by repeated caliper measurement. We found that tumor growth was slowed in mice treated with OP449 compared with controls (Fig. 6A). For MDA-MB-231 tumors, all mice were killed when control tumors reached 2 cm in diameter, and tumors were harvested and fixed in formalin for histological analysis. Consistent with our data in cell lines, this analysis revealed that the OP449 tumors had an increased number of apoptotic cells as measured by TUNEL assay and a decreased number of proliferating cells as measured by the cellular marker for proliferation, Ki67, staining (Fig. 6 B and C). Following on these results, we performed a second study with MDA-MB-231 tumors, treating them three times per week for 40 d and again observed decreased tumor growth with OP449 treatment (Fig. S5A). On the last day, we injected mice with OP449 or the vehicle control 2 h before they were killed. Tumors were dissected, flash frozen, and used for Western blot analysis of OP449 in the tumors and for PP2A activity assays. We observed OP449 specifically in the tumors of treated mice (Fig. 6D) and a significant increase in PP2A activity in the OP449-treated tumors compared with PBS-treated tumors (Fig. 6E).
Fig. 6.
OP449 suppresses breast tumor growth in vivo associated with increased PP2A activity. (A) Tumor growth curve for MDA-MB-231, MDA-MB-436, and MDA-MB-468 xenografts in the fourth mammary gland of NSG mice following treatment with OP449 or PBS. (B) TUNEL assay and (C) IF for Ki67 for MDA-MB-231 harvested xenografts from A. The mean and SD of total apoptotic cells in 75 random fields or Ki67 positive cells in 25 fields for three control mice (six tumors) and four OP449-treated mice (eight tumors) are graphed. (D) Western blot analysis of OP449 with peptide-specific antibody in lysates from MDA-MB-231 xenografts (Fig. S5A). (E) Tumor lysates from D were used to measure PP2A activity as described previously (22).
To further address the pharmacokinetic, distribution, and plasma stability of OP449, studies were conducted to detect OP449 protein in plasma isolated from OP449-infused rats after 1-h infusion of 2 mg/kg. Western blotting indicated that OP449 is immediately detectable and then rapidly cleared from the blood, without the appearance of a degradation product (Fig. S5B). We extended this analysis by incubating OP449 in rat plasma in vitro for 12, 24, and 48 h at 37 °C followed by Western blotting and observed that OP449 was stable in isolated plasma with a half-life of >24 h (Fig. S5C). The rapid clearance of OP449 in vivo from the blood stream was associated with its detection in tissues, which persists for more than 6 h. Together these results suggest that OP449 can decrease the growth and the tumorigenic potential of breast cancer cells in vivo, and this is associated with increased apoptosis and decreased proliferation. In addition, OP449 has in vivo bioavailability sufficient for treating tumors and eliciting a pharmacologically induced increase in PP2A activity in the treated tumors.
Discussion
In analyses of oncogenic mechanisms in breast cancer, accumulation of the MYC protein has been observed in 46% of primary breast tumors (1, 31). Many mechanisms have been proposed to explain the elevated levels of MYC protein in tumors. Previous work from our laboratory has shown that increased levels of MYC can result from disruption of the normal MYC protein degradation pathway rather than increased expression at the mRNA level (4). Our research also revealed that PP2A-B56α is a critical negative regulator of MYC protein stability through its dephosphorylation of serine 62 (10). This is highly relevant because Hahn and coworkers demonstrated that inhibition of PP2A activity is required for cell transformation (14, 15). Furthermore, we have shown that this requirement for PP2A inhibition can be partially replaced by expression of the more stable MYCT58A mutant (6). In this report, we studied the role of two cellular inhibitors of PP2A, SET and CIP2A, in breast cancer to understand if their expression (i) is common in tumors, (ii) regulates tumorigenesis, (iii) affects the level and/or activity of pS62-MYC, and (iv) could be exploited as potential targets for breast cancer therapy.
We found that SET and CIP2A are frequently overexpressed in breast cancer cell lines at both the mRNA and protein levels. In addition, we found that CIP2A, SET, and pS62-MYC, but not pT58-MYC are frequently co-overexpressed in human primary tumor samples relative to matched normal tissue. For CIP2A, these findings confirmed a previous report that CIP2A directly regulates MYC S62 phosphorylation and stability (20) and that CIP2A is overexpressed in human breast tumors (19). In contrast, our finding that SET is frequently overexpressed in breast cancers is novel and suggests that overexpression of PP2A inhibitors may play an important role in the development of human breast cancer. Interestingly, we found that CIP2A overexpression correlated with the triple negative breast cancer subtype, a result that was further supported with RNA-seq data from human breast cancer cell lines. This result is important, as the claudin-low breast cancer subtype associates highly with up-regulation of the MYC/MYC-associated factor X network (24). This correlation suggests that CIP2A could potentially be used as a diagnostic biomarker for more malignant, MYC-driven tumor types. Whereas SET was overexpressed in greater than 50% of all breast cancer samples tested, there was no significant correlation between increased SET levels and any breast cancer subtype, suggesting that SET plays a broader role in sustaining oncogenic signaling in breast cancers.
It has been shown that CIP2A deletion results in a reduction of oncogenic potential for multiple tumors (20, 26, 32). Furthermore, SET knockdown was shown to negatively affect tumorigenesis in blast crisis chronic myelogenous leukemia (33). Here we show that in breast cancer SET and CIP2A, knockdown reduces cell growth in vitro and significantly attenuates tumor growth in vivo. Furthermore, loss of these PP2A inhibitors decreased the level of pS62-MYC in these cells and suppressed global MYC-driven gene expression. These results suggest that inhibiting either SET or CIP2A could be a viable strategy to posttranslationally target MYC and inhibit tumor growth in breast cancer.
Whereas no known inhibitors of CIP2A have been described, previous work has demonstrated that the SET antagonist OP449 (originally named COG449) activates PP2A, is cytotoxic to primary chronic lymphocytic leukemia cells, and decreases lymphoma xenograft tumor growth (22). To explore the therapeutic potential for PP2A activation through SET inhibition as an approach for breast cancer therapy, we treated breast cancer cells with OP449 and measured growth and oncogenic potential in vitro and in vivo. OP449 treatment resulted in increased apoptosis and decreased proliferation in vitro and in vivo. Interestingly, we found that although MYCWT overexpression was additive with OP449 treatment in inhibiting the growth of MCF10A cells, the MYCT58A mutant, which is resistant to PP2A and has constitutively high S62 phosphorylation, was able to rescue this effect. Importantly, OP449 inhibited tumor growth at doses that were not toxic to mice, and other studies using OP449, showed selective inhibition of cancer growth, whereas normal fibroblasts remained unaffected (25). Furthermore, we show here that OP449 has in vivo bioavailability and can be detected in the tumors of treated mice, and treated tumors have a resultant increase in PP2A activity.
In summary, inhibiting SET (and potentially CIP2A) may provide an important therapeutic strategy for the treatment of breast cancer by targeting MYC in a way that has not yet been achieved by other means. Furthermore, SET antagonism with OP449 will facilitate down-regulation of other PP2A-regulated pathways including NFκB, the nucleoside diphosphate kinase nm23-H1, the AKT kinase, and the small GTP binding protein Rac-1 (25), all of which likely contribute to the potent growth inhibitory effects we observed here in breast cancer. This makes OP449 a favorable drug because in addition to targeting MYC, which may have opposing effects on metastasis (34), OP449 can facilitate down-regulation of these other oncogenes. These results further validate the use of SET antagonists and PP2A activators as a novel strategy for breast cancer therapy.
Materials and Methods
Cell Culture, Knockdown, and Cell Population Expansion Assay.
All cell lines were purchased from American Type Culture Collection except SKBR3, which was a gift from Joe Gray (OHSU, Portland, OR). Cell culture and knockdown methods are described in SI Materials and Methods. The cytotoxicity assay was performed in reduced serum (0.1%) due to observed precipitation of OP449 at higher serum concentrations. Acquisition and culturing of primary tissue is described in SI Materials and Methods [Institutional Review Board (IRB) approval no. 3330]. The cell population expansion assay was performed on an IncuCyte Zoom (Essen Bioscience), and detailed information on this assay is described in SI Materials and Methods.
Western Blot, ChIP, and IF.
Western blot, ChIP, and IF analyses were performed as described previously (4, 11, 12) and as detailed in SI Materials and Methods. Patient samples used for IF analysis were obtained from the OHSU Cancer Pathology Shared Resource (IRB approval no. 6478).
qRT-PCR and RNA-Seq.
The TissueScan Breast Cancer cDNA array (array 4), TaqMan primers, and RNA-seq are described in SI Materials and Methods. cDNA from patient samples (Fig. S1B) was provided by Dexi Chen (Department of Infectious Diseases, Capital University of Medical Sciences, You'an Hospital, Beijing) and described in ref. 4.
Orthotopic Xenografts of Breast Cancer Cell Lines and OP449 Pharmacokinetics.
Orthotopic xenograft of breast cancer cell lines is described in SI Materials and Methods. Briefly, cells were xenografted into the fourth mammary gland of NSG mice. For the treatment study, once tumors were palpable, mice were divided into two groups and treated with OP449 or vehicle control (PBS) 3 d/wk. The xenografted tumors were harvested to perform TUNEL, Ki67, PP2A activity, and OP449 detection assays (described in SI Materials and Methods). OP449 pharmacokinetic studies are described in SI Materials and Methods.
Statistics.
SD for all graphs was calculated from three independent experiments (unless otherwise stated in the figure legend) using GraphPad Prism 5. P values were analyzed by Student t test, with a two-tailed method (*P < 0.05, **P < 0.01, and ***P < 0.001).
Supplementary Material
Acknowledgments
We thank Dr. Joe Gray for providing the SKBR3 cell line and the RNA-seq data from a panel of breast cancer cell lines, Dr. Dexi Chen for providing cDNA from breast cancer samples, Dr. Nicholas Wang and Darcie Babcock for help with dose response analysis, Dr. Paul Spellman for helpful suggestions, Dr. Xiaoyan Wang for technical help, and all members of the R.C.S. laboratory for editing the manuscript and other helpful suggestions. Sequencing for the RNA-seq data was performed in the Oregon Health & Science University (OHSU) Integrated Genomics Laboratory and Gene Profiling Shared Resource. This study was supported by R01 CA100855 and CA129040, Department of Defense BC061306, Susan G. Komen BCTR0706821, the Anna Fuller Foundation and the Colson Family Foundation (R.C.S.), and a OHSU Tartar Trust Fellowship (to M.J.).
Footnotes
Conflict of interest statement: D.J.C. is an employee and shareholder of Oncotide Pharmaceuticals, Inc. J.O. is an employee of Oncotide Pharmaceuticals, Inc.
This article is a PNAS Direct Submission.
Data deposition: RNA-seq gene expression data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE48216 and GSE58008).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1317630111/-/DCSupplemental.
References
- 1.Chrzan P, Skokowski J, Karmolinski A, Pawelczyk T. Amplification of c-myc gene and overexpression of c-Myc protein in breast cancer and adjacent non-neoplastic tissue. Clin Biochem. 2001;34(7):557–562. doi: 10.1016/s0009-9120(01)00260-0. [DOI] [PubMed] [Google Scholar]
- 2.Wolfer A, et al. MYC regulation of a “poor-prognosis” metastatic cancer cell state. Proc Natl Acad Sci USA. 2010;107(8):3698–3703. doi: 10.1073/pnas.0914203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malempati S, et al. Aberrant stabilization of c-Myc protein in some lymphoblastic leukemias. Leukemia. 2006;20(9):1572–1581. doi: 10.1038/sj.leu.2404317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang X, et al. Mechanistic insight into Myc stabilization in breast cancer involving aberrant Axin1 expression. Proc Natl Acad Sci USA. 2012;109(8):2790–2795. doi: 10.1073/pnas.1100764108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sears R, et al. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14(19):2501–2514. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yeh E, et al. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat Cell Biol. 2004;6(4):308–318. doi: 10.1038/ncb1110. [DOI] [PubMed] [Google Scholar]
- 7.Wang X, et al. Phosphorylation regulates c-Myc’s oncogenic activity in the mammary gland. Cancer Res. 2011;71(3):925–936. doi: 10.1158/0008-5472.CAN-10-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hann SR. Role of post-translational modifications in regulating c-Myc proteolysis, transcriptional activity and biological function. Semin Cancer Biol. 2006;16(4):288–302. doi: 10.1016/j.semcancer.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 9.Arnold HK, Sears RC. Protein phosphatase 2A regulatory subunit B56alpha associates with c-myc and negatively regulates c-myc accumulation. Mol Cell Biol. 2006;26(7):2832–2844. doi: 10.1128/MCB.26.7.2832-2844.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arnold HK, Sears RC. A tumor suppressor role for PP2A-B56alpha through negative regulation of c-Myc and other key oncoproteins. Cancer Metastasis Rev. 2008;27(2):147–158. doi: 10.1007/s10555-008-9128-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnold HK, et al. The Axin1 scaffold protein promotes formation of a degradation complex for c-Myc. EMBO J. 2009;28(5):500–512. doi: 10.1038/emboj.2008.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farrell AS, et al. Pin1 regulates the dynamics of c-Myc DNA binding to facilitate target gene regulation and oncogenesis. Mol Cell Biol. 2013;33(15):2930–2949. doi: 10.1128/MCB.01455-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Westermarck J, Hahn WC. Multiple pathways regulated by the tumor suppressor PP2A in transformation. Trends Mol Med. 2008;14(4):152–160. doi: 10.1016/j.molmed.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 14.Sablina AA, Hector M, Colpaert N, Hahn WC. Identification of PP2A complexes and pathways involved in cell transformation. Cancer Res. 2010;70(24):10474–10484. doi: 10.1158/0008-5472.CAN-10-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sablina AA, Hahn WC. SV40 small T antigen and PP2A phosphatase in cell transformation. Cancer Metastasis Rev. 2008;27(2):137–146. doi: 10.1007/s10555-008-9116-0. [DOI] [PubMed] [Google Scholar]
- 16.Calin GA, et al. Low frequency of alterations of the alpha (PPP2R1A) and beta (PPP2R1B) isoforms of the subunit A of the serine-threonine phosphatase 2A in human neoplasms. Oncogene. 2000;19(9):1191–1195. doi: 10.1038/sj.onc.1203389. [DOI] [PubMed] [Google Scholar]
- 17.Esplin ED, et al. The glycine 90 to aspartate alteration in the Abeta subunit of PP2A (PPP2R1B) associates with breast cancer and causes a deficit in protein function. Genes Chromosomes Cancer. 2006;45(2):182–190. doi: 10.1002/gcc.20284. [DOI] [PubMed] [Google Scholar]
- 18.Marsh A, et al. kConFab Mutation analysis of five candidate genes in familial breast cancer. Breast Cancer Res Treat. 2007;105(3):377–389. doi: 10.1007/s10549-006-9461-z. [DOI] [PubMed] [Google Scholar]
- 19.Come C, et al. CIP2A is associated with human breast cancer aggressivity. Clin Cancer Res. 2009;15(16):5092–5100. doi: 10.1158/1078-0432.CCR-08-3283. [DOI] [PubMed] [Google Scholar]
- 20.Junttila MR, et al. CIP2A inhibits PP2A in human malignancies. Cell. 2007;130(1):51–62. doi: 10.1016/j.cell.2007.04.044. [DOI] [PubMed] [Google Scholar]
- 21.Li M, Guo H, Damuni Z. Purification and characterization of two potent heat-stable protein inhibitors of protein phosphatase 2A from bovine kidney. Biochemistry. 1995;34(6):1988–1996. doi: 10.1021/bi00006a020. [DOI] [PubMed] [Google Scholar]
- 22.Christensen DJ, et al. SET oncoprotein overexpression in B-cell chronic lymphocytic leukemia and non-Hodgkin lymphoma: A predictor of aggressive disease and a new treatment target. Blood. 2011;118(15):4150–4158. doi: 10.1182/blood-2011-04-351072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agarwal A, et al. Antagonism of SET using OP449 enhances the efficacy of tyrosine kinase inhibitors and overcomes drug resistance in myeloid leukemia. Clin Cancer Res. 2014;20(8):2092–103. doi: 10.1158/1078-0432.CCR-13-2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heiser LM, et al. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc Natl Acad Sci USA. 2012;109(8):2724–2729. doi: 10.1073/pnas.1018854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Switzer CH, et al. Targeting SET/I(2)PP2A oncoprotein functions as a multi-pathway strategy for cancer therapy. Oncogene. 2011;30(22):2504–2513. doi: 10.1038/onc.2010.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khanna A, et al. MYC-dependent regulation and prognostic role of CIP2A in gastric cancer. J Natl Cancer Inst. 2009;101(11):793–805. doi: 10.1093/jnci/djp103. [DOI] [PubMed] [Google Scholar]
- 27.Mao JH, et al. FBXW7 targets mTOR for degradation and cooperates with PTEN in tumor suppression. Science. 2008;321(5895):1499–1502. doi: 10.1126/science.1162981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hydbring P, et al. Phosphorylation by Cdk2 is required for Myc to repress Ras-induced senescence in cotransformation. Proc Natl Acad Sci USA. 2010;107(1):58–63. doi: 10.1073/pnas.0900121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schaefer CF, et al. PID: The Pathway Interaction Database. Nucleic Acids Res. 2009;37(Database issue):D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deming SL, Nass SJ, Dickson RB, Trock BJ. C-myc amplification in breast cancer: A meta-analysis of its occurrence and prognostic relevance. Br J Cancer. 2000;83(12):1688–1695. doi: 10.1054/bjoc.2000.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J, et al. CIP2A is over-expressed in acute myeloid leukaemia and associated with HL60 cells proliferation and differentiation. Int J Lab Hematol. 2011;33(3):290–298. doi: 10.1111/j.1751-553X.2010.01288.x. [DOI] [PubMed] [Google Scholar]
- 33.Neviani P, et al. The tumor suppressor PP2A is functionally inactivated in blast crisis CML through the inhibitory activity of the BCR/ABL-regulated SET protein. Cancer Cell. 2005;8(5):355–368. doi: 10.1016/j.ccr.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 34.Liu H, et al. MYC suppresses cancer metastasis by direct transcriptional silencing of αv and β3 integrin subunits. Nat Cell Biol. 2012;14(6):567–574. doi: 10.1038/ncb2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.