Significance
The spatial distributions of neuronal populations are frequently patterned, but molecular mechanisms underlying their orderliness have received little attention. We used a genetic screen to identify prospective genes participating in the patterning within a population of retinal amacrine cells. We mapped variation in their patterning, assessed using nearest-neighbor analysis across a panel of 25 recombinant inbred strains, to a genomic locus on chromosome 11. There, we identified pituitary tumor-transforming gene 1 (Pttg1) as a candidate. Critically, Pttg1 expression correlated with regularity across these 25 strains, and a variant in Pttg1 was found to modulate expression. By knocking out Pttg1, patterning was selectively disrupted in two types of retinal neuron.
Keywords: nearest neighbor distance, regularity index, recombinant inbred mice, quantitative trait locus
Abstract
Neurons are commonly organized as regular arrays within a structure, and their patterning is achieved by minimizing the proximity between like-type cells, but molecular mechanisms regulating this process have, until recently, been unexplored. We performed a forward genetic screen using recombinant inbred (RI) strains derived from two parental A/J and C57BL/6J mouse strains to identify genomic loci controlling spacing of cholinergic amacrine cells, which is a subclass of retinal interneuron. We found conspicuous variation in mosaic regularity across these strains and mapped a sizeable proportion of that variation to a locus on chromosome 11 that was subsequently validated with a chromosome substitution strain. Using a bioinformatics approach to narrow the list of potential candidate genes, we identified pituitary tumor-transforming gene 1 (Pttg1) as the most promising. Expression of Pttg1 was significantly different between the two parental strains and correlated with mosaic regularity across the RI strains. We identified a seven-nucleotide deletion in the Pttg1 promoter in the C57BL/6J mouse strain and confirmed a direct role for this motif in modulating Pttg1 expression. Analysis of Pttg1 KO mice revealed a reduction in the mosaic regularity of cholinergic amacrine cells, as well as horizontal cells, but not in two other retinal cell types. Together, these results implicate Pttg1 in the regulation of homotypic spacing between specific types of retinal neurons. The genetic variant identified creates a binding motif for the transcriptional activator protein 1 complex, which may be instrumental in driving differential expression of downstream processes that participate in neuronal spacing.
Like many other CNS structures, the retina is composed of recurring modular microcircuits. Distinct neuronal populations are each distributed across the tissue and establish stereotypic patterns of connectivity with one another. To ensure a complete functional coverage by these microcircuits, individual neuronal populations are arranged in regular arrays across the entire retina (1). The regularity of these arrays is achieved by the presence of a zone of exclusion surrounding each cell, minimizing proximity between like-type cells, and although several developmental mechanisms, such as selective cell death and tangential migration, have been shown to mediate formation of such exclusion zones (2), the molecular underpinnings of these processes have received little attention. Recently, however, the EGF-like transmembrane receptor Megf10, which is expressed in only two neuronal populations of the retina, has been shown to mediate recognition and repulsion between like-type cells (3). In another retinal neuronal population, the anti- and proapoptotic factors Bcl2 and Bax have been shown to modulate programmed cell death, a process that yields increased regularity of this mosaic (4, 5). Aside from these examples, relatively little is known about the genetic determinants that coordinate these retinal cell biological processes.
In the present study, we exploited a forward genetic screen to identify such molecular players for a population of retinal interneurons, the cholinergic amacrine cells, for which the regularity and intercellular spacing have been studied extensively (6–9). These neurons are distributed as two populations that reside in different layers of the mature retina, the inner nuclear layer (INL) and ganglion cell layer (GCL), and although both populations exist as nonrandom mosaics, the regularity of the GCL population is known to be corrupted by several cell extrinsic factors unique to this layer (9). We therefore screened for genes that affect the regularity of the INL population. We previously observed natural variation in the regularity of cholinergic amacrine cells across six strains of mice (9). By mapping variation in mosaic regularity to the genomic variation across 25 strains of recombinant inbred (RI) mice, we identified a significant quantitative trait locus (QTL) on chromosome 11. We used bioinformatics databases to narrow down the 54 genes at this locus based on retinal expression, known function, and genetic variants; after this database mining, pituitary tumor-transforming gene 1 (Pttg1) was evident as the top candidate gene. We show differential Pttg1 expression between two parental strains, C57BL/6J and A/J, and identify a six nucleotide deletion in the promoter sequence in the former strain that significantly increases Pttg1 expression. We also show a significant correlation between Pttg1 expression levels and mosaic regularity across the 25 RI strains derived from those same two laboratory strains. Finally, we assess a direct role for Pttg1 in regulating mosaic regularity and intercellular spacing of cholinergic amacrine cells and examine the specificity of its role in this process in three other retinal cell types.
Results
Regularity of the Cholinergic Amacrine Cell Mosaic Varies Across 25 RI Mouse Strains.
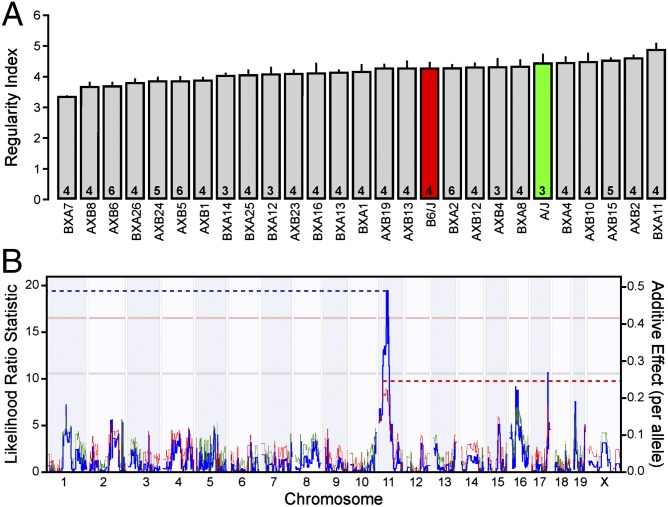
Cholinergic amacrine cells in the INL of the mouse retina are distributed as a patterned array of neurons that is significantly more regular than a random distribution, evidenced by an analysis of their nearest neighbor distances (Fig. S1 A–E). Little difference exists in the regularity index derived from these nearest neighbor distances between the A/J and C57BL/6J (B6/J hereafter) inbred laboratory strains, despite large differences in the total number of cholinergic amacrine cells in these two strains, as previously described (9). Variation in mosaic regularity does not correlate with local density (9), as expected from this scale-invariant measure. Across the 25 RI strains derived from these parental strains (the AXB/BXA strain set), however, there was conspicuous variation in the regularity index, with a 49% increase from the lowest to the highest RI strain (Fig. 1A). Such graded interstrain variation, in the presence of low intrastrain variation [the average coefficient of variation (CoV) for each strain being 0.09], indicates an underlying polygenic source. The regularity of the mosaic in the strain with the lowest regularity index, the BXA7 strain, was still discriminable from random simulations matched for density and constrained by soma size (real: 3.37 ± 0.08 vs. random: 3.02 ± 0.08, P = 0.003; mean ± SEM hereafter), suggesting that cellular mechanisms controlling mosaic regularity are still operating, albeit less effectively in some strains than in others. Additionally, the variation in the regularity index above and below that of the parental strains (Fig. 1A) indicates the presence of variants for which both A/J and B6/J alleles increase the regularity of the cholinergic amacrine cell mosaic, canceling one another’s additive effects within the parental strains.
Fig. 1.
The variation in mosaic regularity across the RI strains maps a significant QTL to chromosome 11. (A) The average regularity index (being the mean nearest neighbor distance divided by the SD) varies considerably across the 2 parental (red and green bars) and 25 RI (gray bars) strains, plotted here from the lowest strain to highest strain. The regularity indices for six of these strains have been reported previously (9). n = number of retinas analyzed per strain, with the mean and SE indicated. (B) Interval mapping reveals a significant QTL on chromosome 11 with a likelihood ratio statistic of 19.23 (blue trace, left axis). Horizontal lines indicate suggestive (gray, P = 0.63) and significant (pink, P = 0.05) thresholds defined through 2,000 random permutations of the strain data. Red and green traces indicate the directionality, as the trait increases with B6/J and A/J alleles, respectively, and the magnitude of the allelic effect (right axis).
Variation in the Regularity of the Cholinergic Amacrine Cell Mosaic Maps a Significant QTL to Chromosome 11.
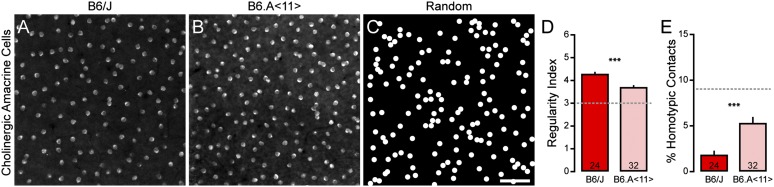
By correlating the variation in the regularity index to the genomic variation across the RI strains, we mapped a significant QTL to the middle of chromosome 11 (Fig. 1B). The peak of the QTL resided at 38.6 Mb, near rs13480975, with a likelihood ratio statistic (LRS) of 19.4. The presence of two B6 alleles at this locus was associated with an increase in the regularity index by 0.50, accounting for 33% of the total variation of this trait across the RI strain set. A chromosome substitution mouse strain, in which an A/J chromosome 11 was introgressed in an otherwise B6/J genome (B6.A <11>), was analyzed to validate the presence of the QTL. As predicted, the presence of A/J alleles on chromosome 11 significantly reduced the regularity of the cholinergic amacrine cell mosaic compared with B6/J mice by 0.58 (P = 0.021; Fig. 2 A, B, and D), roughly a 50% decrease relative to a random distribution of cells (Fig. 2C). Coincident with this decrease in regularity, there was an increased frequency of homotypic cell neighbors being in direct contact (homotypic pairs) in the B6.A<11> retina (Fig. 2E); such pairs of cells were rarely seen the in B6/J retina, although they were far more common in random distributions. These results confirm that a genetic variant (or variants) on chromosome 11 modulates the spacing of cholinergic amacrine cells and reduces the regularity of the mosaic.
Fig. 2.
The chromosome 11 substitution strain exhibits a decrease in the regularity of cholinergic amacrine cells. (A and B) Sample mosaics of cholinergic amacrine cells in the B6/J retina (A) and B6.A<11> retina (B) revealed by immunostaining with antibodies to choline acetyltransferase. Note the decreased regularity in the B6.A<11> retina and the increased frequency of pairs of cells in contact. (C) Sample of a simulated random mosaic, matched in density and constrained by soma size. (D and E) Analysis of these mosaics reveals a significant reduction in the regularity index (D) and an increase in the frequency of homotypic pairs (E) for the population of cholinergic amacrine cells in the B6.A<11> retina relative to the B6/J retina. n = number of fields analyzed, with the mean and SE indicated. The broken gray lines indicate the average value for random simulations matched in density and constrained by soma size. (Scale bar, 50 µm.) ***P < 0.001.
Pttg1 Is the Most Promising Candidate Gene at the Chromosome 11 Locus.
We focused on an interval spanning a 10-Mb region, from 37.5 to 47.5 Mb, on chromosome 11, containing 54 genes. We used several bioinformatic databases to narrow down this list of genes based on the presence of genetic variants between the two parental strains and determined whether these genes were expressed in the retina during development; variants were only included in this analysis if they yielded coding mutations or were found in putative regulatory regions. Of the genes at this interval, 38 contained genetic variants between the two parent strains, and 18 of these 38 genes were expressed during retinal development. These 18 candidate genes were subsequently analyzed using a microarray dataset measuring gene expression in the adult eye across each of the RI strains. The expression of each gene was checked for significant correlation with the regularity index across the RI strains, as well as for the presence of a significant expression QTL localized to that gene’s position on chromosome 11. The presence of such a cis-acting expression QTL (cis-eQTL) would suggest that a genetic variant within the regulatory region of a gene is altering its expression (10), providing a potential mechanism for how the variation between the A/J and B6/J alleles might be influencing the trait of interest. Finally, candidates were screened for known roles in cell recognition, migration, and apoptosis. Through this analysis (summarized in Table S1), Pttg1 emerged as the most promising candidate gene.
Pttg1 Expression Correlates with Regularity Index Across the RI Strains and Maps a Significant cis-eQTL.
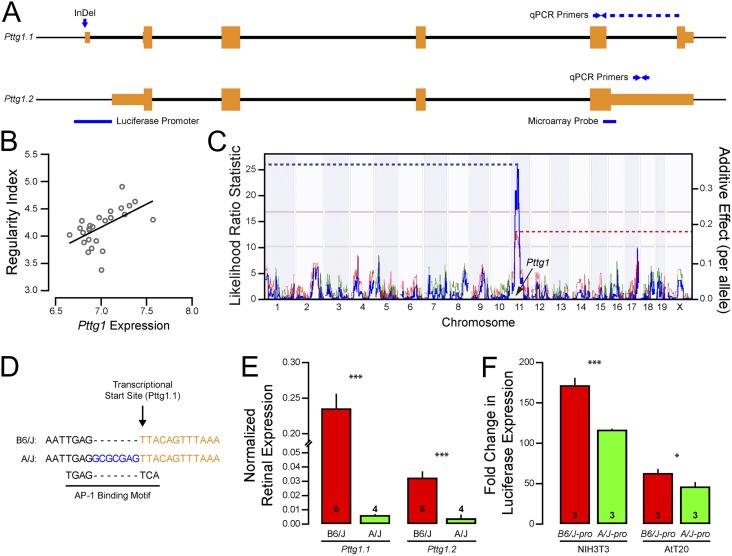
Pttg1 codes for an intracellular protein that is implicated in several diverse functions, acting as the mammalian homolog to yeast securin (11) and as a transcriptional transactivator. Pttg1 was first discovered because of its role in promoting pituitary adenomas (12), as misregulation of Pttg1 expression leads to tumor growth and vascularization (13). Two annotated splice variants of Pttg1 exist (Fig. 3A): a six-exon short variant that corresponds to the canonical isoform of Pttg1 (herein identified as Pttg1.1) and a four-exon long variant (Pttg1.2). Using the SAGE retinal expression database (14), Pttg1 was found to be expressed in the retina throughout embryonic and postnatal development, as well as in adulthood, although spatial expression across the different cellular layers is still unknown. Microarray analysis of purified populations of retinal neurons in development and maturity, however, confirms moderate Pttg1 expression within the cholinergic amacrine cells (3, 15). Most compelling, microarray data, using a probe to Pttg1.2 (Fig. 3A), revealed that the expression of Pttg1 was positively correlated with regularity index across the RI strains (r = 0.471, P = 0.012; Fig. 3B), and its expression mapped a significant QTL to its location in the genome, indicating a cis-eQTL (Fig. 3C). Although no SNP exists between the two parental haplotypes, a seven-nucleotide insertion/deletion (InDel) was found directly upstream of the transcriptional start site of Pttg1.1 using the Sanger-Wellcome Mouse Genome Database (Fig. 3D) and was independently verified in three somatic tissues (kidney, liver, and retina) of A/J and B6/J mice (Fig. S2A). The B6/J variant appears to be nearly unique across mouse strains analyzed to date by the Sanger-Wellcome Mouse Genome project (Fig. S2B), suggesting that the variant arose recently as a deletion in the C57BL genome. The only other substrains known to carry it are C57BLKS, C57BL/6N, and C67BL/6By. Notably, microarray analysis from another recombinant inbred strain set (BXD), derived from the C57BL/6J and DBA/2J inbred strains (the latter carrying the insertion present in A/J), map a strong cis-eQTL for Pttg1 expression as well, using either whole eye mRNA or retinal mRNA, and including a probe targeting an exon sequence common to both isoforms (www.genenetwork.org; accession record ID GN207 and GN302).
Fig. 3.
Pttg1 expression is augmented by a seven-nucleotide deletion in the B6/J allelic variant. (A) Two splice variants of mouse Pttg1 have been annotated, represented here as Pttg1.1 and Pttg1.2. Orange boxes indicate exons, and thick black lines indicate introns. Location of microarray probe used in B and C, InDel variant shown in D, splice variant-specific primers used in E, and promoter cloned for F are indicated as well. (B) Whole eye microarray data across the RI strains revealed a positive correlation (r = 0.471) between Pttg1 expression and the regularity index of cholinergic amacrine cells. (C) This variation in Pttg1 expression also mapped a significant QTL (LRS = 26.0) to the location of Pttg1 on chromosome 11, indicating a putative cis-eQTL. (D) The B6/J genome contains a seven nucleotide deletion in the Pttg1 promoter, which is predicted to create an AP-1 binding motif. (E) Through qPCR, both splice variants of Pttg1 were found to be expressed in greater abundance in B6/J retina than in A/J retinas. n = number of retinas analyzed. (F) The B6/J promoter was significantly more efficient at driving a luciferase reporter than the A/J promoter in mouse fibroblast NIH3T3 and pituitary AtT20 cell lines. n = number of wells transfected. *P < 0.05 and ***P < 0.001. Mean and SE are indicated in E and F.
The Seven-Nucleotide Deletion in the B6/J Variant of the Pttg1 Promoter Increases the Transcriptional Activation of this Gene.
The cis-eQTL resulting from the microarray data suggested that Pttg1 was differentially expressed between the two parental strains. The microarray data used from the AXB/BXA strain set for this analysis, however, measured RNA levels from whole eyes; additionally, this probe failed to recognize the canonical variant Pttg1.1. We therefore assessed Pttg1 expression in adult A/J and B6/J retinas using quantitative PCR (qPCR) with variant-specific primers (Fig. 3A). We found that the canonical Pttg1 variant was expressed in considerably greater abundance than the noncanonical variant, and, confirming the microarray data, both variants showed greater expression in the B6/J retina relative to A/J retina (Fig. 3E). To assess directly whether the seven-nucleotide deletion altered transcriptional activation of Pttg1, we cloned ∼375 bp of the Pttg1 promoter, with or without the deletion (B6/J-pro and A/J-pro, respectively), upstream of a luciferase reporter gene. As predicted, the presence of the deletion in the Pttg1 promoter significantly increased transcriptional activation (∼30% induction) of the promoter in mouse fibroblast NIH3T3 cells and in pituitary AtT20 cells (Fig. 3E), implicating the InDel in either disrupting an existing regulatory element or creating a novel one, ultimately modulating homotypic spacing. To assess the requirement of Pttg1 in modulating the spacing of like-type neurons and, consequently, the regularity of the retinal mosaic, we analyzed the retinas of Pttg1 KO mice.
Pttg1 KO Mice Exhibit Disrupted Regularity of Some, but Not All, Retinal Populations.
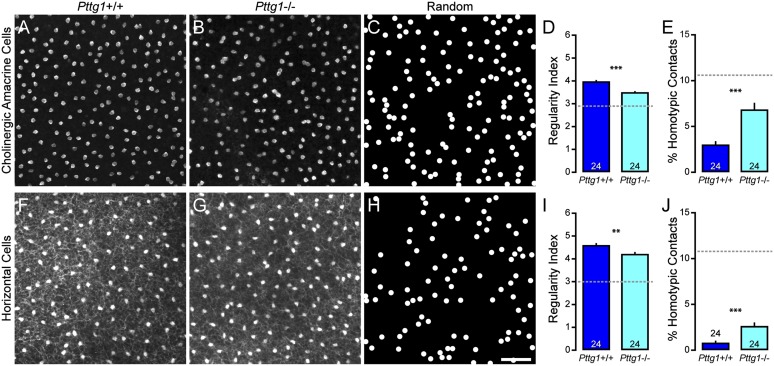
We confirmed a previous report that the architecture of the Pttg1−/− retina was normal (16), extending those findings by showing no obvious alterations in the cellular lamination of the retina or in the stratification patterns of various cell types revealed by labeling retinal sections with antibodies to choline acetyltransferase, calbindin, synaptotagmin 2, vesicular glutamate transporter 3, tyrosine hydroxylase, calsenilin, calretinin, protein kinase C, C-terminal binding protein 2, or receptor expression enhancing protein 6, or by labeling with peanut agglutinin (PNA) lectin (Fig. S3). We then assessed the mosaic organization of cholinergic amacrine cells in the INL of the Pttg1−/− retina (Fig. 4 A and B) and found a significant reduction in the regularity index compared with littermate Pttg1+/+ retinas (Fig. 4D), as well as an increase in the frequency of homotypic pairs (Fig. 4E). The regularity of the cholinergic mosaics in Pttg1−/− retinas was less than that observed in all but one of the RI strains, yet was not as irregular as observed in random distributions (Fig. 4C). Both the decrease in regularity and the increase in the frequency of homotypic pairs were of a comparable magnitude to that seen in B6.A <11> retinas (Fig. 2 A–E), yet neither the B6.A <11> retinas nor the Pttg1−/− retinas contained average densities of cholinergic amacrine cells that were significantly different from their respective WT comparison groups (B6/J = 1,308 ± 28 cells/mm2 vs. B6.A<11> = 1,306 ± 8 cells/mm2; Pttg1+/+ = 1,379 ± 30 cells/mm2 vs. Pttg1−/− = 1,375 ± 14 cells/mm2). To assess the specificity of Pttg1 in regulating the regularity of retinal mosaics, we analyzed the spatial distributions of three additional cell types situated in the INL: the horizontal cells, type 2 bipolar cells, and VGluT3+ amacrine cells. We found similar significant effects in the Pttg1−/− retina for the population of horizontal cells for both regularity and the frequency of homotypic pairs (Fig. 4 F–J). In contrast, neither the other amacrine cell examined (VGluT3+), nor the cone bipolar cell (type 2) in the Pttg1−/− retina, showed any effect on mosaic patterning or homotypic spacing (Fig. S4). The estimated total number for all four of these cell types showed minimal differences between Pttg1+/+ and Pttg1−/− retinas (Fig. S5).
Fig. 4.
Cholinergic amacrine cell and horizontal cell mosaics are disrupted in Pttg1−/− mice. (A and B) Sample mosaics of cholinergic amacrine cells in the Pttg1+/+ retina (A) and Pttg1−/− retina (B). (C) Sample of a simulated random mosaic, matched in density and constrained by soma size. (D and E) Cholinergic amacrine cell mosaics in the KO retina had reduced regularity (D) and an increased frequency of homotypic pairs (E). (F–J) Similar results were found when analyzing the population of horizontal cells. n = number of fields analyzed, with the mean and SE indicated. The broken gray lines indicate the average value for random simulations matched in density and constrained by soma size. (Scale bar, 50 µm.) **P < 0.01 and ***P < 0.001.
One feature that discriminates these four cell types is their susceptibility to Bax-mediated cell death. Both the VGluT3+ amacrine cells and the type 2 cone bipolar cells undergo extensive Bax-mediated cell death (Fig. S5 G and H), although of the two, only the VGluT3+ amacrine cell type yields a mosaic that is significantly more orderly than a random distribution (Fig. S4 D and E). By contrast, neither the cholinergic amacrine cells nor the horizontal cells undergo Bax-mediated cell death (Fig. S5 E and F). Rather, those two cell types are known to generate their mosaic patterning through tangential dispersion as they assemble their mosaics (17), doing so through the actions of Megf10 (3). We consequently asked whether Pttg1 mediates its effects on mosaic order through the activation of Megf10 in a luciferase assay. In two different cell lines (NIH3T3 and mouse neuronal RGC-5 cells), at both 24 and 48 h after transfection, we failed to find evidence for Pttg1 activating the Megf10 promoter (Fig. S6A). These results show that the reduced mosaic patterning in the Pttg1−/− retina does not appear to be a consequence of reduced Megf10 expression, indicating that Pttg1 modulates cholinergic amacrine cell spacing through a distinct molecular pathway. Indeed, neither the variation in mosaic regularity across the RI strains nor the variation in Pttg1 expression correlated with Megf10 expression (Fig. S6B). To understand whether the effects of Pttg1 are, like Megf10, operating as the cholinergic amacrine cell mosaic is being assembled, or whether its role is in the maintenance of this spacing subsequent to its establishment, we examined the regularity of the mosaic during development, toward the end of the first postnatal week, after mosaic assembly mediated by Megf10 is complete (3). The regularity of the mosaics in the Pttg1+/+ and Pttg1−/− retinas at postnatal day 6 were found to be identical (Fig. S6 C–G), and more importantly, were at a level indiscriminable from the mature Pttg1+/+ mosaics (compare with Fig. 4 D and E). Pttg1, therefore, must be acting to maintain the regular spacing of the cholinergic amacrine cells rather than to drive its initial formation.
Discussion
The patterning in the distribution of retinal neurons is a fundamental feature of retinal organization, ensuring that every location on the retina is subserved by each cell type. Although cell types vary in their degree of regularity, nearly all have been documented to be more regular than random distributions (2). The 2D spatial statistics of this patterning have been shown to be better recapitulated by simulations of cells constrained by their proximity to one another rather than by lattice-like simulations that incorporate local variation in positioning (18), suggesting that such patterning within the retina reflects solely the action of local spacing rules between neighboring cells (2). Exactly how such local spacing rules are established during development is becoming increasingly clear; although periodic cell fate assignments would, in principle, be sufficient to create orderly mosaics, several studies have now shown that mosaic cells exhibit mobility and/or die during development, suggesting that tangential dispersion or selective cell death minimizes the proximity between homotypic neighbors. Additionally, cells may also actively maintain their spacing once it is achieved during development (19). The molecular mechanisms underlying these various processes, however, are still poorly understood.
Cholinergic amacrine cells in the INL of the mouse retina display marked regularity in their distribution, the development of which has been characterized for over a decade, making them one of the most well-studied mosaics in the CNS. As newly generated cholinergic amacrine cells migrate, they exhibit no minimal spacing with their homotypic neighbors, indicating that establishment of mosaic regularity occurs only after these cells arrive and integrate into their emerging mosaics (7). Although these cells are modestly overproduced during development, the regularity of their mosaic is not improved after those extraneous cells die (being Bax independent), nor is the regularity disrupted if these cells are maintained experimentally (20, 21). The primary factor in establishing patterning of this mosaic, therefore, is thought to act through homotypic interactions that minimize proximity between like-type cells, driving them apart (2). Cholinergic amacrine cells thus provide an ideal model for discovering molecular factors required for the development and maintenance of a feature that may be common to many cell populations in the CNS.
Although selective cell death and tangential migration are established factors in the development of retinal mosaics, mechanisms of mosaic maintenance remain poorly understood. However, after initial mosaics form, particularly for early generated cells such as the cholinergic amacrine cells or horizontal cells, they must maintain their regularity as later-generated cells are produced within the enlarging retina. That mosaic regularity continues to be actively maintained after its initial formation is demonstrated by the effect of microtubule destabilizing agents in the postnatal rat retina: even after the mosaic has become established, administration of these agents yields a conspicuous collapse of mosaic spacing that can then be regained over subsequent days (19). Exactly how the microtubule network within the cytoskeleton participates in maintaining mosaic regularity remains unclear, but the present results suggest a role for Pttg1 in this maintenance function.
We found that Pttg1 exerts its effects in a cell type-dependent manner, as it plays a role in minimizing homotypic contact within the cholinergic amacrine cell and horizontal cell mosaics, but not within the VGluT3+ amacrine cell or type 2 bipolar cell mosaics. One distinguishing feature of the VGluT3+ amacrine cell and type 2 bipolar cell populations is that each undergoes abundant overproduction during development that is subsequently pared down through Bax-mediated apoptosis, producing a mosaic with little more regularity (if any) relative to a random distribution of cells. The same has been shown for dopaminergic amacrine cells, a cell type that does not evidence tangential dispersion during development (17), but rather undergoes substantial Bax-mediated cell death (22), transforming a random distribution of cells into one with a marginally more regular distribution (4). In contrast, cholinergic amacrine cells and horizontal cells do not use selective cell death as a mechanism to increase mosaic regularity during development while both disperse tangentially via Megf-mediated homotypic interactions in the process of establishing their regularity (6, 23, 24). Perhaps the very cell types that use tangential dispersion during development to produce this regularity become dependent on a Pttg1 signaling pathway to maintain it. Pttg1 is expressed throughout retinal development (14) and by multiple types of retinal cells in maturity and at postnatal day 7 (3, 15), with cholinergic amacrine cells being one of the most heavily expressing cells. Because Pttg1 differentially regulates gene expression through its interaction with different cofactors (13), the specificity in its biological role at a particular postnatal stage, and within only particular types of retinal cells, may be due to the temporally restricted presence of these cofactors in selective cell types.
The variant of the Pttg1 promoter found in the B6/J mouse strain sheds light on potential regulatory mechanisms for Pttg1 expression, suggesting a molecular pathway that may control mosaic maintenance. This seven-nucleotide deletion in the B6/J Pttg1 promoter significantly increases its overall expression in the retina, potentially through the creation of a novel activator protein 1 (AP-1) binding motif (Fig. 3C), a transcriptional activator. AP-1 is heterodimeric protein complex made up of proteins in the Jun and Fos families, well-characterized oncogenes, and acts as a signal transducer by responding to a variety of extracellular stimuli and subsequently regulating gene expression. These genes have been shown to be involved in several cellular processes including proliferation, apoptosis, and migration (25–27). We suggest that the AP-1 complex is activated in cholinergic amacrine cells, increasing Pttg1 expression and its downstream signaling required for maintaining the spacing between neighboring cells. Indeed, Pttg1 overexpression has been positively correlated with tumor invasiveness and the protein has been implicated in activation of other proteins intimately involved in cell motility, such as those of the Akt and Rho families (28–31). While the molecular pathways involved remain to be determined, the expression of Pttg1 is clearly linked to the maintenance of orderly mosaics, regulating the spacing of homotypic neighbors.
Methods
Mice and Tissue Collection.
C57BL/6J, A/J, and B6.A<11> mice were obtained from the Jackson Laboratory, as were mice from 25 strains of the AXB/BXA RI strain set. Pttg1−/− mice were derived as previously described (32) and backcrossed to C57BL/6J for more than 10 generations. Despite such repeated backcrossing, a small region around the Pttg1 gene on chromosome 11 at 43 Mb will still retain the haplotype of the 129 strain ES cells. We analyzed the haplotypes of A, 129, and C57BL/6 genomes and found the congenic interval (between A and 129) likely to extend from about 37–58 Mb. Bax−/− mice were originally obtained from the Jackson Laboratory and were subsequently backcrossed onto a C57BL/6J background for more than 10 generations. Mice were deeply anesthetized and perfused with fixative, and the retinas were subsequently dissected, as described in further detail in SI Methods.
Immunofluorescence.
Cholinergic amacrine cells, horizontal cells, type 2 cone bipolar cells, and VGluT3+ amacrine cells were labeled in adult whole mount preparations with primary antibodies raised against choline acetyltransferase, calbindin, synaptotgmin 2, and vesicular glutamate transporter 3, respectively. Various cell types and synaptic structures were labeled in adult retinal sections with 11 primary antibodies, as well as a PNA lectin conjugated to a fluorescent Alexa Fluor dye (1:1,000; Life Technologies). To identify cholinergic amacrine cells in the INL of postnatal day 6 retinas, rabbit-anti-SRY-box2 primary antibodies were used (33). Primary antibodies were visualized using secondary antibodies raised in donkey against goat, rabbit, mouse, sheep, or guinea pig IgG and conjugated to fluorescent Alexa Fluor (1:200; Life Technologies) or cyanine dyes (1:200; Jackson ImmunoReasearch Labs). Further details are provided in SI Methods, and a list of antibodies is provided in Table S2.
Imaging and Spatial Analysis.
For the B6/J and A/J parental strains, AXB/BXA RI strains, and B6.A <11> chromosome substitution strain, the population of cholinergic amacrine cells in the INL was visualized using an Olympus BH2 fluorescence microscope equipped with a video camera and stage encoders. Four central and four peripheral fields were sampled for each retina (Fig. S1A), and the position of each cholinergic amacrine cell was recorded using BioQuant Nova software (BIOQUANT Image Analysis). For Pttg1+/+ and Pttg1−/− mice, confocal image stacks were taken of each cell population using an Olympus FV1000 laser scanning confocal microscope following the same sampling protocol. Cell position was determined using ImageJ image analysis software (rsbweb.nih.gov/ij/). For each field, the regularity and intercellular spacing were assessed using custom software designed in Matlab (MathWorks). Similar analyses were conducted for the mosaics of cholinergic amacrine cells, the horizontal cells, the VGluT3+ amacrine cells, and the type 2 cone bipolar cells in Pttg1+/+ and Pttg1−/− retinas, as described in further detail in SI Methods.
Quantification of Cell Number.
The total number of cholinergic amacrine cells, horizontal cells, VGluT3+ amacrine cells, and type 2 bipolar cells was estimated by determining an average density of each cell type and then multiplying by total retinal area for Bax+/+ and Bax−/− retinas, as well as for Pttg1+/+ and Pttg1−/− retinas, as described in detail in SI Methods.
QTL Mapping and Bioinformatics.
Interval mapping was conducted using the regularity index for each RI strain, with the mapping module in GeneNetwork (www.genenetwork.org; accession record ID #10259). All megabase positions are relative to C57BL6/J genome, assembly NCBI37/mm9. Permutation testing was performed to determine statistical significance of any QTL, generating a suggestive (P = 0.67) and significant (P = 0.05) threshold, indicated by the gray and pink horizontal lines, respectively, in Figs. 1B and 3C. Genes within the interval were further interrogated as described in SI Methods.
qPCR and Luciferase Analyses.
Sets of retinas from six B6/J and four A/J adult mice were collected under RNase-free conditions, from which total RNA was extracted using an RNeasy Plus Mini Kit (Qiagen). Single-strand cDNA was synthesized using an iScript cDNA Synthesis Kit (Bio-Rad), and the levels of Pttg1 expression were measured using real-time qPCR using a MyiQ Single Color Real-Time PCR Detection System (Bio-Rad). All samples were run simultaneously and in triplicate, and the abundance of RNA was corrected for PCR product size, primer melting temperature, and primer efficiency (34). Levels of Pttg1 expression were normalized to the geometric mean of three internal control genes. All primers used for qPCR are listed in SI Methods.
Two different mouse Pttg1 promoters, B6/J-pro and A/J-pro (Table S3), were synthesized by GENEWIZ and subsequently cloned into a pGL3-Basic luciferase reporter vector (Promega). The mouse Megf10 promoter (−1,271/+30; Table S3) was synthesized by GENEWIZ and subsequently cloned into pGL3-Basic luciferase reporter vector, whereas mouse Pttg1 cDNA was subcloned into pcDNA3.1 vector (Invitrogen). Transfections were performed in triplicate wells and repeated twice for reproducibility. Further details are described in SI Methods.
Statistics.
All statistical comparisons between groups were made using Student two-tailed t tests. For those statistical comparisons including total cell number, regularity index, and homotypic pairs, all data collection was conducted blind to condition.
Supplementary Material
Acknowledgments
This research was supported by the National Institutes of Health Grants EY19968 (to B.E.R.), CA75979 (to S.M.), and EY21200 (to L.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The regularity index data for each strain reported in this paper have been deposited in GeneNetwork (www.genenetwork.org) in the AXB/BXA Published Phenotype Data Set, accession no. 10259. The gene expression data referred to herein are available in GeneNetwork in their respective databases, accession nos. GN207, GN210, and GN302.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1323543111/-/DCSupplemental.
References
- 1.Wässle H, Riemann HJ. The mosaic of nerve cells in the mammalian retina. Proc R Soc Lond B Biol Sci. 1978;200(1141):441–461. doi: 10.1098/rspb.1978.0026. [DOI] [PubMed] [Google Scholar]
- 2.Reese BE. Mosaics, tiling and coverage by retinal neurons. In: Masland RH, Albright T, editors. The Senses: Vision. Oxford, UK: Elsevier; 2008. pp. 439–456. [Google Scholar]
- 3.Kay JN, Chu MW, Sanes JR. MEGF10 and MEGF11 mediate homotypic interactions required for mosaic spacing of retinal neurons. Nature. 2012;483(7390):465–469. doi: 10.1038/nature10877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raven MA, Eglen SJ, Ohab JJ, Reese BE. Determinants of the exclusion zone in dopaminergic amacrine cell mosaics. J Comp Neurol. 2003;461(1):123–136. doi: 10.1002/cne.10693. [DOI] [PubMed] [Google Scholar]
- 5.Keeley PW, et al. Neuronal clustering and fasciculation phenotype in Dscam- and Bax-deficient mouse retinas. J Comp Neurol. 2012;520(7):1349–1364. doi: 10.1002/cne.23033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galli-Resta L, Resta G, Tan SS, Reese BE. Mosaics of islet-1-expressing amacrine cells assembled by short-range cellular interactions. J Neurosci. 1997;17(20):7831–7838. doi: 10.1523/JNEUROSCI.17-20-07831.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galli-Resta L, Novelli E, Volpini M, Strettoi E. The spatial organization of cholinergic mosaics in the adult mouse retina. Eur J Neurosci. 2000;12(10):3819–3822. doi: 10.1046/j.1460-9568.2000.00280.x. [DOI] [PubMed] [Google Scholar]
- 8.Farajian R, Raven MA, Cusato K, Reese BE. Cellular positioning and dendritic field size of cholinergic amacrine cells are impervious to early ablation of neighboring cells in the mouse retina. Vis Neurosci. 2004;21(1):13–22. doi: 10.1017/s0952523804041021. [DOI] [PubMed] [Google Scholar]
- 9.Whitney IE, Keeley PW, Raven MA, Reese BE. Spatial patterning of cholinergic amacrine cells in the mouse retina. J Comp Neurol. 2008;508(1):1–12. doi: 10.1002/cne.21630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilad Y, Rifkin SA, Pritchard JK. Revealing the architecture of gene regulation: The promise of eQTL studies. Trends Genet. 2008;24(8):408–415. doi: 10.1016/j.tig.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zou H, McGarry TJ, Bernal T, Kirschner MW. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science. 1999;285(5426):418–422. doi: 10.1126/science.285.5426.418. [DOI] [PubMed] [Google Scholar]
- 12.Pei L, Melmed S. Isolation and characterization of a pituitary tumor-transforming gene (PTTG) Mol Endocrinol. 1997;11(4):433–441. doi: 10.1210/mend.11.4.9911. [DOI] [PubMed] [Google Scholar]
- 13.Vlotides G, Eigler T, Melmed S. Pituitary tumor-transforming gene: Physiology and implications for tumorigenesis. Endocr Rev. 2007;28(2):165–186. doi: 10.1210/er.2006-0042. [DOI] [PubMed] [Google Scholar]
- 14.Blackshaw S, et al. Genomic analysis of mouse retinal development. PLoS Biol. 2004;2(9):E247. doi: 10.1371/journal.pbio.0020247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siegert S, et al. Transcriptional code and disease map for adult retinal cell types. Nat Neurosci. 2012;15(3):487–495, S1–S2. doi: 10.1038/nn.3032. [DOI] [PubMed] [Google Scholar]
- 16.Yetemian RM, Craft CM. Characterization of the pituitary tumor transforming gene 1 knockout mouse retina. Neurochem Res. 2011;36(4):636–644. doi: 10.1007/s11064-010-0334-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reese BE, Necessary BD, Tam PP, Faulkner-Jones B, Tan SS. Clonal expansion and cell dispersion in the developing mouse retina. Eur J Neurosci. 1999;11(8):2965–2978. doi: 10.1046/j.1460-9568.1999.00712.x. [DOI] [PubMed] [Google Scholar]
- 18.Hore VR, Troy JB, Eglen SJ. Parasol cell mosaics are unlikely to drive the formation of structured orientation maps in primary visual cortex. Vis Neurosci. 2012;29(6):283–299. doi: 10.1017/S0952523812000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galli-Resta L, Novelli E, Viegi A. Dynamic microtubule-dependent interactions position homotypic neurones in regular monolayered arrays during retinal development. Development. 2002;129(16):3803–3814. doi: 10.1242/dev.129.16.3803. [DOI] [PubMed] [Google Scholar]
- 20.Galli-Resta L, Novelli E. The effects of natural cell loss on the regularity of the retinal cholinergic arrays. J Neurosci. 2000;20(3):RC60. doi: 10.1523/JNEUROSCI.20-03-j0005.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Resta V, Novelli E, Di Virgilio F, Galli-Resta L. Neuronal death induced by endogenous extracellular ATP in retinal cholinergic neuron density control. Development. 2005;132(12):2873–2882. doi: 10.1242/dev.01855. [DOI] [PubMed] [Google Scholar]
- 22.Whitney IE, Raven MA, Ciobanu DC, Williams RW, Reese BE. Multiple genes on chromosome 7 regulate dopaminergic amacrine cell number in the mouse retina. Invest Ophthalmol Vis Sci. 2009;50(5):1996–2003. doi: 10.1167/iovs.08-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reese BE, Harvey AR, Tan SS. Radial and tangential dispersion patterns in the mouse retina are cell-class specific. Proc Natl Acad Sci USA. 1995;92(7):2494–2498. doi: 10.1073/pnas.92.7.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raven MA, Stagg SB, Nassar H, Reese BE. Developmental improvement in the regularity and packing of mouse horizontal cells: Implications for mechanisms underlying mosaic pattern formation. Vis Neurosci. 2005;22(5):569–573. doi: 10.1017/S095252380522504X. [DOI] [PubMed] [Google Scholar]
- 25.Mechta-Grigoriou F, Gerald D, Yaniv M. The mammalian Jun proteins: Redundancy and specificity. Oncogene. 2001;20(19):2378–2389. doi: 10.1038/sj.onc.1204381. [DOI] [PubMed] [Google Scholar]
- 26.Jochum W, Passegué E, Wagner EF. AP-1 in mouse development and tumorigenesis. Oncogene. 2001;20(19):2401–2412. doi: 10.1038/sj.onc.1204389. [DOI] [PubMed] [Google Scholar]
- 27.Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002;4(5):E131–E136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 28.Heaney AP, et al. Expression of pituitary-tumour transforming gene in colorectal tumours. Lancet. 2000;355(9205):716–719. doi: 10.1016/S0140-6736(99)10238-1. [DOI] [PubMed] [Google Scholar]
- 29.Wondergem B, et al. Expression of the PTTG1 oncogene is associated with aggressive clear cell renal cell carcinoma. Cancer Res. 2012;72(17):4361–4371. doi: 10.1158/0008-5472.CAN-11-2330. [DOI] [PubMed] [Google Scholar]
- 30.Ito T, et al. Pituitary tumor-transforming 1 increases cell motility and promotes lymph node metastasis in esophageal squamous cell carcinoma. Cancer Res. 2008;68(9):3214–3224. doi: 10.1158/0008-5472.CAN-07-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoon CH, et al. PTTG1 oncogene promotes tumor malignancy via epithelial to mesenchymal transition and expansion of cancer stem cell population. J Biol Chem. 2012;287(23):19516–19527. doi: 10.1074/jbc.M111.337428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Z, Moro E, Kovacs K, Yu R, Melmed S. Pituitary tumor transforming gene-null male mice exhibit impaired pancreatic beta cell proliferation and diabetes. Proc Natl Acad Sci USA. 2003;100(6):3428–3432. doi: 10.1073/pnas.0638052100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fischer AJ, Zelinka C, Scott MA. Heterogeneity of glia in the retina and optic nerve of birds and mammals. PLoS ONE. 2010;5(6):e10774. doi: 10.1371/journal.pone.0010774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vandesompele J, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.