Significance
The canonical degradation pathway for plasma membrane proteins is through endocytosis and targeting to lysosomes. In some cases, plasma membrane proteins undergo retrograde trafficking to the endoplasmic reticulum (ER) for extraction through an endoplasmic reticulum-associated degradation pathway. We identified a novel proteasome-mediated pathway for the degradation of a plasma membrane iron transporter, ZIP14. Extraction and degradation of ZIP14 occurred after endocytosis, but did not occur by retrograde trafficking to the ER. Both ubiquitination and deglycosylation of ZIP14 were identified prior to degradation. To our knowledge, ZIP14 is the first identified multispanning plasma membrane protein to undergo such a pathway. In addition, we showed that membrane extraction is regulated by cellular iron content, providing insight into the disorders of iron metabolism.
Keywords: SLC39A14, hereditary hemochromatosis
Abstract
Protein degradation is instrumental in regulating cellular function. Plasma membrane proteins targeted for degradation are internalized and sorted to multivesicular bodies, which fuse with lysosomes, where they are degraded. ZIP14 is a newly identified iron transporter with multitransmembrane domains. In an attempt to dissect the molecular mechanisms by which iron regulates ZIP14 levels, we found that ZIP14 is endocytosed, extracted from membranes, deglycosylated, and degraded by proteasomes. This pathway did not depend on the retrograde trafficking to the endoplasmic reticulum and thus did not involve the well-defined endoplasmic reticulum-associated protein degradation pathway. Iron inhibited membrane extraction of internalized ZIP14, resulting in higher steady-state levels of ZIP14. Asparagine-linked (N-linked) glycosylation of ZIP14, particularly the glycosylation at N102, was required for efficient membrane extraction of ZIP14 and therefore is necessary for its iron sensitivity. These findings highlight the importance of proteasomes in the degradation of endocytosed plasma membrane proteins.
Zrt/IRT-like protein 14 (ZIP14) is a metal transporter containing multiple transmembrane domains encoded by the gene, SLC39A14 (1, 2). It mediates both nontransferrin-bound and transferrin-bound iron uptake into cells (1, 3–5). In keeping with its functions, ZIP14 is localized at the plasma membrane and in transferrin-containing endosomal compartments (1, 2, 4). Recent studies demonstrate that the level of ZIP14 protein is increased in the liver of rats fed a high iron diet and in iron-loaded human hepatoma cells, suggesting that ZIP14 contributes to tissue iron loading under high-iron conditions (6). A tissue expression array shows that ZIP14 mRNA is ubiquitously expressed with high levels in the liver, pancreas, and heart (2). These tissues accumulate most of the iron in disorders with iron overload, such as hereditary hemochromatosis, resulting in liver cirrhosis, diabetes, and heart failure (7, 8). The most prevalent form of hereditary hemochromatosis is due to mutations in HFE (9). We have shown that overexpression of HFE in HepG2 cells decreases iron and ZIP14 levels (3). Moreover, HFE expression decreases the stability of ZIP14, suggesting the involvement of protein degradation and intracellular iron in ZIP14 regulation.
Protein degradation is a very effective way to regulate cellular function. Plasma membrane proteins targeted for degradation are internalized as vesicles and transported to early endosomal compartments where they are sorted to multivesicular bodies, which in turn fuse with lysosomes (10–12). Ubiquitin often plays a critical role in this pathway because ubiquitin modification of the target protein serves as an essential determinant for endocytosis (13). Tight regulation of the degradation of endocytosed plasma membrane proteins enables cells to adapt to their environment. In cases of nutrient receptors, such as the transferrin receptor 1 (TfR1), ubiquitination is able to reroute TfR1 from the recycling pathway to lysosomal degradation (14). Recently, Tachiyama and colleagues showed that increased cellular iron stimulates the ubiquitination and promotes lysosomal degradation of TfR1 (15). Consequently, reduced TfR1 limits further iron accumulation. The low-density lipoprotein receptor (LDLR) is another example of a receptor controlled by the nutrient that it transports. Under conditions of elevated intracellular cholesterol, the transcription of the E3 ubiquitin ligase, inducible degrader of the LDLR, is up-regulated, leading to increased ubiquitination and lysosomal degradation of LDLR (16–19). As a result, cells limit further uptake of LDL cholesterol.
In contrast, our present study demonstrates that iron deficiency promotes the degradation of ZIP14 and that the degradation of ZIP14 is mediated by proteasomes, rather than lysosomes. The proteasomal degradation of plasma membrane ZIP14 was through a pathway that involves endocytosis, membrane extraction, and deglycosylation. This pathway did not depend on the retrograde trafficking to the endoplasmic reticulum (ER). Mechanistically, we showed that iron supplementation prevents the extraction of ZIP14 into the cytosol and thus prevents its proteasomal destruction. Surprisingly, glycosylation at asparagine 102 (N102) was necessary for the membrane extraction and iron sensitivity of ZIP14.
Results
ZIP14 Is Down-Regulated by Iron Deficiency and Up-regulated by Iron Overload.
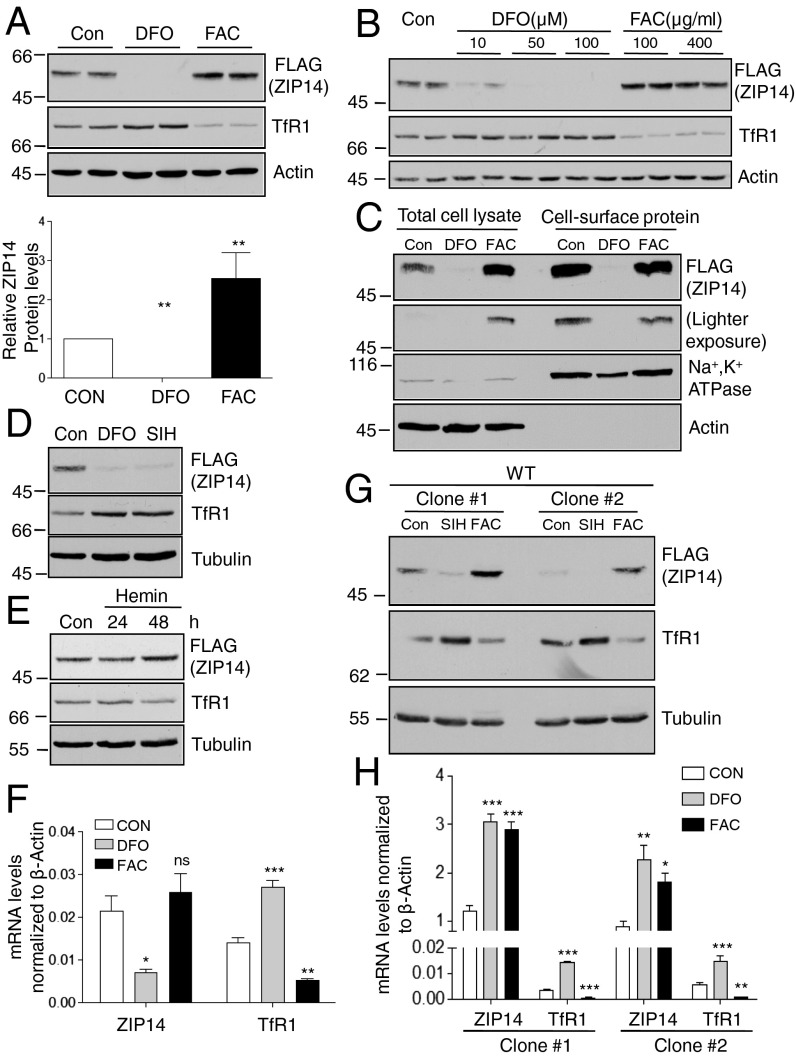
To determine the mechanisms by which iron regulates ZIP14, we used a HepG2 cell line where recombination was used to insert a FLAG epitope onto the C terminus of endogenous ZIP14 (HepG2-ZIP14 cells) (4). Thus, the endogenous gene-regulatory machinery remains intact. We found that iron depletion by desferrioxamine (DFO) abolished detectable ZIP14 protein, whereas iron supplementation with ferric ammonium citrate (FAC) increased its level (Fig. 1 A and B) similar to what was previously reported (3–5). TfR1 was used as a control for the iron loading of cells. Under high-intracellular-iron conditions TfR1 is down-regulated to prevent further uptake of iron-laden Tf, and under low-iron conditions TfR1 levels increase. Cell-surface proteins were biotinylated with biotin disulfide N-hydroxysulfosuccinimide ester (NHS-SS-biotin). Isolation of biotinylated proteins revealed that FAC did not increase the amount of ZIP14 on the cell surface, suggesting that iron supplementation raised the amount of intracellular ZIP14 (Fig. 1C). The enrichment of Na+, K+ ATPase and the lack of detectable actin in the biotin-labeled fractions indicate that biotin labeled the plasma membrane proteins and that the cells remained intact during the labeling procedure (Fig. 1C). To assess the role of intracellular iron in the regulation of ZIP14, a cell-membrane–permeable iron chelator, salicylaldehyde isonicotinyl hydrazone (SIH), and an iron-containing porphyrin, hemin, were used to iron-deplete and to iron-load cells, respectively. Hemin was used to bypass iron-mediated iron uptake through ZIP14. Similar to DFO treatment, incubation with SIH down-regulated ZIP14 (Fig. 1D). Hemin increased ZIP14 and reduced TfR1. The down-regulation of TfR1 indicated that the cells were iron-loaded (Fig. 1E). The latter is expected because hemin is degraded by heme oxygenase in the cytoplasm, releasing iron, and because intracellular iron decreases the stability of TfR1 mRNA (20). These results suggest that intracellular iron but not ZIP14-mediated iron transport regulates ZIP14 levels in HepG2 cells. ZIP14 mRNA levels decreased about 60% after iron depletion and had a trend of slight increase after iron supplementation (Fig. 1F). However, the protein levels of ZIP14 were undetectable with iron chelation and increased about twofold with iron supplementation (Fig. 1A). The differences in ZIP14 mRNA levels were unable to explain the large differences in ZIP14 protein seen in immunoblots (Fig. 1 A and F).
Fig. 1.
Iron deficiency decreases cellular levels of ZIP14, whereas iron overload increases ZIP14. HepG2-ZIP14 cells were used in A–F. (A) Immunoblots of ZIP14 by using FLAG antibody after cells were treated with DFO (100 μM) or FAC (100 µg/mL) for 24 h. The intensities of ZIP14 bands were quantified by Image J and presented as the relative levels to controls (Lower). Con: control. (B) Immunoblots of ZIP14 after cells were treated for 24 h with DFO or FAC. (C) Cells were treated with DFO (100 μM) or FAC (100 µg/mL) for 24 h, and then cell-surface proteins were labeled with NHS-SS-biotin. Samples were analyzed by immunoblotting. Blots were probed for P-type Na+, K+ ATPase as a marker for plasma membrane proteins and actin as a control for cytosolic proteins. (D) Cells were treated with 100 µM DFO and 100 µM SIH or (E) 50 µM hemin to alter intracellular iron. Immunoblotting was performed to examine ZIP14 levels. (F) ZIP14 and TfR1 mRNA after DFO or FAC treatment were quantified by qRT-PCR (n = 3 independent experiments). CON: control. HEK293 cells stably expressing ZIP14-FLAG were used afterward. Clone #1 and clone #2 indicate different stable cell clones (Table S1). Cells were treated with 100 µM SIH or 25 µg/mL FAC for 24 h. (G) Immunoblots of ZIP14 and TfR1. (H) ZIP14 and TfR1 mRNA were analyzed by qRT-PCR (n = 3 independent experiments). *P < 0.05, **P < 0.001, ***P < 0.0001, compared with control.
To further investigate the mechanisms by which ZIP14 protein is controlled, we generated stably transfected HEK293 cells, expressing ZIP14 with a C-terminal FLAG epitope and lacking the 5′ and 3′ untranslated region of ZIP14 mRNA (HEK293-ZIP14 cells, Table S1). We observed that iron depletion reduced, whereas iron repletion increased, ZIP14 protein (Fig. 1G), indicating that introduction of the ZIP14 transcript under an exogenous promoter has little discernible impact on ZIP14 regulation by iron. ZIP14 mRNA levels measured by specific amplification of the ZIP14-FLAG sequence failed to correlate with intracellular iron levels (Fig. 1H), reinforcing the importance of the regulation of ZIP14 protein by iron. TfR1 was used as an indicator of intracellular iron levels. Its mRNA and, consequently, its protein levels increase with iron deficiency and decrease with iron loading (20). Iron depletion resulted in a decrease in ZIP14 protein independent of changes in ZIP14 mRNA levels, suggesting that elevated protein degradation is involved in the down-regulation of ZIP14.
Iron Deficiency Promotes the Proteasomal Degradation of ZIP14.
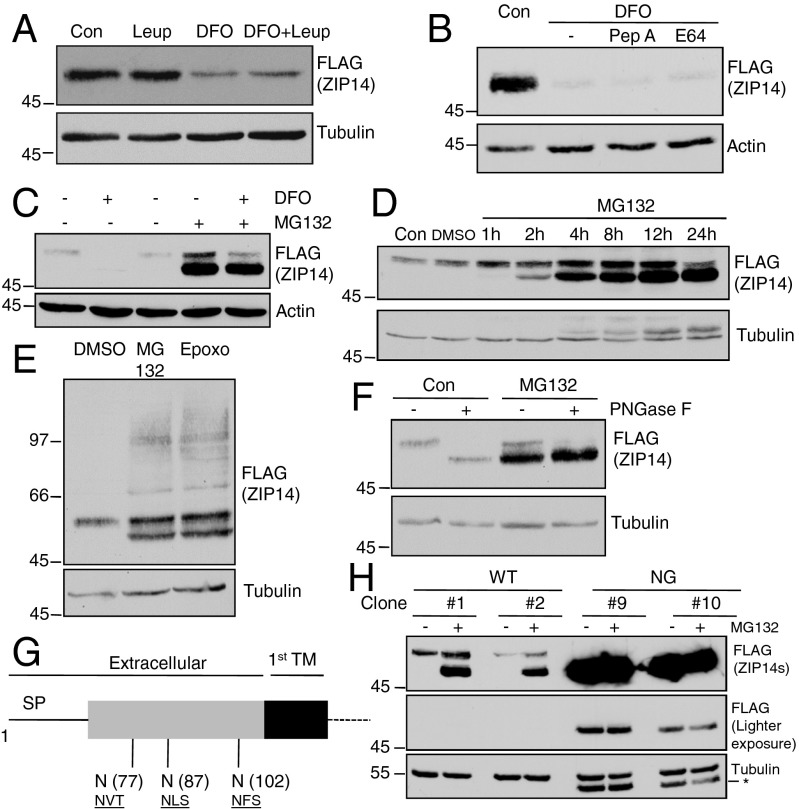
Inhibitors of two main eukaryotic proteolytic systems, lysosomal or proteasomal degradation, were used to examine whether the reduced ZIP14 after iron depletion was due to increased protein degradation. Leupeptin, pepstatin A, and E64, which block protein degradation in lysosomes, did not prevent the iron chelation-induced ZIP14 down-regulation (Fig. 2 A and B). Inhibitors that block lysosomal degradation of proteins by interfering with the acidification of vesicles, such as bafilomycin A, were not used because they inhibit iron uptake into cells. In contrast, the proteasomal inhibitor MG132 abolished the down-regulation of ZIP14 in iron-depleted HepG2 cells (Fig. 2C). Inhibition of proteasomal activity also increased the steady-state level of ZIP14 within an hour (Fig. 2D). Strikingly, MG132 treatment led to a rapid accumulation of a lower molecular weight (MW) ZIP14 species detected in immunoblots (Fig. 2 C and D). To further confirm the involvement of proteasomes in ZIP14 degradation, epoxomicin (Epoxo), another selective proteasome inhibitor, was used. Similar to MG132 treatment, Epoxo treatment enhanced the amount of mature ZIP14 (Fig. 2E, Upper) and resulted in an accumulation of the lower MW ZIP14 species (Fig. 2E). We also observed that both MG132 and Epoxo treatment resulted in a high molecular smear detected by anti-FLAG antibody (Fig. 2E), suggesting that protein ubiquitination is involved. Further analysis by immunoprecipitation of ZIP14 followed by detection with anti-ubiquitin antibody revealed that iron depletion increased ubiquitinated ZIP14, which is consistent with increased proteasomal degradation (Fig. S1).
Fig. 2.
ZIP14 degradation induced by iron deficiency is proteasome-dependent and is preceded by deglycosylation. HepG2-ZIP14 cells were used in A–F. (A) Cells were incubated with 100 μM DFO, DMSO (as vehicle-treated control), and 10 μM leupeptin (Leup) individually or in combination with DFO for 12 h. ZIP14 levels were analyzed by immunoblotting. (B) Cells were incubated with DFO, DMSO, 10 μM pepstatin A (Pep A), or 10 μM E64 in combination with DFO for 24 h, and ZIP14 levels were analyzed by immunoblotting. (C) Cells were incubated with DFO, DMSO (as vehicle-treated control), and 10 μM MG132 individually or in combination with DFO for 12 h. ZIP14 levels were analyzed by immunoblotting. (D) The time-course effect of MG132 was examined. (E) Cells were treated with epoxomicin (Epoxo, 10 μM). (F) Lysates collected from DMSO-treated (Con) or MG132-treated cells were digested with PNGase F. (G) Diagram of ZIP14 glycosylation sites on its extracellular amino terminus. SP: signal peptide; TM: transmembrane domain. (H) HEK293 stable transgenic cells were treated with MG132 (12 h) before harvesting for immunoblotting. Numbers with “#” indicate different stable cell clones (Table S1). An asterisk (*) indicates unstripped signal from anti-FLAG antibody. Note that the endogenous ZIP14-3XFLAG travels with a higher MW than that of the ZIP14-FLAG-Myc construct transfected into HEK293 cells.
The identity of the lower MW ZIP14 species stimulated by proteasomal inhibition was tested by treating the cell lysates with the N-linked deglycosidase, PNGase F. PNGase F treatment converted wild-type (WT) ZIP14 to a deglycosylated form (DG) and did not change the electrophorectic mobility of the lower MW species generated by MG132 treatment of cells (Fig. 2F). The finding that newly accumulated ZIP14 species after proteasomal inhibition had a similar electrophoretic mobility of the DG ZIP14 and was insensitive to PNGase F suggested that the lower MW species is a DG ZIP14 (Fig. 2F). Analysis of the amino acid sequence of ZIP14 for consensus sequences for N-linked glycosylation indicated three potential sites (N77, N87, and N102) all located in the extracellular N terminus (Fig. 2G). To identify which consensus sequences were used, each Asn was replaced by Ala (A) individually through site-directed mutagenesis. In addition, a mutant lacking all three N-linked glycosylation sites was generated. The mutated ZIP14s were examined first by transient transfection of each mutant individually into HEK293 cells and then by establishing HEK293 cell lines that stably express each mutant (Table S1). Immunoblot analysis of the cell lysates was performed to assess the electrophoretic mobilities of WT and single-mutant ZIP14s. In cells transiently transfected with ZIP14, two species were detected: the higher MW species represented the glycosylated form and a minor lower MW species had a similar mobility as DG ZIP14. PNGase F digestion of the lysates from WT ZIP14-transfected cells resulted in the disappearance of the higher MW ZIP14 species and an accumulation of the lower MW ZIP14 species that corresponds to the DG ZIP14 (Fig. S2 A–C). Moreover, the higher MW species of the single Asn mutants (N77A, N87A, or N102A) had a lower MW than that of the WT ZIP14, suggesting that each site was used for glycosylation (Fig. S2A). Only one band representing the glycosylated ZIP14 was detected in stable HEK293 cell lines expressing WT and Asn-to-Ala mutants. We also observed that single replacement of Asn with Ala at positions 77, 87, and 102 led to a decrease in the MW of the mature ZIP14 (Fig. S2B), indicating that each site is glycosylated. ZIP14 from WT cell lysates shifted to a lower MW DG species after PNGase F digestion, indicating removal of N-linked oligosaccharides. This lower MW species had a similar electrophorectic mobility as the nonglycosylated (NG) ZIP14 where all three N-linked glycosylation sites were mutated. Moreover, PNGase F treatment did not result in any shift in MW of the NG ZIP14, confirming that the NG ZIP14 has no other N-linked oligosaccharides (Fig. S2C). HEK293-ZIP14 cells preincubated with MG132 for 2 h resulted in an increase of the steady-state levels of the mature ZIP14 and a robust enhancement of the lower MW species, whereas HEK293-NG-ZIP14 cells similarly treated failed to increase the amount of ZIP14 or to initiate an accumulation of an even lower MW species, demonstrating that the lower MW species induced by proteasomal inhibition indeed represents the DG ZIP14 rather than a partially cleaved form of ZIP14 (Fig. 2H). Taken together, these results indicated that ZIP14 is degraded through a proteasomal rather than a lysosomal pathway and that ZIP14 undergoes deglycosylation before proteasomal processing.
Endocytosis Is Required for the Degradation of Plasma Membrane ZIP14.
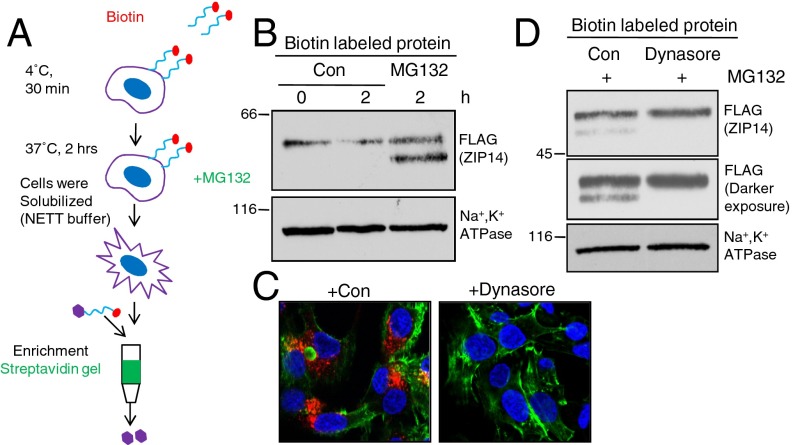
Plasma membrane proteins are degraded as part of a quality control mechanism to allow only properly folded proteins to exit the ER or as a regulatory mechanism to downregulate ion transport, signaling or cell–cell communication. Deglycosylation of ZIP14 could occur in either pathway. To examine whether ZIP14 deglycosylation happens after it reaches the plasma membrane, we used a biotin pulse-chase experiment to examine the fate of the cell surface ZIP14. Cell surface proteins were biotinylated with an impermeable form of biotin, followed by warming of the cells to 37 °C to allow internalization. Streptavidin beads were used to isolate the biotinylated proteins (Fig. 3A). Inhibition of proteasomal activity by MG132 prevented the degradation of plasma membrane ZIP14 and resulted in an accumulation of the biotin-labeled DGZIP14, suggesting that the deglycosylation step happens after and not before ZIP14 reaches the plasma membrane (Fig. 3B). To determine whether endocytosis is required before ZIP14 deglycosylation, cells were pretreated with dynasore, an inhibitor of endocytosis (21, 22). Consistent with inhibition of clathrin-dependent endocytosis, the uptake of a control protein, transferrin, was significantly inhibited after dynasore treatment (Fig. 3C). Importantly, pretreatment with dynasore strongly suppressed the MG132-induced accumulation of the DG ZIP14, indicating that ZIP14 endocytosis is required before the deglycosylation of ZIP14 (Fig. 3D).
Fig. 3.
Plasma membrane ZIP14 is internalized before proteasomal processing. HepG2-ZIP14 cells were used. (A) Schematic illustration of experiment. Cell-surface proteins were biotinylated at 4 °C for 30 min, and cells were then treated with 10 μM MG132 in complete medium at 37 °C for 2 h. Cell lysates were collected and cell-surface proteins were isolated by streptavidin gel. (B) Cell-surface proteins were labeled with cell-membrane–impermeable NHS-SS-biotin at 4 °C and then chased for 2 h at 37 °C with or without the addition of MG132. (C) Cells were incubated with or without dynasore (80 µM) for 15 min followed by the addition of transferrin. After 30 min the cells were placed on ice, washed extensively to remove unbound surface transferrin, and fixed with paraformaldahyde. Cells stained for transferrin (red), actin (green), and nuclei (blue). (D) Immunoblots of cell-surface protein after preincubation with dynasore.
Iron Prevents Internalized ZIP14 from Being Extracted into the Cytosol.
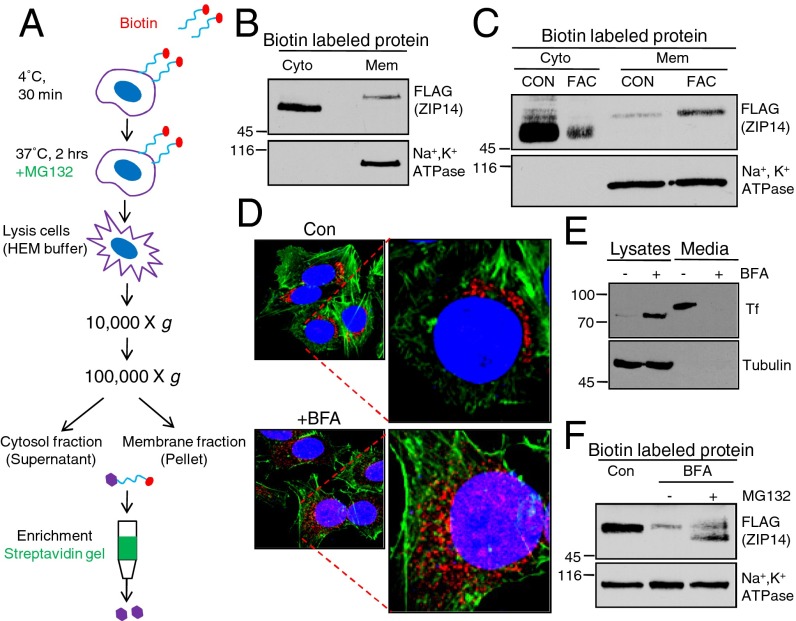
Membrane proteins can undergo retrograde transport to the ER and degradation through the endoplasmic reticulum-associated protein degradation (ERAD) pathway (23, 24). Degradation of membrane proteins in the ER by proteasomes is preceded by extraction of the protein from the membrane, followed by deglycosylation. We analyzed whether the internalized and deglycosylated ZIP14 was still associated with cellular membranes (Fig. 4A). Interestingly, the DG ZIP14 induced by MG132 was detected only in the cytosolic fraction, whereas the mature form of ZIP14 was associated mainly with cellular membranes (Fig. 4 B and C), suggesting that ZIP14 is internalized and extracted from the membrane before undergoing the proteasomal degradation. This finding, linked with our data that iron supplementation did not enhance the cell-surface ZIP14 but raised intracellular ZIP14 levels (Fig. 1C), led us to examine the effect of iron on the membrane extraction of endocytosed ZIP14. Preloading HepG2-ZIP14 cells with iron for 24 h resulted in an increased level of membrane-associated ZIP14 and a decreased amount of cytosolic DG ZIP14 (Fig. 4C), suggesting that increased cellular iron content prevents internalized ZIP14 from being extracted from the membrane. Consequently, less ZIP14 was degraded by proteasomes. To test the possibility that ZIP14 undergoes retrograde trafficking to the ER for degradation through the ERAD pathway, cell-surface proteins were biotinylated at 4 °C and cells were incubated with brefeldin A (BFA) before and during warming them to 37 °C. BFA blocks retrograde transport from the Golgi to the ER and anterograde transport to the plasma membrane, resulting in the vesiculation of the Golgi and trans-Golgi network (TGN) (25). Notably, BFA did not prevent the degradation or the deglycosylation of internalized ZIP14 (Fig. 4F). Golgi 97 staining and the secretion of transferrin were used as controls to demonstrate the efficacy of BFA. BFA treatment of cells indeed resulted in the fragmentation of the TGN (Fig. 4D and Fig. S3). HepG2 cells normally secrete detectable levels of transferrin. BFA inhibited transferrin (Tf) secretion and trapped a lower-molecular-weight form of Tf inside cells (Fig. 4E). The lack of effect of BFA on the deglycosylation of internalized ZIP14 suggests that ZIP14 does not undergo retrograde translocation to the ER to be extracted (Fig. 4F).
Fig. 4.
Iron prevents internalized plasma membrane ZIP14 from being extracted from the cytosol. HepG2-ZIP14 cells were used. (A) Schematic illustration of the protocol to analyze plasma membrane protein associated with cytosolic or membrane fractions. (B) Total membrane (Mem) and cytosolic (Cyto) fractions were isolated by two-step centrifugations. Biotin-labled cell-surface ZIP14 in each fraction was isolated by streptavidin gel and analyzed by immunoblotting. (C) Cells were treated with PBS (CON) or FAC for 24 h, and then the internalized ZIP14 in the cytosol and the membrane fractions were compared by immunoblotting. (D) Cells treated with BFA (10 µg/mL) for 1 h were stained for Golgi97 (red), actin (green), and nuclei (blue). (E) Cells were treated with BFA (10 µg/mL) for 12 h. Total cell lysates and cell media from control and BFA-treated cells were collected and analyzed by immunoblotting for transferrin. (F) Cell-surface proteins were labeled with NHS-SS-biotin at 4 °C. Cells were then incubated with BFA for 1 h followed by 2 h of incubation with MG132. Biotin-labled cell-surface proteins were isolated by streptavidin gel and analyzed by immunoblotting.
N-Linked Glycosylation but Not Intracellular Loop2 Is Required for the Iron Sensitivity of ZIP14.
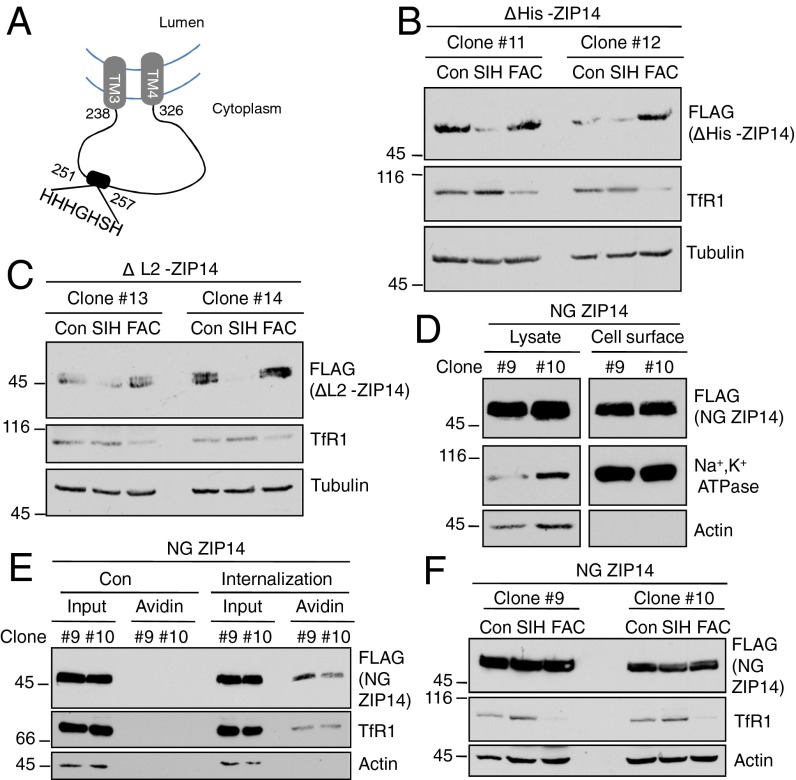
We determined which structural features of ZIP14 might be necessary for its extraction from the membrane. In silico analysis of ZIP14 membrane topology revealed that its second intracellular loop (L2) located between TM3 and TM4 contains a histidine-rich region (His) from H251 to H257 that may function as a metal-binding motif (Fig. 5A). Iron binding to His could lead to a change in ZIP14 conformation, which, in turn, would result in decreased membrane extraction. We reasoned that if the His region is important for extraction of ZIP14 from the membrane, then ZIP14 lacking this region (ΔHis-ZIP14, Table S1) would not be sensitive to cellular iron status. Interestingly, the response of ΔHis-ZIP14 to iron chelation or iron supplementation was not disturbed compared with that of WT ZIP14 (Fig. 5B). When the entire L2 domain of ZIP14 (residues 238–326) (ΔL2-ZIP14, Table S1) was deleted, it was still sensitive to regulation by iron (Fig. 5C). Therefore, the iron sensitivity of ZIP14 is not attributable to the His region or any residues localized within the L2 domain.
Fig. 5.
The iron sensitivity of ZIP14 depends on N-linked oligosaccharides, but not on the histidine-rich motif-containing intracellular loop. (A) Topology of the ZIP14 fragment between TM3 and TM4 within the endocytic compartment. (B) HEK293 stable transgenic cells expressing ZIP14 without its histidine-rich region or (C) the predicted second intracellular loop were treated with 100 µM SIH or 25 µg/mL FAC for 24 h, followed by immunoblotting. (D) Cell-surface proteins were labeled with NHS-SS-biotin. Samples were analyzed by immunoblotting. (E) Cells were labeled with NHS-SS-biotin at 4 °C, and then cells were warmed to 37 °C to induce endocytosis (under control conditions, cells were kept at 4 °C to prevent endocytosis). Subsequently, cells were incubated again at 4 °C and were treated with the cell-impermeable reducing agent MesNa to remove surface biotinylation. Cells were lysed, immunoprecipitated with streptavidin–agarose, and analyzed by immunoblotting to measure internalization. Biotinylated cell-surface NG ZIP14 was detected when cells were allowed to undergo endocytosis. Cells maintained at 4 °C were used as control. Under these conditions, no biotinylated protein was detected, indicating efficient stripping of the cell-surface biotinylation by MesNa treatment. (F) Cells stably transfected with NG ZIP14 were treated with 100 µM SIH or 25 µg/mL FAC for 24 h, followed by immunoblotting. Numbers with “#” indicate different stable cell clones (Table S1).
Protein glycosylation, especially the N-linked oligosaccharides, has been implicated in modulating protein degradation and metabolism (26). We noted that, in the clonal HEK293 cell lines stably transfected with ZIP14, the level of the NG ZIP14 was always higher than that of the WT ZIP14 (Fig. 2H), even though both were driven by the same exogenous CMV promoter and transfected at the same concentration of plasmid. This observation raised the possibility that the NG ZIP14 might be more stable. Biotinylation of NG ZIP14 indicated that ZIP14 traffics to the cell surface (Fig. 5D) and undergoes endocytosis similar to WT ZIP14 (Fig. 5E). Notably, the levels of NG ZIP14 did not change with iron chelation or iron supplementation (Fig. 5F), suggesting that N-linked oligosaccharides on the N-terminal domain of ZIP14 play an essential role in regulating the steady-state levels of ZIP14 and its sensitivity to iron.
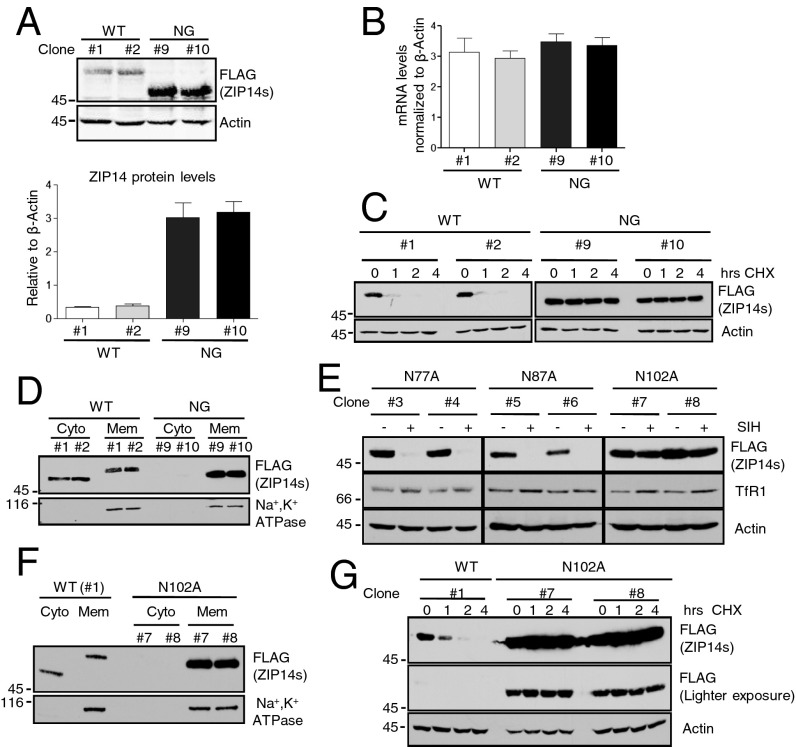
The steady-state level of NG ZIP14 was about eight times higher than that of the WT protein (Fig. 6A). The increased ZIP14 was not due to differences in mRNA levels (Fig. 6B), but due to significantly enhanced protein stability (Fig. 6C). To investigate whether the increased stability was due to a defect in the extraction of ZIP14 from membranes, we compared the levels of internalized plasma membrane proteins in cytosolic and membrane-bound fractions of the cell lysates from MG132-treated WT and NG ZIP14 cells. Significantly less NG ZIP14 was detected in the cytosol (Fig. 6D), indicating that N-linked glycosylation is required for efficient membrane extraction of ZIP14. This result explains why the NG ZIP14 does not respond to SIH or FAC treatment. In the absence of its N-linked oligosaccharides, ZIP14 is not efficiently extracted from the cytosol for proteasomal processing; therefore, changes in cellular iron status did not have a significant effect on regulating NG ZIP14.
Fig. 6.
Glycosylation at asparagine 102 is required for the membrane extraction and iron sensitivity of ZIP14. HEK293 stable transgenic cells expressing WT or NG ZIP14 were used in A–D. (A) Immunoblotting analysis by Licor ODYSSEY scanner and quantification of the blots. n = 3 independent experiments. (B) The expression levels of the WT and the NG ZIP14 mRNA were quantified and normalized to β-actin. n = 3 independent experiments. (C) The HEK293 WT ZIP14 and HEK293 NG ZIP14 cells were incubated with cycloheximide (CHX, 100 µg/mL) for 0, 1, 2, and 4 h, followed by immunoblotting analysis. (D) Comparison of the internalized plasma membrane proteins in the membrane (Mem) and cytosolic (Cyto) fractions from the WT and NG ZIP14 cell lysates. (E) HEK293 stably transfected cells expressing single Asn-to-Ala mutant ZIP14 were treated with SIH (100µM) and analyzed by immunoblotting. (F) Comparison of the internalized plasma membrane proteins in the Mem and Cyto fractions from the WT and N102A ZIP14 cell lysates. Numbers with “#” indicate different stable cell clones (Table S1). (G) Cells were treated with CHX, and then the stability of the N102A ZIP14 was compared with that of the WT ZIP14 by immunoblotting.
Glycosylation at Asparagine 102 Is Sufficient to Determine the Iron Sensitivity and Membrane Extraction of ZIP14.
The role of each N-linked glycosylation site in determining the iron sensitivity of ZIP14 was assessed. HEK293 cells stably expressing ZIP14 with single Asn-to-Ala mutations were treated with SIH to deplete cellular iron. The N77A or N87A ZIP14 responded to SIH treatment similarly to WT ZIP14. However, the N102A ZIP14 did not change with iron chelation, indicating that Asn102 is important for ZIP14 to sense the cellular iron levels (Fig. 6E). Further analysis indicated that, similar to the triple Asn mutation (NG ZIP14), N102A ZIP14 resulted in a defect in membrane extraction (Fig. 6F). Consequently, it was much more stable than WT ZIP14 (Fig. 6G). Thus, glycosylation at N102 is critical in determining its membrane extraction and the iron sensitivity of ZIP14.
Discussion
Our results implicate an important role for proteasomes in the down-regulation of ZIP14, a plasma membrane protein. Proteins residing at the cell surface participate in regulating cellular activity. They can transport nutrients, ions, and metabolites; act as receptors for various extracellular signals; anchor macromolecules on either side of the membrane; catalyze specific reactions; and function in cell-to-cell or cell-to-matrix interactions (27). Targeting plasma membrane proteins to lysosomes is a major mechanism to down-regulate the function of these proteins (13, 28). Several groups have reported the involvement of proteasomes in the degradation of single-membrane–spanning proteins. Most notably are the Met tyrosine kinase receptor (29) and the ErbB-4 receptor (30). Both the Met tyrosine kinase receptor and the ErbB-4 receptor are cleaved either on the luminal side proximal to the transmembrane domain or within the transmembrane domain. The cytosolic domains are degraded by the proteasome. However, the detailed mechanisms underlying proteasomal processing of endocytosed membrane proteins remain poorly understood.
We show that an internalized plasma membrane protein with multiple transmembrane domains undergoes proteasomal degradation and that ZIP14 is extracted from the membrane, deglycosylated in the cytosol, and degraded through a proteasomal pathway that is related to the ERAD extraction of proteins from the ER. Importantly, we find that iron and glycosylation are two contributing factors in regulating the membrane extraction and proteasomal processing of this multipass membrane protein. The demonstration that ZIP14 becomes a substrate for proteasomes suggests the existence of retro-translocation from endocytic vesicles to the cytosol. The extraction of many ER membrane proteins for proteasomal degradation depends on the cytosolic ATPase p97 (31, 32). Recent studies show that p97 is also involved in extracting the mitochondrial outer membrane proteins, Mcl1 and Mfn1 (33, 34). The identification of proteins that compose the machinery for extracting proteins from endocytic compartments and the questions of whether this machinery requires the function of p97 and how iron regulates this process for ZIP14 are the subject of future experiments.
Glycosylation at N102 is crucial for the membrane extraction and iron sensitivity of ZIP14. Oligosaccharides are known to be involved in determining the 3D configuration of proteins (35), which could play an important role in protein–protein interactions. Removal of N-linked oligosaccharides at position N102 might directly or indirectly disrupt the interaction between ZIP14 and the retro-translocation machinery required for ZIP14 membrane extraction. Distinguishing WT and N102A mutant ZIP14 preferential binding partners may provide a platform to identify the minimal set of components required for the extraction machinery from the endocytic vesicles to the cytosol.
Our study unveils the mechanism by which iron regulates ZIP14. We find that iron deficiency stimulates the ubiquitination and proteasomal degradation of ZIP14 and that excess iron prevents endocytosed ZIP14 from being extracted from the membrane, avoiding proteasomal degradation. These findings have implications in the regulation of the distribution of iron in the body and could have potential clinical applications. The proteasome is a possible therapeutic target for regulating the amount of iron stored in the liver and other ZIP14-containing organs when the standard iron deficiency treatment, such as dietary replacement or exogenous iron supplements, is inadequate to restore body iron or when patients need rapid restitution of iron stores. Proteasome inhibition has been developed and tested broadly for antitumor activities (36–39), but the potential to treat iron deficiency has not been explored. By inhibiting the proteasomal activity, iron-depleted cells might be able to preserve the function of ZIP14 to obtain iron for maintaining proper cellular function.
Materials and Methods
SI Materials and Methods describes in detail cell culture conditions, cell lines used, immunoblotting, immunofluorescence, and lysate preparations including reagents and buffers used. It also describes the procedure for isolation of plasma membrane proteins, separation of cytosol and membrane fractions, and endocytosis analysis with mercaptoethanesulfonic acid (MesNa) treatment. Information about plasmids, antibodies, and primers is also provided (Tables S2 and S3).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants DK054488 (to C.A.E.), DK072166 (to C.A.E.), DK080765 (to A.-S.Z.), T32 (GM071338-08) (to C.W.), and DK080706 (to M.D.K.) and by the Collins Medical Trust Award (to N.Z.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1405355111/-/DCSupplemental.
References
- 1.Liuzzi JP, Aydemir F, Nam H, Knutson MD, Cousins RJ. Zip14 (Slc39a14) mediates non-transferrin-bound iron uptake into cells. Proc Natl Acad Sci USA. 2006;103(37):13612–13617. doi: 10.1073/pnas.0606424103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taylor KM, Morgan HE, Johnson A, Nicholson RI. Structure-function analysis of a novel member of the LIV-1 subfamily of zinc transporters, ZIP14. FEBS Lett. 2005;579(2):427–432. doi: 10.1016/j.febslet.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 3.Gao J, Zhao N, Knutson MD, Enns CA. The hereditary hemochromatosis protein, HFE, inhibits iron uptake via down-regulation of Zip14 in HepG2 cells. J Biol Chem. 2008;283(31):21462–21468. doi: 10.1074/jbc.M803150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao N, Gao J, Enns CA, Knutson MD. ZRT/IRT-like protein 14 (ZIP14) promotes the cellular assimilation of iron from transferrin. J Biol Chem. 2010;285(42):32141–32150. doi: 10.1074/jbc.M110.143248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pinilla-Tenas JJ, et al. Zip14 is a complex broad-scope metal-ion transporter whose functional properties support roles in the cellular uptake of zinc and nontransferrin-bound iron. Am J Physiol Cell Physiol. 2011;301(4):C862–C871. doi: 10.1152/ajpcell.00479.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nam H, et al. ZIP14 and DMT1 in the liver, pancreas, and heart are differentially regulated by iron deficiency and overload: Implications for tissue iron uptake in iron-related disorders. Haematologica. 2013;98(7):1049–1057. doi: 10.3324/haematol.2012.072314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hentze MW, Muckenthaler MU, Andrews NC. Balancing acts: Molecular control of mammalian iron metabolism. Cell. 2004;117(3):285–297. doi: 10.1016/s0092-8674(04)00343-5. [DOI] [PubMed] [Google Scholar]
- 8.Ganz T, Nemeth E. Hepcidin and disorders of iron metabolism. Annu Rev Med. 2011;62:347–360. doi: 10.1146/annurev-med-050109-142444. [DOI] [PubMed] [Google Scholar]
- 9.Feder JN, et al. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet. 1996;13(4):399–408. doi: 10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- 10.Hochstrasser M. Introduction to intracellular protein degradation. Chem Rev. 2009;109(4):1479–1480. doi: 10.1021/cr900054t. [DOI] [PubMed] [Google Scholar]
- 11.Luzio JP, Parkinson MD, Gray SR, Bright NA. The delivery of endocytosed cargo to lysosomes. Biochem Soc Trans. 2009;37(Pt 5):1019–1021. doi: 10.1042/BST0371019. [DOI] [PubMed] [Google Scholar]
- 12.Piper RC, Katzmann DJ. Biogenesis and function of multivesicular bodies. Annu Rev Cell Dev Biol. 2007;23:519–547. doi: 10.1146/annurev.cellbio.23.090506.123319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacGurn JA, Hsu PC, Emr SD. Ubiquitin and membrane protein turnover: From cradle to grave. Annu Rev Biochem. 2012;81:231–259. doi: 10.1146/annurev-biochem-060210-093619. [DOI] [PubMed] [Google Scholar]
- 14.Raiborg C, et al. Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat Cell Biol. 2002;4(5):394–398. doi: 10.1038/ncb791. [DOI] [PubMed] [Google Scholar]
- 15.Tachiyama R, et al. Proteome of ubiquitin/MVB pathway: Possible involvement of iron-induced ubiquitylation of transferrin receptor in lysosomal degradation. Genes Cells. 2011;16(4):448–466. doi: 10.1111/j.1365-2443.2011.01499.x. [DOI] [PubMed] [Google Scholar]
- 16.Zelcer N, Hong C, Boyadjian R, Tontonoz P. LXR regulates cholesterol uptake through Idol-dependent ubiquitination of the LDL receptor. Science. 2009;325(5936):100–104. doi: 10.1126/science.1168974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong C, et al. The E3 ubiquitin ligase IDOL induces the degradation of the low density lipoprotein receptor family members VLDLR and ApoER2. J Biol Chem. 2010;285(26):19720–19726. doi: 10.1074/jbc.M110.123729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scotti E, et al. Targeted disruption of the idol gene alters cellular regulation of the low-density lipoprotein receptor by sterols and liver x receptor agonists. Mol Cell Biol. 2011;31(9):1885–1893. doi: 10.1128/MCB.01469-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L, Xu M, Scotti E, Chen ZJ, Tontonoz P. Both K63 and K48 ubiquitin linkages signal lysosomal degradation of the LDL receptor. J Lipid Res. 2013;54(5):1410–1420. doi: 10.1194/jlr.M035774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muckenthaler MU, Galy B, Hentze MW. Systemic iron homeostasis and the iron-responsive element/iron-regulatory protein (IRE/IRP) regulatory network. Annu Rev Nutr. 2008;28:197–213. doi: 10.1146/annurev.nutr.28.061807.155521. [DOI] [PubMed] [Google Scholar]
- 21.Nankoe SR, Sever S. Dynasore puts a new spin on dynamin: A surprising dual role during vesicle formation. Trends Cell Biol. 2006;16(12):607–609. doi: 10.1016/j.tcb.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 22.Macia E, et al. Dynasore, a cell-permeable inhibitor of dynamin. Dev Cell. 2006;10(6):839–850. doi: 10.1016/j.devcel.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 23.Bonifacino JS, Rojas R. Retrograde transport from endosomes to the trans-Golgi network. Nat Rev Mol Cell Biol. 2006;7(8):568–579. doi: 10.1038/nrm1985. [DOI] [PubMed] [Google Scholar]
- 24.Ghosh P, Dahms NM, Kornfeld S. Mannose 6-phosphate receptors: New twists in the tale. Nat Rev Mol Cell Biol. 2003;4(3):202–212. doi: 10.1038/nrm1050. [DOI] [PubMed] [Google Scholar]
- 25.Mallard F, et al. Direct pathway from early/recycling endosomes to the Golgi apparatus revealed through the study of shiga toxin B-fragment transport. J Cell Biol. 1998;143(4):973–990. doi: 10.1083/jcb.143.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Helenius A, Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem. 2004;73:1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- 27.Josic D, Clifton JG. Mammalian plasma membrane proteomics. Proteomics. 2007;7(16):3010–3029. doi: 10.1002/pmic.200700139. [DOI] [PubMed] [Google Scholar]
- 28.Piper RC, Luzio JP. Ubiquitin-dependent sorting of integral membrane proteins for degradation in lysosomes. Curr Opin Cell Biol. 2007;19(4):459–465. doi: 10.1016/j.ceb.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeffers M, Taylor GA, Weidner KM, Omura S, Vande Woude GF. Degradation of the Met tyrosine kinase receptor by the ubiquitin-proteasome pathway. Mol Cell Biol. 1997;17(2):799–808. doi: 10.1128/mcb.17.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vecchi M, Carpenter G. Constitutive proteolysis of the ErbB-4 receptor tyrosine kinase by a unique, sequential mechanism. J Cell Biol. 1997;139(4):995–1003. doi: 10.1083/jcb.139.4.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 2004;429(6994):841–847. doi: 10.1038/nature02656. [DOI] [PubMed] [Google Scholar]
- 32.Ye Y, Meyer HH, Rapoport TA. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414(6864):652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- 33.Tanaka A, et al. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol. 2010;191(7):1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu S, Peng G, Wang Y, Fang S, Karbowski M. The AAA-ATPase p97 is essential for outer mitochondrial membrane protein turnover. Mol Biol Cell. 2011;22(3):291–300. doi: 10.1091/mbc.E10-09-0748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shental-Bechor D, Levy Y. Folding of glycoproteins: Toward understanding the biophysics of the glycosylation code. Curr Opin Struct Biol. 2009;19(5):524–533. doi: 10.1016/j.sbi.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 36.Moreau P, et al. Proteasome inhibitors in multiple myeloma: 10 years later. Blood. 2012;120(5):947–959. doi: 10.1182/blood-2012-04-403733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Campello L, Esteve-Rudd J, Cuenca N, Martín-Nieto J. The ubiquitin-proteasome system in retinal health and disease. Mol Neurobiol. 2013;47(2):790–810. doi: 10.1007/s12035-012-8391-5. [DOI] [PubMed] [Google Scholar]
- 38.Schmidt M, Finley D. Regulation of proteasome activity in health and disease. Biochim Biophys Acta. 2014;1843(1):13–25. doi: 10.1016/j.bbamcr.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gallerani E, et al. A first in human phase I study of the proteasome inhibitor CEP-18770 in patients with advanced solid tumours and multiple myeloma. Eur J Cancer. 2013;49(2):290–296. doi: 10.1016/j.ejca.2012.09.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.