Significance
Atrial Fibrillation (AF) is the most common sustained cardiac arrhythmia in the human population. It is critical to elucidate the molecular mechanisms underlying AF, given that the prevalence of AF is expected to dramatically increase as the human population ages. We identified a microRNA (miR)-regulated genetic pathway that delimits sinoatrial node development and inhibits AF. To our knowledge, our data are the first genetic evidence showing that miR deletion results in AF predisposition. Moreover, to our knowledge, our data are the first demonstration that sinoatrial node regulatory genes are regulated by miRs. Our findings suggest attractive therapeutic targets to treat AF given that miR-based therapeutics are feasible using miR antagonists and mimics.
Keywords: irregular heart rate, single nucleotide variant, mouse genetics
Abstract
The molecular mechanisms underlying atrial fibrillation, the most common sustained cardiac arrhythmia, remain poorly understood. Genome-wide association studies uncovered a major atrial fibrillation susceptibility locus on human chromosome 4q25 in close proximity to the paired-like homeodomain transcription factor 2 (Pitx2) homeobox gene. Pitx2, a target of the left-sided Nodal signaling pathway that initiates early in development, represses the sinoatrial node program and pacemaker activity on the left side. To address the mechanisms underlying this repressive activity, we hypothesized that Pitx2 regulates microRNAs (miRs) to repress the sinoatrial node genetic program. MiRs are small noncoding RNAs that regulate gene expression posttranscriptionally. Using an integrated genomic approach, we discovered that Pitx2 positively regulates miR-17-92 and miR-106b-25. Intracardiac electrical stimulation revealed that both miR-17-92 and miR-106b-25 deficient mice exhibit pacing-induced atrial fibrillation. Furthermore electrocardiogram telemetry revealed that mice with miR-17-92 cardiac-specific inactivation develop prolonged PR intervals whereas mice with miR-17-92 cardiac-specific inactivation and miR-106b-25 heterozygosity develop sinoatrial node dysfunction. Both arrhythmias are risk factors for atrial fibrillation in humans. Importantly, miR-17-92 and miR-106b-25 directly repress genes, such as Shox2 and Tbx3, that are required for sinoatrial node development. Together, to our knowledge, these findings provide the first genetic evidence for an miR loss-of-function that increases atrial fibrillation susceptibility.
Atrial fibrillation (AF), the most common arrhythmia in adult patients, increases in prevalence with age to almost 5% of the population over 65. Patients with AF have an increased risk of stroke, dementia, and heart failure (1). Electrical impulses that are critical for a coordinated, physiologic heartbeat originate in the sinoatrial node (SAN). In AF, abnormal fibrillatory atrial impulses override normal SAN function, with resultant irregular conduction to the ventricles. Many cases of ectopic electrical activity originate in the pulmonary vein (2). Other sites of ectopy include the left atrial posterior wall, superior vena cava, interatrial septum, crista terminalis, and coronary sinus myocardium (3, 4).
Multiple approaches have been used to uncover genes that may contribute to the AF phenotype in adult patients. A seminal genome-wide association study (GWAS), subsequently replicated by multiple studies, uncovered a single nucleotide variant (SNV) on human 4q25 that was strongly associated with familial AF (5). Patients with the 4q25 variant exhibited early onset AF that was independent from other risk factors such as hypertension and diabetes, suggesting a novel biologic mechanism involving genes located on chromosome 4q25. The presence of the 4q25 SNV also had prognostic value because patients with the SNV are prone to cardioembolic stroke and AF recurrence after ablation therapy (6, 7).
MicroRNAs (miRs) are 21- to 25-nucleotide noncoding RNAs that function in biologic processes by posttranscriptional gene silencing (8, 9). Mouse studies have revealed multiple roles for miRs in heart development (9, 10). MiR-17-92 and its two homologous clusters, miR-106a-363 and miR-106b-25, encode polycistronic miRs that are processed from a common primary miR and are grouped into different families based on their seed sequences (Fig. S1A) (11, 12). Germ-line miR-17-92 loss-of-function results in heart defects including ventricular septal defects that result from abnormal differentiation of second heart field cardiac progenitors (13, 14).
Paired-like homeodomain transcription factor 2 (Pitx2) is the gene in closest proximity to the 4q25 SNV that has biologic relevance for AF (5). Pitx2 is expressed on the left side of multiple organ primordia and mature organs including the heart. Within the developing heart, Pitx2 is highly expressed in multiple sites that are prone to ectopic electrical activity, such as the left atrium, pulmonary vein, interatrial septum, crista terminalis, and left caval vein myocardium (15, 16). Pitx2 haploinsufficient (Pitx2null/+) adult mice are prone to AF under intracardiac electrical stimulation, revealing a direct functional connection between AF and reduced Pitx2 levels (16, 17).
Here, we discovered that miR-17-92 and miR-106b-25 are novel downstream Pitx2 target genes and are AF susceptibility genes. MiR-17-92 and miR-106b-25 inhibit the SAN genetic program in the coronary sinus and left atrium. To our knowledge, our findings are the first genetic evidence that miRs regulate SAN development and are implicated in AF predisposition.
Results
Pitx2 Positively Regulates miR-17-92 and miR-106b-25.
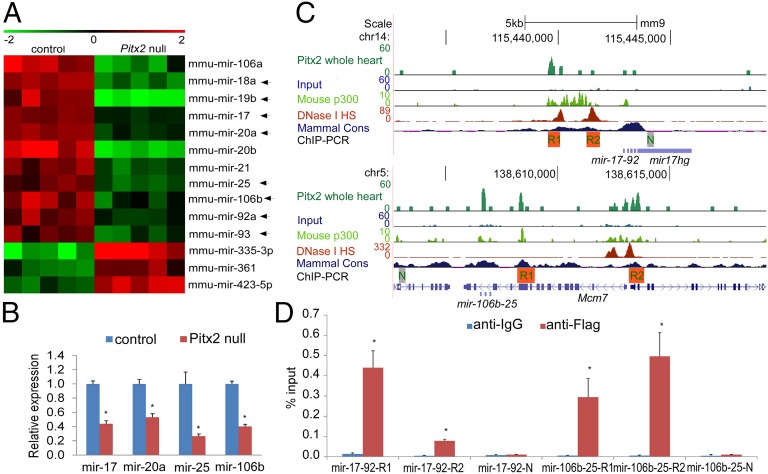
To gain insight into miRs involved in AF pathogenesis, we performed miR expression profiling using hearts from Pitx2null/null mutant and wild type, and ChIP-sequencing (ChIP-Seq) using hearts from the Pitx2-Flag allele (16, 18), which express Pitx2 with a C-terminal Flag epitope tag (Fig. 1 A and C). The miR profiling data at embryonic day 13.5 (E13.5) reveal that multiple individual miRs encoded by miR-17-92 and miR-106b-25 are reduced in Pitx2null/null mutant hearts (Fig. 1A). These findings are further confirmed by real-time RT-PCR (Fig. 1B). ChIP-Seq analysis using chromatin from adult mouse heart of the Pitx2Flag allele (GEO accession no. GSE50401) (18) revealed that Pitx2 directly binds conserved chromatin upstream of miR-17-92 and miR-106b-25 (Fig. 1C). The Pitx2-occupied chromatin also are DNase I hypersensitive sites (ENCODE, 8-wk heart) and are occupied by p300 (ChIP-Seq data from GEO accession no. GSE32587) (19), suggesting Pitx2 actively regulates miR-17-92 and miR-106b-25. The Pitx2 binding regions were further confirmed by real-time PCR using E13.5 Pitx2-Flag mouse heart ChIP chromatin, indicating that Pitx2 directly binds miR-17-92 and miR-106b-25 chromatin during embryogenesis (Fig. 1D). In addition, several miRs such as miR-335 and miR-423 were up-regulated in Pitx2null mutant hearts (Fig. 1A) and ChIP-Seq data suggest that they are also potential direct Pitx2 target genes (Fig. S1 B and C).
Fig. 1.
Pitx2 positively regulates miR-17-92 and miR-106b-25. (A) Heat map of miR array with wild-type control and Pitx2-null mutant hearts at E13.5. Arrows designate individual miRs from mir-17-92 and mir-106b-25. (B) Real-time PCR validation of miR array data at E13.5. *P < 0.05. (C) Three-month-old mouse whole-heart ChIP-Seq enrichment profiles (GSE50401) (18) for Pitx2-bound loci at upstream of miR-17-92 and miR-106b-25. The peaks from different datasets including Pitx2 ChIP-Seq, input track, P2 mouse heart p300 ChIP-Seq (GSE32587) (19), and 8-wk-old heart DNase I Hypersensitive Site (HS) Seq (ENCODE), as well as mammal conservation (cons) track, are aligned for comparison. Locations of real-time PCR validation of ChIP enrichment are indicated under datasets. For Pitx2 ChIP-Seq, input, and P300 ChIP-Seq, the scale indicates read count that was normalized to 10 million reads. For the DNase I HS track, we used the ENCODE default scale. N, negative control; R1, region1; R2, region 2. (D) In vivo real-time ChIP PCR indicated that miR-17-92 and miR-106b-25 were bound by Pitx2 in E13.5 mouse hearts.
mir-17-92 Expression Overlaps with Pitx2 Expression.
To determine the miR-17-92 expression pattern in heart, we generated an miR-17-92 bacterial artificial chromosome transgenic LacZ reporter line (20) and compared it with a Pitx2 LacZ knock-in allele (16). In E12.5 hearts, LacZ staining indicated that miR-17-92 is expressed in the outflow tract (OFT), left atrium (LA), coronary sinus, and atrioventricular canal (Fig. S2 A–C), where Pitx2 is also endogenously expressed (Fig. S2 D–F). Together, these data indicate that miR-17-92 and Pitx2 have overlapping expression domains.
miR-17-92 and miR-106b-25 Mutant Mice Have Normal Baseline Electrophysiological Parameters.
We analyzed baseline cardiac electrophysiological parameters of miR-17-92 and miR-106b-25 deficient adult mice. Four genotypes including wild-type, miR-17-92null/+, miR-106b-25null/+, and miR-106b-25null/null mice were studied. Only miR-17-92null/+ adult mice were compared in the study because miR-17-92null/null mutants are embryonic lethal. For each genotype, we evaluated heart rate (HR), the interval from the beginning of the P wave to the peak of the R wave (PR), the duration of the interval between the beginning of the Q wave to the peak of the S wave (QRS), the duration of the Q-T interval corrected for the heart rate atrial effective refractory period (QTc), the sinus node recovery time (SNRT), atrial effective refractory period (AERP), and the atrioventricular nodal effective refractory period (AVERP) (Table S1).
All baseline electrophysiologic parameters were similar in miR-17-92null/+, miR-106b-25null/+, and miR-106b-25null/null mice compared with wild-type mice. There were no episodes of spontaneous AF observed in any studied mice.
miR-17-92 and miR-106b-25 Deficient Mice Are Susceptible to Pacing-Induced Atrial Fibrillation.
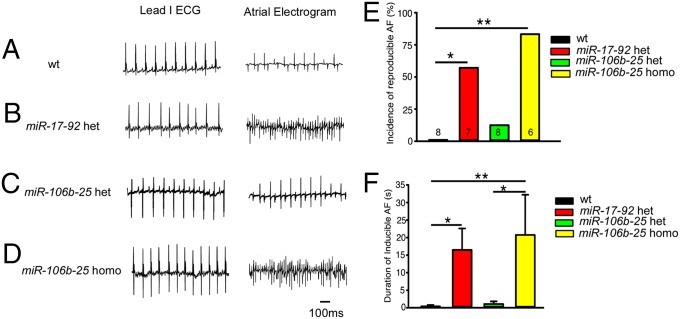
To evaluate whether the loss of miR-17-92 and miR-106b-25 increases susceptibility to AF initiation, we performed intracardiac electrical stimulation studies. An overdriving pacing protocol was used to induce AF, and three pacing trials were applied in each mouse. Simultaneous surface ECG and intracardiac electrograms revealed absent P-waves and irregular R-wave to R-wave (RR) intervals in miR-17-92 null/+ (Fig. 2B) and miR-106b-25null/null (Fig. 2D) mice after burst pacing, which are suggestive of AF. In contrast, wild-type (Fig. 2A) and miR-106b-25null/+ (Fig. 2C) mice showed sinus rhythm instead of reproducible AF following rapid pacing.
Fig. 2.
MiR-17-92 and miR-106b-25 deficient mice are susceptible to atrial fibrillation. Simultaneous recordings of simultaneous surface ECG (lead I) and intracardiac electrograms in wild-type (wt) (A), miR-17-92 null/+ (miR-17-92 het) (B), miR-106b-25 null/+ (miR-106b-25 het) (C), and miR-106b-25null/null (miR-106b-25 homo) (D) mice after burst pacing. Absent P-waves and irregular RR intervals suggest pacing-induced AF in miR-17-92 het (B) and miR-106b-25 homo (D) mice. (E) The incidence of inducible AF in different genotypes compared with wt mice are summarized in the bar graph. (F) The duration of the longest inducible AF episode in different genotypes compared with wt mice is summarized in the bar graph. *P < 0.05, **P < 0.01.
MiR-17-92 null/+ and miR-106b-25null/null showed a much higher incidence of reproducible pacing-induced AF: 57.4% of miR-17-92 null/+ mice (4 of 7) and 83.3% of miR-106b-25null/null mice (5 of 6) had reproducible pacing-induced AF after atrial pacing whereas 14.3% of miR-106b-25null/+ (1 of 8) and no wild-type (0 of 8) exhibited reproducible AF (Fig. 2E). Moreover, we quantified the duration of the longest AF episode in each tested animal and compared the differences between different genotypes. The miR-17-92 null/+ and miR-106b-25null/null mice also developed significantly longer duration of inducible AF compared with wild-type mice (Fig. 2F). Taken together, our findings indicate that loss of miR-17-92 and miR-106b-25, like Pitx2, increases susceptibility to AF.
Sinoatrial-Node Dysfunction in Mice with miR-17-92 Cardiac-Specific Knockout and miR-106b-25 Haploinsufficiency.
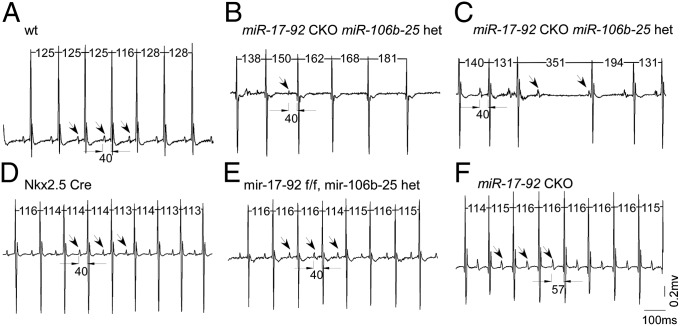
To determine whether compound mutants for miR-17-92 and miR-106b-25 develop atrial arrhythmias, we conditionally inactivated miR-17-92 using the Nkx2.5 Cre driver that directs cre activity in heart. Continuous ambulatory ECGs were recorded using telemetry in mice aged from 6 wk to 4 mo old. We first evaluated Nkx2.5Cre;miR-17-92flox/flox adult mice and found that they all had prolonged PR intervals or first-degree atrioventricular (AV) block (Fig. 3F, n = 6), which is an AF risk factor in the human population (1). Importantly, long-term follow-up of patients in the Framingham cohort found that prolonged PR interval commonly progressed to more severe arrhythmias requiring pacemaker implantation that included SAN dysfunction, high-grade AV block, and AF (1).
Fig. 3.
Mice with miR-17-92 cardiac-specific knockout and miR-106b-25 haploinsufficiency have spontaneous arrhythmias. Continuous ambulatory ECGs were recorded in mice using telemetry. Arrows designate measured PR intervals. Continuous ambulatory ECGs indicated SAN dysfunction in a 6-wk-old Nxk2.5Cre; miR-17-92flox/flox; miR-106b-25null/+ (miR-17-92 CKO miR-106b-25 het) mouse (B) but not in WT mouse (A), Nxk2.5Cre (D) miR-17-92flox/flox; miR-106b-25null/+ (E), and Nxk2.5Cre; miR-17-92flox/flox (F). Although no episode of SAN dysfunction was detected, Nxk2.5Cre; miR-17-92flox/flox showed prolonged PR intervals. Four-month-old miR-17-92 CKO miR-106b-25 het mice had not only SAN dysfunction, but also type 2 second-degree atrioventricular block (an example shown in C). RR interval and PR interval measurements are labeled, and arrows designate P waves.
The Nkx2.5Cre;miR-17-92flox/flox;miR-106b-25null/null double mutant mice were runted, for reasons that are under investigation and could not be used for these studies. However, Nkx2.5Cre;miR-17-92flox/flox;miR-106b-25null/+ (n = 10) mice developed SAN dysfunction, also called sick-sinus syndrome, which was revealed by irregular sinus rhythm with low amplitude P waves and variable ventricular response (Fig. 3B). SAN dysfunction, which frequently coexists with AF in human patients (21), is a known AF risk factor (22) and significantly associated with prolonged atrial fibrillatory cycle length (23). Both 6-wk-old (n = 4) and 4-mo-old (n = 6) Nkx2.5Cre;miR-17-92flox/flox;miR-106b-25null/+ mice had SAN dysfunction, with more frequent and longer arrhythmias in 4-mo-old mice. Moreover, some 4-mo-old Nkx2.5Cre;miR-17-92flox/flox;miR-106b-25null/+ mice had, in addition to SAN dysfunction, type 2 second-degree atrioventricular block (AV block) (3 out of 6) (Fig. 3C). Together these data suggest that progressive removal of miR-17-92 and miR-106b-25 complexes results in atrial arrhythmias of increasing severity that in human patients are known AF risk factors. Moreover, our findings support the hypothesis that miR-17-92 and miR-106b-25 have overlapping functions in atrial homeostasis. Notably, the Nkx2.5Cre;miR-17-92flox/flox;miR-106b-25null/+ failed to develop sustained AF perhaps because of the presence of the single allele of miR-106b-25.
Because it has been reported that aged Nkx2.5Cre mice have background-dependent phenotypes including arrhythmias (24), we performed a number of control experiments. Importantly, SAN dysfunction was not observed in mice of other genotypes including wild-type (6 wk old, n = 4) (Fig. 3A), Nkx2.5Cre (6 wk old, n = 4; 4 mo old, n = 4) (Fig. 3D), miR-17-92flox/flox; miR-106b-25null/+ (6 wk old, n = 4; 4 mo old, n = 4) (Fig. 3E), and Nkx2.5Cre; miR-17-92flox/flox (6 wk old, n = 3; 4 mo old, n = 3) (Fig. 3F). In addition, one 4-mo-old Nkx2.5Cre mice had prolonged PR interval that is similar as detected in 6-wk-old Nkx2.5Cre; miR-17-92flox/flox mice.
Normal Cardiac Structure in Mice with miR-17-92 Cardiac-Specific Knockout and miR-106b-25 Haploinsufficiency.
To examination whether the cardiac arrhythmias in Nkx2.5Cre; miR-17-92flox/flox; miR-106b-25null/+ mice were caused by cardiac structural abnormalities, we first did echocardiography to examine the cardiac function of Nkx2.5Cre; miR-17-92flox/flox; miR-106b-25null/+ mutants (n = 6) compared with miR-17-92flox/flox; miR-106b-25null/+ and Nkx2.5Cre controls (miR-17-92flox/flox; miR-106b-25null/+, n = 4; Nkx2.5Cre n = 3). Compared with control mice, no obvious changes in cardiac contractile function, dimension and wall thickness were found in Nkx2.5Cre; miR-17-92flox/flox; miR-106b-25null/+ mutant mice at the age of 4 mo (Fig. S3 and Table S2). We also performed hematoxylin/eosin (H&E) staining in Nkx2.5Cre; miR-17-92flox/flox; miR-106b-25null/+ mutants (n = 10) and miR-17-92flox/flox; miR-106b-25null/+ control mice (n = 4) at the age of 4 mo. We didn’t detect any obvious defect of cardiac structure in Nkx2.5Cre; miR-17-92flox/flox; miR-106b-25null/+ mutants (n = 10) compared with miR-17-92flox/flox; miR-106b-25null/+ controls (n = 4) (Fig. S4 A and B) except for one mutant that had a small membranous ventricular septal defect. There were no atrial septal defects and no evidence for atrial enlargement in the Nkx2.5Cre; miR-17-92flox/flox; miR-106b-25null/+ mutants either histologically or by measuring atrial weights (Fig. S4 A, B, and J). These findings are consistent with a recent report from Chen et al., who also didn’t find structure changes in Nkx2.5Cre; miR-17-92flox/flox mutant mice (25).
Wheat germ agglutinin (WGA) staining data indicated that there was no obvious change in cardiomyocyte cell size of ventricular myocardium of Nkx2.5Cre; miR-17-92flox/flox; miR-106b-25null/+ mutants (n = 3) compared with miR-17-92flox/flox; miR-106b-25null/+ controls (n = 3) at 4 mo old (Fig. S4 C–E). Notably, although Chen et al. found that 10-mo-old Nkx2.5Cre; miR-17-92flox/flox mutant mice exhibited cardiomyocyte hypertrophy (25), we did not detect such changes in younger mice. Masson's Trichrome staining was used to detect the level of fibrosis, which revealed no significant difference between Nkx2.5Cre; miR-17-92flox/flox; miR-106b-25null/+ mutants (n = 3) and miR-17-92flox/flox; miR-106b-25null/+ controls (n = 3) (Fig. S4 F–I). Moreover, we evaluated the ratio of atrial weight to body weight (AW/BW) in Nkx2.5Cre; miR-17-92flox/flox; miR-106b-25null/+ mutants (n = 4) compared with miR-17-92flox/flox; miR-106b-25null/+ and Nkx2.5Cre controls (Nkx2.5Cre, n = 3; miR-17-92flox/flox; miR-106b-25null/+, n = 3) and found there was no significant difference in AW/BW between mutants and controls (Fig. S4J). Taken together, we conclude that there were no obvious cardiac structural defects in all but one of the 4-mo-old Nkx2.5Cre; miR-17-92flox/flox; miR-106b-25null/+ mutants.
miR-17-92 Represses Shox2 and Tbx3.
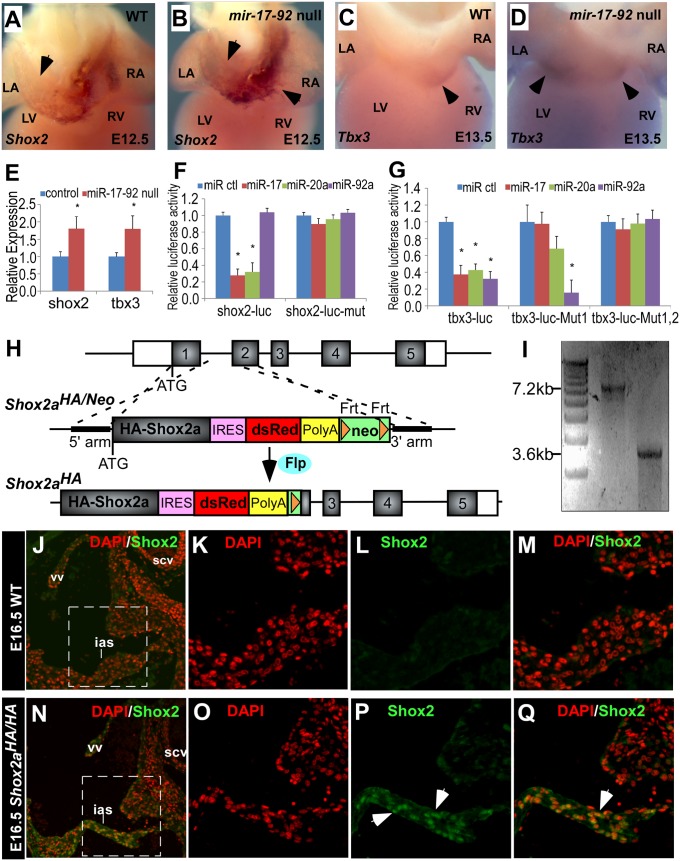
The short stature homeobox gene Shox2 and T-Box gene Tbx3, SAN gene program components, are required for normal SAN development (26, 27). Whole-mount in situ hybridization followed by transverse sections through inflow of the heart revealed that Shox2 expression is up-regulated in left superior caval vein and coronary sinus of miR-17-92null/null mutants (Fig. 4 A and B and Fig. S5 A and B). Immunofluorescence data using a Shox2 antibody also indicated the up-regulation of Shox2 expression in miR-17-92null/null mutant left superior caval vein (Fig. S5 I–L) compared with wild-type control (Fig. S5 E–H). Tbx3, that enables SAN myocardium to separate from working atrial myocardium is expanded and up-regulated in the left atrioventricular canal of miR-17-92null/null mutant whereas Tbx3 expression is more prominent in the right atrioventricular canal of wild-type controls (Fig. 4 C and D and Fig. S5 C and D). To quantify the expression level of Shox2 and Tbx3 in miR-17-92null/null mutants compared with wild type, whole-heart total RNA were isolated for quantitative RT-PCR (qRT-PCR). qRT-PCR data revealed significant Shox2 and Tbx3 up-regulation in miR-17-92 mutants compared with wild type (Fig. 4E).
Fig. 4.
MiR-17-92 directly represses Shox2 and Tbx3. (A–D) Whole-mount in situ hybridization with indicated probes in embryonic mouse hearts. Black arrows designate expression pattern. (E) qRT-PCR data indicate that expression of Shox2 and Tbx3 elevate in miR-17-92 knockout mutants. (F and G) Luciferase reporter assays with reporters and miRs as labeled. Mean ± SEM, *P < 0.05. (H) Schematic diagram of Shox2aHA allele. The mouse Shox2 gene contains six exons as indicated by numbered blocks (coding sequence indicated in gray and noncoding sequence indicated in white). Partial of the endogenous Shox2 sequence was replaced by DNA sequence encoding N-terminal FLAG-HA-tagged, full-length Shox2a, IRES-DsRed, SV40 early polyA signal and Frt flanking neomycin-resistance cassette driven by PGK promoter (neo), which is named Shox2aHA/Neo allele. Neo is further removed by crossing with germ-line Flp deleter strain to obtain Shox2aHA allele. (I) Long range PCR showing targeted clones of Shox2aHA/Neo allele. Forward primer on 5′ flanking region out of the 5′ homology arm and reverse primer on DsRed yield a 7.2-kb band; forward primer on neo and reverse primer on 3′ flanking region out of 3′ homology arm yield a 3.6-kb band. (J–Q) Immunofluorescence shows Shox2 is highly expressed in E16.5 atrial septum of Shox2aHA/HA embryos but not in wild-type embryos. Boxed areas in J and N are correspondingly shown at higher magnification in K–M and O–Q. Nuclei (red) stained with DAPI. White arrows designate Shox2-expressing cells. ias, interatrial septum; scv, surperior caval vein; vv, venous valve.
miR-17-92 and miR-106b-25 Directly Regulate Shox2 and Tbx3.
The Shox2 3′ UTR contains conserved miR-17/20a/106b family seed sequence (Fig. S6A), and the Tbx3 3′ UTR contains both a conserved miR-17/20a/106b family seed site and a conserved miR-92a/25 family seed site (Fig. S6 B and C). We constructed luciferase reporter plasmids with the 3′ UTRs of Shox2 and Tbx3 to test whether those miR seed sites function in vitro. There are drastic reductions in luciferase activity for both Shox2 and Tbx3 luciferase reporters when cotransfected with respective miR mimics (Fig. 4 F and G and Fig. S6 D and E). When we mutate the respective miR seed sites within the 3′ UTRs of Shox2 and Tbx3, the repression caused by the corresponding miR is ablated (Fig. 4 F and G and Fig. S6 D and E).
Given that Shox2 3′ UTR contains four conserved seed sites of different miRs/miR families (Fig. S7A) and that only miR-17 family is expressed in the embryonic heart whereas the other miRs/miR families are barely detectable (Fig. S7B), we hypothesized that miR-17 family exerts the dominant miR regulation on the Shox2 3′ UTR. To obtain genetic evidence supporting Shox2 regulation by miR-17 family, we generated a Shox2aHA knock-in allele that replaced the endogenous 3′ UTR with a heterologous 3′ UTR to release Shox2 expression from endogenous miR regulation. Notably the Shox2aHA allele still keeps the majority of endogenous transcriptional regulatory sequences, including intron 2 that contains a Pitx2 binding region (Fig. 4 H and I, Fig. S8A, Methods, and SI Methods). Homozygous Shox2aHA mice (Shox2aHA/HA) are neonatal lethal: 81.25% (13 out 16 from nine litters) of Shox2aHA/HA mice die within postnatal day1 without obvious morphological defects. To determine whether Shox2 expression is expanded in the Shox2aHA/HA embryos, we performed immunofluorescence studies with a Shox2 antibody. At E16.5, Shox2 is highly expressed in interatrial septum of Shox2aHA/HA (Fig. 4 N–Q) but not in wild type (Fig. 4 J–M). Moreover, expression of Shox2 in SAN is dramatically higher in Shox2aHA/HA (Fig. S8 D and E) compared with wild type (Fig. S8 B and C). Although we cannot exclude the possibility that other features of the Shox2HA allele, such as loss of splicing, may contribute to its abnormal expression, our findings are consistent with Shox2 regulation by miR-17-92.
In addition, we further performed in situ hybridization in E12.5 miR-17-92null/null mutants and wild-type controls using Bmp4 probe, which was previously shown as a downstream target of Shox2 (28). The in situ hybridization and heart-sectioning data indicated that Bmp4 expression is up-regulated in miR-17-92null/null mutants (Fig. S9 D–F) in a similar pattern of Shox2 up-regulation in miR-17-92null/null mutants. Bmp4 is highly expressed in the left superior caval vein and coronary sinus of miR-17-92null/null mutants (Fig. S9 D–F) whereas it is only moderately expressed in the coronary sinus of wild-type controls (Fig. S9 A–C). Together, these data further support the model that the miR-17-92 and miR-106b-25 directly inhibit Shox2.
Discussion
The SAN develops from a larger pool of competent cells that is progressively refined through the function of important developmental regulatory genes such as Pitx2. Progenitors within the sinus venosus give rise to sinus horn myocardium and SAN myocardium. In chick, discrete cells within lateral plate also contribute to the pacemaker (29). Recent work indicates that a dominant pacemaker activity is found in the E8.5 left inflow tract, where Pitx2c is expressed, but switches to the right SAN by E12.5 (15, 30). Our data are consistent with the notion that Pitx2 directly regulates this developmental switch in part by regulating miR-17-92 and miR-106b-25.
Our findings indicate that miR-17-92 and miR-106b-25 directly target SAN genes Shox2 and Tbx3. Previous studies have shown that both Shox2 and Tbx3 promote SAN specification while inhibiting working myocardium specification. Although Shox2 is required for SAN development in mice, it has not yet been implicated in human heart disease. However, there are data indicating that Shox2 is a target for Tbx5 that has been implicated in human AF and prolonged PR interval through GWAS studies (31, 32). Tbx3 has also been implicated in human PR interval prolongation in GWAS studies (32) and induces pacemaker properties in adult myocardium when misexpressed (33). Other than the novel targets we identified here, miR-17-92 and miR-106b-25 have other targets. A transgenic mouse study using conditional miR-17-92 overexpression suggested that miR-17-92 directly targets Pten and Cx43 (34). It will be necessary to investigate other miR-17-92 target genes to thoroughly understand these two miR clusters in AF.
Mutations in miRs have not yet been uncovered in AF patients. Before our study, genetic loss-of-function studies in mice have failed to show a requirement for miRs in AF. A number of profiling studies suggested potential roles for miRs in AF. For example, miR-328 expression was elevated in human AF patients and in a canine AF model although direct functional analysis of miR-328 awaits further experimentation (35). MiR-21 expression was increased in AF patients, a mouse model of spontaneous AF, and age-induced atrial fibrosis whereas miR-21 knockdown suppressed atrial fibrosis and AF promotion in rats (36, 37). A recent report indicated that miR-29b plasma levels were decreased in patients with AF or congestive heart failure (38). Similarly, miR-26 was down-regulated in the atria of an AF dog model, and experimental miR-26 knockdown reproduced AF-induced fibroblast activation, a central event in AF-promoting remodeling (39). Expression of miR-26 family members was reduced in human AF patients, and Kcnj2 was shown to be a direct miR-26 target. Moreover, miR-26 transcription was repressed by nuclear factor of activated T cells making a direct connection between calcium signaling and miR-26 in AF (40).
Experiments looking at miR-17-92 showed that miR-19a expression was reduced in serum of patients with persistent AF (41). In patients with AF and mitral stenosis, several individual members of the miR-17-92 cluster, including miR-17, miR-19a, miR-19b, and miR-20a, were significantly down-regulated compared with healthy individuals (42). Cardiac and smooth-muscle conditional overexpression of miR-17-92 in mice developed dilated, hypertrophic cardiomyopathy with arrhythmias (34). Our data along with the human expression data indicate that loss-of-function of all miRs in the miR-17-92 cluster predisposes to AF, suggesting that both loss and gain of miR-17-92 may stimulate cardiac arrhythmias.
The Nkx2.5Cre;miR-17-92flox/flox mutant mice showed prolonged PR interval whereas the Nkx2.5Cre;miR-17-92flox/flox;miR-106b-25null/+ mutants had a more severe phenotype, including SAN node dysfunction and second degree AV block, suggesting functional redundancy between miR-17-92 and miR-106b-25. These phenotypes are likely to be part of a continuous clinical spectrum of conduction disorders in human patients. Data from the Framingham study have shown that many patients that initially present with prolonged PR, also called first-degree AV block, commonly progress to SAN dysfunction with higher-degree AV block. These patients also commonly progress to atrial fibrillation (1). Moreover, a recent report revealed that miR-25 regulates calcium handling by directly regulating Serca2 in adult heart failure (43), suggesting that miR-17-92 and miR-106b-25 function in adult calcium homeostasis. Our data along with recent findings suggest that miR-17-92 and miR-106b-25 have multiple gene targets that function at different stages of AF predisposition.
Previous studies revealed that Pitx2 directly represses Shox2 in left inflow tract working myocardium. We have now found that this repressive mechanism also includes Shox2 repression by Pitx2-regulated miRs miR-17-92 and miR-106b-25. Both Tbx3 and Shox2 expression domains were expanded in Pitx2 null/+ and Pitx2 null/null mutant embryos. The working model that we previously developed (16) holds that Pitx2 functions within a gene regulatory network (GRN) on the left side of the inflow tract to directly repress Shox2 and the SAN genetic program in left atrium and left caval veins.
Our results, indicating that Pitx2 directly transactivates miR-17-92 and miR-106b-25 transcription, uncover a developmental Pitx2-miR pathway that suppresses AF predisposition. The finding that miR-17-92 and miR-106b-25 deficient mice are prone to AF and directly repress the SAN regulatory genes Shox2 and Tbx3 further supports our model (Fig. S10). The notion that ectopic expression of the SAN genetic program can contribute to AF predisposition is an important area for future study. Moreover, it is likely that this proposed developmental mechanism works together with other mechanisms, such as defective calcium handling, in AF predisposition. Although reduced Pitx2 levels have been observed in some adult human AF patients, Pitx2 also functions during development. In addition, recent findings indicate that Pitx2 mRNA levels are elevated in some patients that are in active AF at the time of tissue collection, indicating that Pitx2 transcriptional regulation is complex and requires further study (44). Because our work is limited to rodent studies, definitive insight into the role of miR-17-92 and miR-106b-25 in human AF await further human genetic studies.
Methods
See details in SI Methods.
Mouse Alleles and Transgenic Lines.
The Pitx2Flag, miR-17-92–null, miR-17-92 Flox, miR-106b-25–null, and Nkx2.5Cre alleles were previously described (14, 24, 16). To generate the Shox2HA allele, which is free of endogenous 3′ UTR regulation, we replaced part of the endogenous Shox2 sequence with a Flag-HA-mouseShox2a-DsRed-polyA sequence that included an Frt flanked neomycin-resistance cassette (Fig. 4E) in G4 embryonic stem (ES) cells (45). G418 selected ES clones were screened by long-range PCR followed by sequencing of PCR products. Targeted clones showed a 7.2-kb band and a 3.6-kb band on gel (Fig. 4F). Correctly targeted clone was injected into blastocysts to create chimera mice. Chimeras were crossed with CD-1 females (Charles River) to generate F1 mice and were subsequently genotyped by long-range PCR.
Chromatin Immunoprecipitation, ChIP-Sequencing, and ChIP-Seq Analysis.
E13.5 hearts were collected from Pitx2Flag allele and followed by ChIP analysis as previously described (16), and DNA obtained from ChIP were used for real-time PCR to quantify enrichment. ChIP-Seq was performed using 3-mo-old mouse whole hearts of Pitx2Flag allele, and data are available through the Gene Expression Omnibus (GEO) data repository under accession number GSE50401 (18). For ChIP-Seq analysis, Ion torrent PGM reads were aligned to the mm9 (NCBI Build 37) assembly using Torrent Suite (2.0.1) Ion-alignment (2.0.3–1). A total 1.9 million reads were uniquely mapped to mouse genome mm9. Total reads from input were 1.29 × 107. ChIP-Seq peaks were called using Homer package using threshold FDR effective Poisson 3.7 × 10−8, minimum read number 5. Then, 12,417 significant peaks were called; 8.97% of the reads were enriched in significant peaks. The ChIP-Seq data were converted to bedGraph file and visualized in the University of California, Santa Cruz genome browser. The total read number was normalized to 10 million; therefore, the y axis value indicated the normalized number of reads in a 10-bp window. The Pitx2 ChIP-Seq dataset was also aligned to postnatal day 2 (P2) mouse heart p300 ChIP-Seq (GSE32587) (19) and 8-wk-old mouse heart DNase I Hypersensitive Site Seq (ENCODE) datasets to show correlation.
Transthoracic Echocardiography.
Mice were anesthetized using 1.5% (vol/vol) isoflurane mixed with 95% (vol/vol) O2 and placed on a heated platform where all four limbs were taped to copper ECG electrodes. Cardiac function was assessed using a VisualSonics VeVo 770 Imaging System (VisualSonics) equipped with a high-frequency 30-MHz probe, as described (46).
Cardiac Electrophysiology.
In vivo electrophysiology in mice was conducted as previously described (47). Briefly, intracardiac electrograms were recorded using a 1.1F octapolar catheter (EPR-800; Millar Instruments) inserted via the right jugular vein. Atrial fibrillation (AF) inducibility was determined by previous protocol (48) and defined as overdriving pacing and defined as the occurrence of rapid and fragmented atrial electrograms with irregular AV-nodal conduction and ventricular rhythm for at least 1 s. Three pacing trials were applied in each mouse. Inducibility of AF was considered positive if at least two of three pacing trials induced AF. Moreover, ambulatory ECGs were recorded in mice using telemetry transmitters (Data Sciences International) as previously described (49). Briefly, telemetry transmitters were implanted in the abdominal cavity of mice with s.c. electrodes in a lead II configuration. ECGs were continuously recorded by telemetry using Dataquest software, version 4.1 (Data Sciences International) and analyzed using Dataquest software.
Supplementary Material
Acknowledgments
This work was supported by a 2012–2013 Michael Bilitch Fellowship in Cardiac Pacing and Electrophysiology from the Heart Rhythm Society (to J.W.), American Heart Association (AHA) Grant 12PRE11720003 (to Y.B.), AHA Grant 12BGIA12050207 (to N.L.), AHA Grant 13PRE13750003 (to W.Y.), National Institutes of Health (NIH) Grant 1F32HL105041 (to Y.T.), AHA Grant 13EIA14560061, NIH Grants R01-HL089598 and R01-HL091947, the Fondation Leducq Alliance for CaMKII Signaling in Heart (X.H.T.W.), NIH Grant 5R01HL118761 (to J.F.M.), NIH Grant R01DE17792 (to Y.C.), and the Vivian L. Smith foundation (J.F.M.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE50401).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1405411111/-/DCSupplemental.
References
- 1.Cheng S, et al. Long-term outcomes in individuals with prolonged PR interval or first-degree atrioventricular block. JAMA. 2009;301(24):2571–2577. doi: 10.1001/jama.2009.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haïssaguerre M, et al. Spontaneous initiation of atrial fibrillation by ectopic beats originating in the pulmonary veins. N Engl J Med. 1998;339(10):659–666. doi: 10.1056/NEJM199809033391003. [DOI] [PubMed] [Google Scholar]
- 3.Katritsis DG, et al. Conduction patterns in the cardiac veins: Electrophysiologic characteristics of the connections between left atrial and coronary sinus musculature. J Interv Card Electrophysiol. 2004;10(1):51–58. doi: 10.1023/B:JICE.0000011485.98197.df. [DOI] [PubMed] [Google Scholar]
- 4.Lin WS, et al. Catheter ablation of paroxysmal atrial fibrillation initiated by non-pulmonary vein ectopy. Circulation. 2003;107(25):3176–3183. doi: 10.1161/01.CIR.0000074206.52056.2D. [DOI] [PubMed] [Google Scholar]
- 5.Gudbjartsson DF, et al. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007;448(7151):353–357. doi: 10.1038/nature06007. [DOI] [PubMed] [Google Scholar]
- 6.Husser D, Adams V, Piorkowski C, Hindricks G, Bollmann A. Chromosome 4q25 variants and atrial fibrillation recurrence after catheter ablation. J Am Coll Cardiol. 2010;55(8):747–753. doi: 10.1016/j.jacc.2009.11.041. [DOI] [PubMed] [Google Scholar]
- 7.Bellenguez C, et al. International Stroke Genetics Consortium (ISGC) Wellcome Trust Case Control Consortium 2 (WTCCC2) Genome-wide association study identifies a variant in HDAC9 associated with large vessel ischemic stroke. Nat Genet. 2012;44(3):328–333. doi: 10.1038/ng.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Rooij E, Olson EN. MicroRNAs: Powerful new regulators of heart disease and provocative therapeutic targets. J Clin Invest. 2007;117(9):2369–2376. doi: 10.1172/JCI33099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montgomery RL, et al. Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure. Circulation. 2011;124(14):1537–1547. doi: 10.1161/CIRCULATIONAHA.111.030932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Concepcion CP, Bonetti C, Ventura A. The microRNA-17-92 family of microRNA clusters in development and disease. Cancer J. 2012;18(3):262–267. doi: 10.1097/PPO.0b013e318258b60a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133(2):217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J, et al. Bmp signaling regulates myocardial differentiation from cardiac progenitors through a MicroRNA-mediated mechanism. Dev Cell. 2010;19(6):903–912. doi: 10.1016/j.devcel.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ventura A, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132(5):875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu C, et al. Pitx2c patterns anterior myocardium and aortic arch vessels and is required for local cell movement into atrioventricular cushions. Development. 2002;129(21):5081–5091. doi: 10.1242/dev.129.21.5081. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, et al. Pitx2 prevents susceptibility to atrial arrhythmias by inhibiting left-sided pacemaker specification. Proc Natl Acad Sci USA. 2010;107(21):9753–9758. doi: 10.1073/pnas.0912585107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirchhof P, et al. PITX2c is expressed in the adult left atrium, and reducing Pitx2c expression promotes atrial fibrillation inducibility and complex changes in gene expression. Circ Cardiovasc Genet. 2011;4(2):123–133. doi: 10.1161/CIRCGENETICS.110.958058. [DOI] [PubMed] [Google Scholar]
- 18.Tao Y, et al. Pitx2, an atrial fibrillation predisposition gene, directly regulates ion transport and intercalated disc genes. Circ Cardiovasc Genet. 2014;7(1):23–32. doi: 10.1161/CIRCGENETICS.113.000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.May D, et al. Large-scale discovery of enhancers from human heart tissue. Nat Genet. 2011;44(1):89–93. doi: 10.1038/ng.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, et al. MicroRNA-17-92, a direct Ap-2α transcriptional target, modulates T-box factor activity in orofacial clefting. PLoS Genet. 2013;9(9):e1003785. doi: 10.1371/journal.pgen.1003785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dobrzynski H, Boyett MR, Anderson RH. New insights into pacemaker activity: Promoting understanding of sick sinus syndrome. Circulation. 2007;115(14):1921–1932. doi: 10.1161/CIRCULATIONAHA.106.616011. [DOI] [PubMed] [Google Scholar]
- 22.Lee JM, Kalman JM. 2013. Sinus node dysfunction and atrial fibrillation: Two sides of the same coin? Europace 15(2):161–162.
- 23.Sairaku A, et al. Prediction of sinus node dysfunction in patients with long-standing persistent atrial fibrillation using the atrial fibrillatory cycle length. J Electrocardiol. 2012;45(2):141–147. doi: 10.1016/j.jelectrocard.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 24.Risebro CA, et al. Epistatic rescue of Nkx2.5 adult cardiac conduction disease phenotypes by prospero-related homeobox protein 1 and HDAC3. Circ Res. 2012;111(2):e19–e31. doi: 10.1161/CIRCRESAHA.111.260695. [DOI] [PubMed] [Google Scholar]
- 25.Chen J, et al. mir-17-92 cluster is required for and sufficient to induce cardiomyocyte proliferation in postnatal and adult hearts. Circ Res. 2013;112(12):1557–1566. doi: 10.1161/CIRCRESAHA.112.300658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoogaars WM, et al. Tbx3 controls the sinoatrial node gene program and imposes pacemaker function on the atria. Genes Dev. 2007;21(9):1098–1112. doi: 10.1101/gad.416007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blaschke RJ, et al. Targeted mutation reveals essential functions of the homeodomain transcription factor Shox2 in sinoatrial and pacemaking development. Circulation. 2007;115(14):1830–1838. doi: 10.1161/CIRCULATIONAHA.106.637819. [DOI] [PubMed] [Google Scholar]
- 28.Puskaric S, et al. Shox2 mediates Tbx5 activity by regulating Bmp4 in the pacemaker region of the developing heart. Hum Mol Genet. 2010;19(23):4625–4633. doi: 10.1093/hmg/ddq393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bressan M, Liu G, Mikawa T. Early mesodermal cues assign avian cardiac pacemaker fate potential in a tertiary heart field. Science. 2013;340(6133):744–748. doi: 10.1126/science.1232877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yi T, et al. Electrophysiological mapping of embryonic mouse hearts: Mechanisms for developmental pacemaker switch and internodal conduction pathway. J Cardiovasc Electrophysiol. 2012;23(3):309–318. doi: 10.1111/j.1540-8167.2011.02191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu H, et al. The role of Shox2 in SAN development and function. Pediatr Cardiol. 2012;33(6):882–889. doi: 10.1007/s00246-012-0179-x. [DOI] [PubMed] [Google Scholar]
- 32.Pfeufer A, et al. Genome-wide association study of PR interval. Nat Genet. 2010;42(2):153–159. doi: 10.1038/ng.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bakker ML, et al. T-box transcription factor TBX3 reprogrammes mature cardiac myocytes into pacemaker-like cells. Cardiovasc Res. 2012;94(3):439–449. doi: 10.1093/cvr/cvs120. [DOI] [PubMed] [Google Scholar]
- 34.Danielson LS, et al. Cardiovascular dysregulation of miR-17-92 causes a lethal hypertrophic cardiomyopathy and arrhythmogenesis. FASEB J. 2012;27(4):1460–1467. doi: 10.1096/fj.12-221994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu Y, et al. MicroRNA-328 contributes to adverse electrical remodeling in atrial fibrillation. Circulation. 2010;122(23):2378–2387. doi: 10.1161/CIRCULATIONAHA.110.958967. [DOI] [PubMed] [Google Scholar]
- 36.Adam O, et al. Role of miR-21 in the pathogenesis of atrial fibrosis. Basic Res Cardiol. 2012;107(5):278. doi: 10.1007/s00395-012-0278-0. [DOI] [PubMed] [Google Scholar]
- 37.Cardin S, et al. Role for MicroRNA-21 in atrial profibrillatory fibrotic remodeling associated with experimental postinfarction heart failure. Circ Arrhythm Electrophysiol. 2012;5(5):1027–1035. doi: 10.1161/CIRCEP.112.973214. [DOI] [PubMed] [Google Scholar]
- 38.Dawson K, et al. MicroRNA29: A mechanistic contributor and potential biomarker in atrial fibrillation. Circulation. 2013;127(14):1466–1475, 1475e1-28. doi: 10.1161/CIRCULATIONAHA.112.001207. [DOI] [PubMed] [Google Scholar]
- 39.Harada M, et al. Transient receptor potential canonical-3 channel-dependent fibroblast regulation in atrial fibrillation. Circulation. 2012;126(17):2051–2064. doi: 10.1161/CIRCULATIONAHA.112.121830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo X, et al. MicroRNA-26 governs profibrillatory inward-rectifier potassium current changes in atrial fibrillation. J Clin Invest. 2013;123(5):1939–1951. doi: 10.1172/JCI62185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Z, et al. The expression levels of plasma micoRNAs in atrial fibrillation patients. PLoS ONE. 2012;7(9):e44906. doi: 10.1371/journal.pone.0044906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiao J, et al. MicroRNA expression signature in atrial fibrillation with mitral stenosis. Physiol Genomics. 2011;43(11):655–664. doi: 10.1152/physiolgenomics.00139.2010. [DOI] [PubMed] [Google Scholar]
- 43.Wahlquist C, et al. Inhibition of miR-25 improves cardiac contractility in the failing heart. Nature. 2014;508(7497):531–535. doi: 10.1038/nature13073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gore-Panter SR, et al. Atrial Fibrillation associated chromosome 4q25 variants are not associated with PITX2c expression in human adult left atrial appendages. PLoS ONE. 2014;9(1):e86245. doi: 10.1371/journal.pone.0086245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.George SH, et al. Developmental and adult phenotyping directly from mutant embryonic stem cells. Proc Natl Acad Sci USA. 2007;104(11):4455–4460. doi: 10.1073/pnas.0609277104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Respress JL, et al. Role of RyR2 phosphorylation at S2814 during heart failure progression. Circ Res. 2012;110(11):1474–1483. doi: 10.1161/CIRCRESAHA.112.268094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li N, et al. Inhibition of CaMKII phosphorylation of RyR2 prevents induction of atrial fibrillation in FKBP12.6 knockout mice. Circ Res. 2012;110(3):465–470. doi: 10.1161/CIRCRESAHA.111.253229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sood S, et al. Intracellular calcium leak due to FKBP12.6 deficiency in mice facilitates the inducibility of atrial fibrillation. Heart Rhythm. 2008;5(7):1047–1054. doi: 10.1016/j.hrthm.2008.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chelu MG, et al. Calmodulin kinase II-mediated sarcoplasmic reticulum Ca2+ leak promotes atrial fibrillation in mice. J Clin Invest. 2009;119(7):1940–1951. doi: 10.1172/JCI37059. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.