Significance
Degradation of cellulosic biomass is a key bottleneck in the development of plant-based bioenergies. Studies of cellulolytic microorganisms led to the characterization of a large range of enzymatic systems usable for cellulose breakdown, but current technologies are not efficient enough to warrant their large-scale implementation. This study aims at combining two paradigms in a single system, using a synthetic biology approach. Polysaccharide monooxygenases (redox enzymes capable of boosting cellulose degradation) were engineered to gain the ability to assemble in cellulosomal complexes. As a result of their integration in complexes, these enzymes were able to degrade cellulose at a significantly higher rate compared with the free enzymes, suggesting that this strategy could lead to more efficient technologies for lignocellulose conversion.
Keywords: enzyme synergy, biomass conversion
Abstract
Efficient conversion of cellulose into soluble sugars is a key technological bottleneck limiting efficient production of plant-derived biofuels and chemicals. In nature, the process is achieved by the action of a wide range of cellulases and associated enzymes. In aerobic microrganisms, cellulases are secreted as free enzymes. Alternatively, in certain anaerobic microbes, cellulases are assembled into large multienzymes complexes, termed “cellulosomes,” which allow for efficient hydrolysis of cellulose. Recently, it has been shown that enzymes classified as lytic polysaccharide monooxygenases (LPMOs) were able to strongly enhance the activity of cellulases. However, LPMOs are exclusively found in aerobic organisms and, thus, cannot benefit from the advantages offered by the cellulosomal system. In this study, we designed several dockerin-fused LPMOs based on enzymes from the bacterium Thermobifida fusca. The resulting chimeras exhibited activity levels on microcrystalline cellulose similar to that of the wild-type enzymes. The dockerin moieties of the chimeras were demonstrated to be functional and to specifically bind to their corresponding cohesin partner. The chimeric LPMOs were able to self-assemble in designer cellulosomes alongside an endo- and an exo-cellulase also converted to the cellulosomal mode. The resulting complexes showed a 1.7-fold increase in the release of soluble sugars from cellulose, compared with the free enzymes, and a 2.6-fold enhancement compared with free cellulases without LPMO enhancement. These results highlight the feasibility of the conversion of LPMOs to the cellulosomal mode, and that these enzymes can benefit from the proximity effects generated by the cellulosome architecture.
Cellulose is the most abundant form of organic carbon on Earth. Given its widespread availability and renewability, it is considered a future alternative and sustainable source of energy. The industrial conversion of this polymer in various fuels and chemicals has the potential to replace the consumption of petroleum-based products and, thus, to reduce the environmental and economical costs of fossil fuels (1, 2). However, because of its highly ordered, crystalline structure, cellulose is extremely recalcitrant to depolymerization, thus limiting its cost-effective use as a feedstock for biofuel and chemicals production. In an effort to address the issue of recalcitrance, a large number of studies have focused on the diversity of cellulolytic microorganisms and their cellulose-degradation enzymatic pathways (3, 4).
The consensus model of enzyme-mediated cellulose degradation involves the concerted action of a range of different endo- and exo-acting glucanases, collectively termed “cellulases,” with both enzyme classes performing hydrolysis of the β(1–4) glycosidic bond (5, 6). These enzymes often bear one or more carbohydrate-binding modules (CBM) that can contribute to their activity by promoting binding to the substrate. Interestingly, questions on how these glycoside hydrolases (GH) gain access to the cellulose chain in its crystalline conformation have remained unanswered and have led to the hypothesis that additional factors could modify substrate to make it more accessible (7).
Recently, a series of studies demonstrated that enzymes, termed lytic polysaccharide monooxygenases (LPMOs), were capable of cleaving polysaccharide chains in their crystalline regions through an oxidative mechanism (8–12). Several studies reported that the addition of the enzymes greatly improved the activity of hydrolytic cellulases, resulting in a more efficient conversion of cellulose to fermentable sugars. LPMOs have been described as “cellulase boosters” (13) and likely hold major potential for industrial decomposition of cellulosic materials. These newly described enzymes are classified as “auxiliary activities (AA)” in three families of the Carbohydrate Active Enzyme (CAZy) database: the fungal AA9, formerly known as GH61; the bacterial AA10, formerly known as CBM33 (14, 15); and the recently characterized AA11 (16). Genomic data show that these enzymes are widespread, although they appear to be absent from anaerobic organisms, which coincide with the requirement of molecular oxygen in the catalytic mechanism of the LPMOs. Both families share structural similarities, including a flat substrate-binding surface, a conserved N-terminal histidine residue involved in the coordination of an essential copper ion, and the dependency of the activity on the presence of an electron donor. The reaction mechanism of the majority of the characterized LPMOs involves the oxidation of the C1 carbon of a glucose molecule, leading to the formation of an aldonic acid and a break in the cellulose chain, whereas some members of the AA9 family have been shown to generate oxidation at C4 (17).
Whereas the large majority of the aerobic cellulolytic systems are based on the secretion of individual, free enzymes, a small number of anaerobic bacteria perform efficient degradation of lignocellulose through the secretion of unique, large multienzyme complexes termed cellulosomes (18). These complexes are built around a noncatalytic subunit called the scaffoldin that binds to the insoluble substrate via a cellulose-specific CBM. The scaffoldin also contains a set of subunit-binding modules termed cohesins that mediate the specific incorporation and organization of catalytic subunits through a complementary binding module (the dockerin, carried by each enzymatic subunit) (19). Previous studies have shown that cellulosomal complexes are extremely efficient for the degradation of cellulose and hemicellulose, due to the spatial proximity of synergistically acting enzymes and to the limited diffusion of the enzymes and their substrates (20, 21).
Based on the interlocking modular nature of the cellulosome complex and its components, the concept of “designer cellulosome” was introduced to further manipulate the cellulosomal architecture and incorporate noncellulosomal enzymes to the complexes (22). By taking advantage of the high specificity of the interaction between a cohesin and a dockerin originating from the same species, one can design a chimeric scaffoldin, bearing several cohesin modules from multiple origins, and chimeric enzymes, bearing the corresponding dockerin modules, thus enabling self-assembly of a “tailor-made” enzymatic complex. This concept and its “Lego-like” flexibility has been used to test various enzymatic compositions, the impact of enzyme positioning, and the role of spatial proximity in the synergistic action of the cellulosomal enzyme components (20, 23, 24).
Over the past years, various studies have converted to the cellulosomal mode an array of cellulolytic and hemicellulolytic free enzymes, including endoglucanases, exoglucanases, processive endoglucanases, endoxylanases, and β-glucosidases. In this study, we examined the conversion to the cellulosomal mode of two LPMOs from the aerobic, thermophilic cellulolytic bacterium Thermobifida fusca by attachment of a dockerin module to their respective C termini. We assessed the impact of the conversion procedure on the activity of the chimeric LPMOs, their incorporation into a designer cellulosome, and their contribution toward the enhancement of the enzymatic degradation of crystalline cellulose.
Results
Conversion of T. fusca LPMOs to the Cellulosomal Mode.
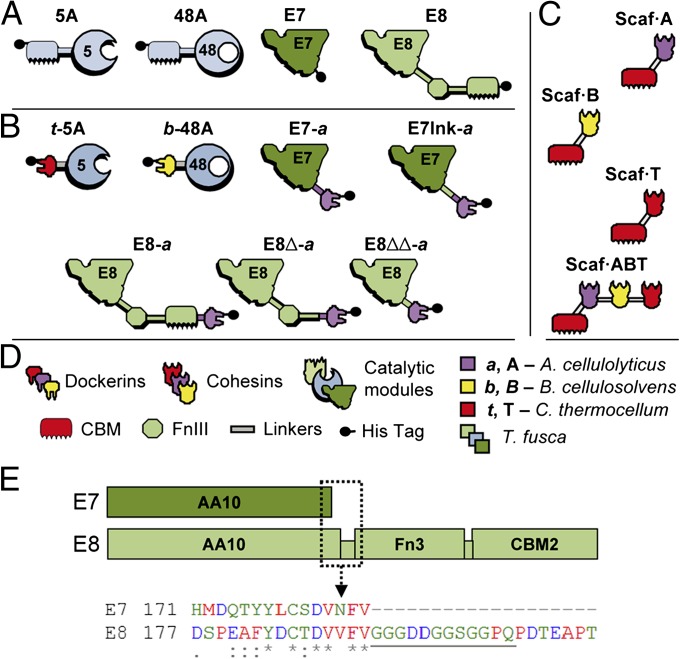
T. fusca has been used as a model organism for the study of thermostable cellulases and their conversion from the free enzymes state to the cellulosomal mode (25). Two LPMOs are encoded in T. fusca genome: TfLPMO10A (hereafter called E7), a 22.7-kDa single-domain AA10; and TfLPMO10B (hereafter called E8), a 44.7-kDa three-domain protein consisting of an N-terminal AA10 domain, followed by a fibronectin 3 and a Family 2 CBM domain (accession nos. YP_289329 and YP_289723). Both enzymes were converted to the cellulosomal mode by fusion to a dockerin domain “DocA” from the bacterium Acetivibrio cellulolyticus (Fig. 1A).
Fig. 1.
Recombinant proteins used in this study. Schematic diagram of the wild-type enzymes (A), chimeric enzymes (B), engineered scaffoldins (C), and key to the diagram (D). Each protein is color coded according to the source of the different modules, as follows: light green, dark green, and light blue, Thermobifida fusca; purple, Acetovibrio cellulolyticus; red, Clostridium thermocellum; yellow, Bacteroides cellulosovens. The numbers 5 and 48 refer to the corresponding CAZY family classification of the catalytic modules (GH5 and GH48). (E, Upper) Diagram of the modular architecture of E7 (Upper, dark green) and E8 (Lower, light green), to scale. (E, Lower) An excerpt of the amino acid sequence alignment (ClustalW) between E7 and E8 is provided, corresponding to the boxed section (dashed line) of the diagram. The linker segment is underlined. The color coding of the residues and the consensus symbols follow the standard ClustalW schemes.
Because the N-terminal histidine residue conserved in LPMOs is essential for their function, the dockerin module can only be fitted at the C-terminal end of the enzymes. Several chimeras were constructed for both LPMOs to assess the impact of different modular architectures (Fig. 1A). Three variants of E8 were produced: E8-a with DocA positioned at the end of the enzyme, E8∆-a where the CBM is replaced by the dockerin, and E8∆∆-a where DocA replaced both the Fn3 and the CBM domains. In the case of E7, two variants were constructed: E7-a, where DocA is fused directly at the C terminus of the AA10 domain, and E7lnk-a, where DocA is fused to E7 by a 12-aa linker. This linker corresponds to that between the AA10 and Fn3 domains of E8 (Fig. 1B). Indeed, E7 and E8 AA10 domains share a high degree of sequence similarity (50% identity and 30% similarity over the 30 C-terminal residues), thus suggesting that the glycine-rich linker could be appropriate to separate the AA10 and dockerin modules. To express the different variants of the LPMOs in Escherichia coli, the coding sequences of the mature proteins were cloned in frame with the pelB signal peptide in the vector pET27b(+). After extraction of the periplasmic fraction, the (His)6-tagged enzymes were purified by Ni-NTA affinity, leading to a purity of >95%, as detected by SDS/PAGE.
Cellulolytic Activity of the Native and Chimeric LPMOs.
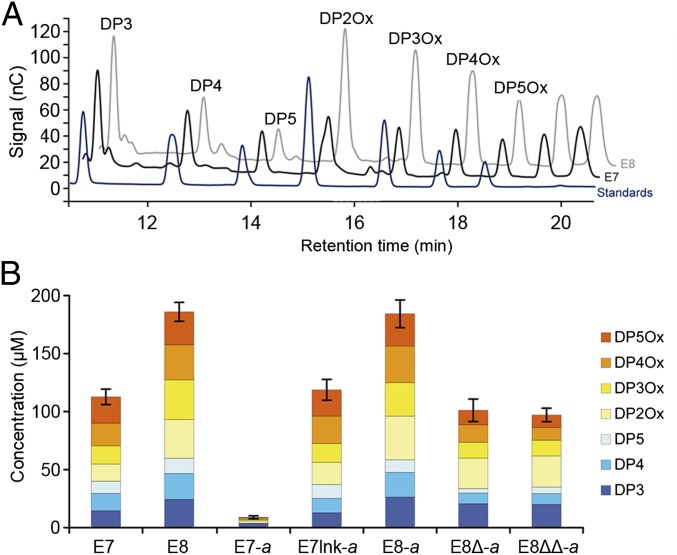
To check their functionality, the different variants of E7 and E8 were assayed for their ability to cleave microcrystalline cellulose in the presence of an electron donor (ascorbic acid). High performance anion exchange chromatography (HPAEC) analysis indicated that the action of E7 and E8 on Avicel led to the release of a mixture of soluble sugars (Fig. 2A). The different peaks were identified based on their measured retention times, and the results indicate that both enzymes released mixtures with similar composition, comprising reduced and oxidized cellooligosaccharides. The degree of polymerization of the released oligosaccharides ranged from 3 to 5 for the reduced products and from 2 to 5 for the oxidized products. The HPAEC analysis also contains two additional peaks with late retention times that probably correspond to aldonic acids cellooligosaccharides with higher degrees of polymerization, but for which no standards were available.
Fig. 2.
Oxidative cleavage of cellulose by E7, E8, and their dockerin variants. (A) HPAEC analysis of the soluble sugars released by the action of E7 (black) and E8 (gray) on Avicel, compared with reduced and oxidized standards (dark blue). Peak annotation was performed according to the standards’ retention times (DP2Ox, cellobionic acid; DP3, cellotriose; DP3Ox, cellotrionic acid; DP4, cellotetraose; DP4Ox, cellotetraonic acid; DP5, cellopentaose; DP5Ox, cellopentaonic acid). Two unlabeled peaks were visible at retention times greater than 19 min for which no standards were available (presumably cellohexaonic and celloheptaonic acids). (B) Quantification by HPAEC analysis of the soluble sugar released from the cleavage of microcrystalline cellulose (Avicel, 10 mg/mL) by 1 µM LPMO, at 50 °C for 72 h. The concentration (in micromolars) of each sugar was determined by integration of the peak area and comparison with a standard curve. Unlabeled peaks were not quantified. Values are the mean of three biological replicates (n = 3). Error bars correspond to one cumulated SD (error bar = ±SDtot; with SDtot = √(SD12 + SD22 + …). For clarity, data are only shown for retention times between 10.5 and 20.5 min. No peak was observed for longer retention times. Detailed concentrations and associated SDs are available in Fig. S1.
Using HPAEC standard curves plotted for each standard, the concentration of the various sugars was determined. This quantitative analysis was used to assess the impact of the conversion to the cellulosomal mode on the LPMOs (Fig. 2B). For the wild-type enzymes, E8 showed activity levels noticeably higher (65%) than those of E7. For the chimeras, E7-a showed a strongly reduced activity compared with wild-type E7, conversely to E7lnk-a and E8-a, which exhibited activity levels similar to their respective wild-type counterparts. The CBM-less variants E8∆-a and E8∆∆-a showed equivalent activity levels, but strongly reduced (46% and 48%, respectively) compared with wild-type E8. Interestingly, this level of activity is similar to what was observed for the other CBM-less LPMOs: E7 and E7lnk-a. This result suggests that the loss in activity observed for E8∆-a and E8∆∆-a is probably caused by the loss of substrate targeting following the removal of the CBM domain, and not by a detrimental effect of the DocA fusion, as observed for E7-a.
Specific Binding of Dockerin-Bearing Enzymes to Cohesins and Incorporation into Designer Cellulosome Complexes.
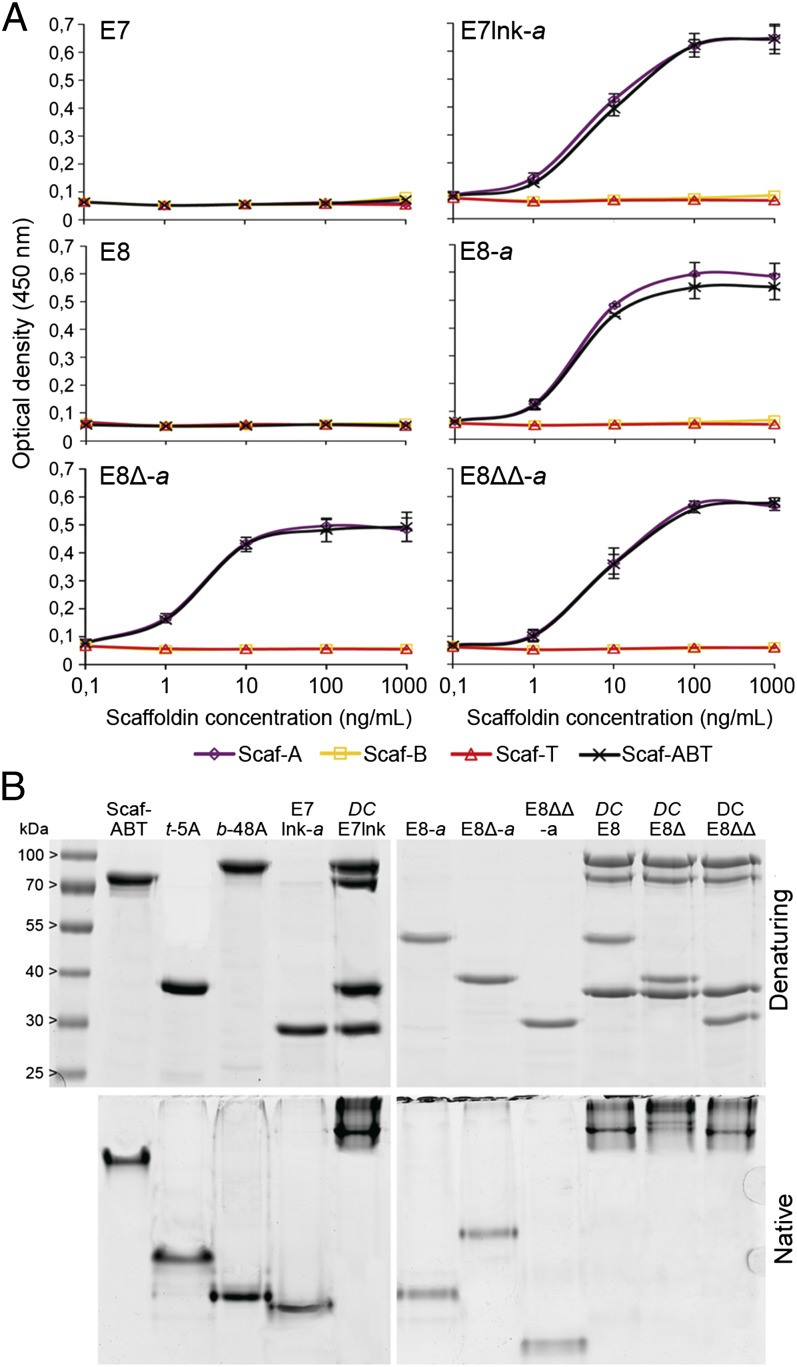
After assessing the activity of the AA10 moiety of the LPMOs variants, the functionality of their dockerin module has to be confirmed. It is especially important to check that the DocA domain is binding specifically to its partner cohesin (CohA). To do so, an affinity-based ELISA was used to measure the binding between the LPMO variants and various engineered scaffoldins (26). Two types of scaffoldins were used (Fig. 1A) in this study: monovalent, bearing a single cohesin domain linked to a CBM module; and trivalent, bearing three different cohesin domains attached to a CBM. Each of the three monovalent scaffoldin bears a cohesin from a different bacterial species, matching the dockerin fused to the chimeric LPMOs and chimeric cellulases.
As expected, neither wild-type E7 nor E8 were able to bind to the scaffoldin (Fig. 3A). Furthermore, the chimeric LPMOs were only able to bind to the CohA-bearing scaffoldins. For each of them, the affinity to the monovalent and the trivalent scaffoldins was identical. These results confirm the specific binding of all of the chimeric LPMOs to scaffoldins bearing a CohA domain. Alongside the LPMOs and trivalent Scaf-ABT, two hydrolytic cellulases were chosen to be part of the final designer cellulosome complex: the endo-glucanase TfCel5A (hereafter called 5A) and the exo-glucanase TfCel48A (hereafter called 48A), both from T. fusca (Fig. 1A). Their chimeric forms, t-5A and b-48A, were constructed by replacing their CBM domains by a dockerin module originating, respectively, from the bacterium Clostridium thermocellum and Bacteroides cellulosolvens. The enzymatic activities and the specific binding properties of these components were determined in previous studies (27).
Fig. 3.
Cohesin–dockerin interactions. (A) Affinity-based ELISA analysis of the interaction between the LPMO variants and the engineered scaffoldins. The LPMOs are individually coated in a plate well. The amount of bound scaffoldin is detected by using HRP-labeled antibodies directed specifically against their CBM modules. Higher amounts of scaffoldin bound are indicated by higher optical densities. Each data point is the mean value of three replicates (n = 3). Error bars correspond to one SD. (B) Electrophoretic mobility of individual components and assembled complexes in denaturing and native PAGE. Equimolar amounts of each protein were analyzed either individually or in combination (DCE7lnk, DCE8, DCE8∆, and DCE8∆∆).
The correct assembly of the various components in a single complex was examined by comparing their electrophoretic mobility in native and denaturing conditions (Fig. 3B). The four distinct designer–cellulosome complexes that were assembled are the following: DCE7lnk, DCE8, DCE8∆, and DCE8∆∆, each containing the scaffoldin Scaf-ABT, the cellulases t-5A and b-48A, and one of the chimeric LPMOs in predetermined equimolar ratios. The electrophoretic profiles of each individual component showed a single major band under both native and denaturing conditions. However, the profiles of the complexes all showed a single band under native conditions but four bands under denaturing conditions, indicating that the designer cellulosomes are all able to self-assemble.
Restoration of LPMOs’ Substrate Targeting by CBM-CohA.
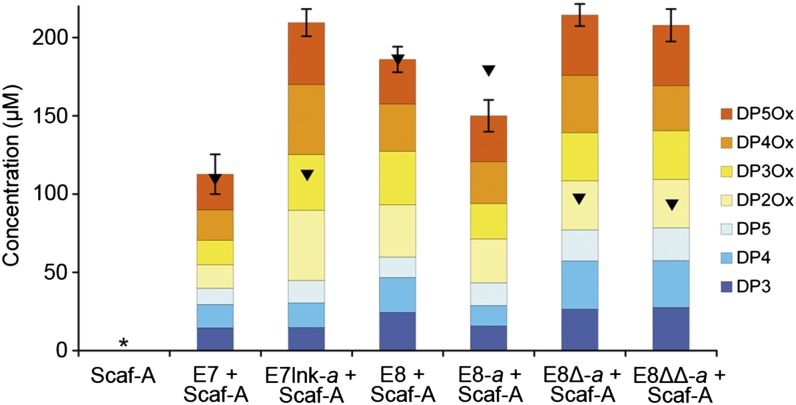
The results of the activity assays showed that the CBM-less LPMOs studied here exhibited lower activity levels than their CBM-bearing counterparts. To confirm that this observation is the result of a loss of substrate targeting, a CBM restoration experiment was performed. The cleavage of Avicel by LPMOs bound to a monovalent scaffoldin Scaf-A was assayed and compared with that of the free enzymes (Fig. 4). As a control, the cellulolytic activity of Scaf-A alone was assayed and yielded no detectable amount of soluble sugars. The activity levels of wild-type E7 and E8 (lacking dockerins) in the presence of Scaf-A, but not bound to it, were similar to those observed previously for the LPMOs alone, confirming that Scaf-A had little or no effect on their enzymatic activity. As anticipated, the activity of E7lnk-a, E8∆-a, and E8∆∆-a coupled to Scaf-A was strongly enhanced compared with the free enzymes, reaching levels of soluble sugars released comparable to that of E8. Interestingly, when bound to Scaf-A, the CBM-bearing E8-a showed a reduction in its activity (−19%), suggesting that the presence of two CBM modules in close proximity in the complex could be detrimental as shown earlier for other types of designer cellulosomes (28).
Fig. 4.
Restoration of the substrate targeting by scaffoldin-borne CBM. Quantification by HPAEC analysis of the soluble sugar released from the cleavage of microcrystalline cellulose (Avicel, 10 mg/mL) by 1 µM of LPMO + 1 µM Scaf-A, at 50 °C for 72 h. Black triangles indicate the total amount of sugars released under the same conditions without the scaffoldin (Fig. 2). See Fig. 2B for additional information. Detailed concentrations and associated SDs are available in Fig. S2.
Cellulolytic Activity of LPMO-Containing Cellulosomes.
Because they only produce small amounts of soluble sugars by themselves, the main interest of using LPMOs for microcrystalline cellulose degradation resides in their synergistic action when paired with one or more hydrolytic cellulases. Having obtained functional LPMO-Dockerin chimeras for E7 and E8, their effect on cellulose hydrolysis by the hydrolytic cellulases 5A and 48A was assayed (Fig. 5). Overall, the three types of geometries assessed were the following: free chimeric enzymes (Free), chimeric enzymes bound to their respective monovalent scaffoldin (CBM), and chimeric enzymes complexed in a designer cellulosome (Designer).
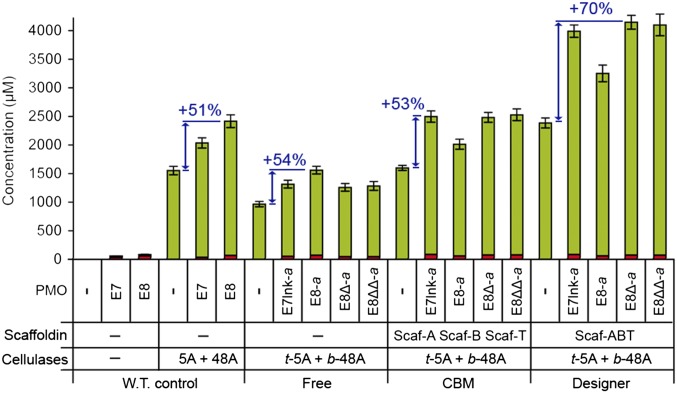
Fig. 5.
Degradation of cellulose by LPMO-containing designer cellulosomes. Quantification by HPAEC analysis of the soluble sugars released from the cleavage of microcrystalline cellulose (Avicel, 10 mg/mL), at 50 °C for 72 h. Different architectures were assayed: free enzymes (W.T. control and Free), enzymes bound to monovalent scaffoldins (CBM), and enzymes bound to trivalent scaffoldin (Designer). Each protein was added at a final concentration of 0.5 µM. Concentrations of reducing sugars (green) and oxidized sugars (red) are shown. Values for reducing sugars were obtained by adding the concentrations of DP1–DP5 (where DP1 = glucose, DP2 = cellobiose, DP3 = cellotriose, DP4 = cellotetraose, and DP5 = cellopentaose). Values for oxidized sugars were obtained by adding the concentrations of DP1Ox-DP5Ox. The detailed concentrations are available in SI Materials and Methods. Blue arrows and the associated values indicate the percent increase (“boost effect”) in reducing sugars, related to the addition of a LPMO, compared to that of the control. Detailed concentrations and associated SDs are available in Fig. S3A. Values for oxidized sugars were obtained by adding the concentrations of DP1Ox-DP5Ox. The detailed concentrations of oxidized sugars and the associated SDs are available in Fig. S3B.
In control experiments (W.T. control), the addition of either wild-type E7 or E8 to a mixture of the wild-type hydrolases led to an increase in soluble sugars released compared with that of the hydrolases in the absence of LPMO (respectively, +28% and +51%). The activity of the hydrolase mixture in different geometries followed a classical pattern for converted enzymes: The free dockerin-bearing chimeras are less active than the wild-type enzymes (here: −38%), CBM restoration via monovalent scaffoldin reestablishes wild-type level of activity (here: −3%), and the assembly in designer cellulosome leads to strong synergy and improved activity compared with the wild type (here: +54%). For each geometry, the addition of a LPMO to the hydrolases systematically led to an increase in the release of soluble sugars compared with the hydrolases alone. In free state, E8-a provided the highest improvement (+54%), whereas E7lnk-a, E8∆-a, and E8∆∆-a had a less beneficial effect (+25–28%). Conversely, in the CBM-restored state, the opposite was observed, with E7lnk-a, E8∆-a, and E8∆∆-a warranting the highest increase (+50–53%), whereas E8-a led to a lower improvement (+20%). The same pattern was observed for the designer cellulosomes, but with superior increases in activity (+63–68% and +34%, respectively). These results demonstrate that the incorporation of LPMOs in designer cellulosome can increase their beneficial impact on hydrolytic cellulases. Overall, by cumulating the benefits of cellulosomal proximity effects and LPMO enhancement of hydrolysis, the most efficient designer cellulosome led to a 2.6-fold increase compared with the free hydrolases.
Discussion
The development of efficient and cost-effective processes for the breakdown of lignocellulose is a key step for implementation of plant-derived biofuels and chemicals. Important progress has been made toward the biological degradation of plant biomass through the study of bacterial and fungal enzymatic systems (4). Over the last few years, a new group of redox enzymes involved in the degradation of cellulose has been described and heralded as a key technological advance in the field of cellulose degradation (29). Indeed, LPMOs, through their oxidative cleavage of crystalline cellulose, allow for a more efficient hydrolysis of cellulose by hydrolytic cellulases (8, 12). In parallel, several studies have shown that highly efficient plant biomass degradation could also be achieved through the use of large multienzyme complexes called cellulosomes (18). The goals of this study were to assess whether LPMOs could be converted to the cellulosomal mode and whether they would retain its cellulolysis enhancement ability while incorporated in a designer cellulosome complex.
To do so, we have chosen to study two LPMOs from the bacterium T. fusca, because our group has already performed the cellulosomal conversion of a dozen biomass-degrading enzymes from this model organism (21, 27, 30, 31). The LPMOs E7 and E8 were initially described in an earlier study (32) but were then considered to be CBMs able of slightly enhancing the activity of other T. fusca cellulases, because their classification as LPMOs was not yet evident. E7 and E8 are both under the control of the CelR regulator, like the majority of the other cellulases and hemicellulases from T. fusca, and their expression is up-regulated when the bacterium is grown on lignocellulosic substrates (33). Recently, they have been characterized as LPMOs, reclassified in the AA10 family, and their cellulolysis enhancement effects were redefined (34). In terms of its modularity, E7 is representative of the most abundant types of architecture of bacterial LPMOs identified to date, because 67% of the known AA10s are single-domain proteins (29). However, E8 has a rare type of modularity, only shared by three known bacterial LPMOs (0.25%), with an Fn3 domain bridging a CBM2 module to the AA10 domain. Interestingly, another architecture comprising two Fn3 domains between the AA10 and CBM2 domains is much more abundant in the CAZY database (10%). Our data suggest that the Fn3 domain of E8 is not essential to the enzyme functionality, confirming the results recently obtained by another group (35). The function of the Fn3 domains remains poorly understood. It is interesting to note that they are found in various cellulosomal cellulases where they are thought to act as a compact form of peptide linker that could be extended when needed, or as solubilization factors for large protein complexes (36).
A mixture of T. fusca endo- and exo-acting cellulases was selected to integrate the complexes alongside the LPMOs, because their complementary activities lead to strong synergistic effects (37). TfCel5A and TfCel48A were chosen because these enzymes were converted successfully to the cellulosome mode in previous studies (27). The insertion of noncellulosomal enzymes into designer cellulosome complexes, although conceptually simple, is not yet a trivial or streamlined process. In most instances, the preferred way of conversion to the cellulosomal mode is by domain swapping of a native CBM for a dockerin. In the case of single-domain enzymes, the addition of a dockerin module is more intricate, because it can be inserted at the N-terminal or C-terminal extremity, either directly or through a linker of various length and composition (30), without a priori knowledge of which solution will be the most efficient. Furthermore, obtaining a functional chimeric enzyme does not necessarily warrant that it will be beneficial when incorporated into a cellulosome complex. For instance, our group reported that the exoglucanase Cel6B from T. fusca could be successfully converted to the cellulosome mode but, upon insertion in a complex, would cause a strong antiproximity effect (31).
In the current study, we obtained one functional chimera for E7 and three for E8. Notwithstanding the loss of substrate targeting effect caused by the removal of the CBM module, it is interesting to note that in free state, the four LPMO-Docs all exhibited similar activity levels. However, when bound to a scaffoldin, E8-a showed a significant loss of activity compared with the other variants of E8. Because it is the only LPMO-Doc that includes a CBM domain, it is possible that adverse interplay may have occurred between the enzyme-borne and the scaffoldin-borne CBMs. A previous study, documenting a similar phenomenon, suggested that CBM modules should only be retained on the enzyme if they have a different substrate specificity than the CBM present on the scaffoldin (23).
The addition of a LPMO to the cellulase mixture had the expected effect, strongly increasing the amount of soluble sugar released, as described (9, 29). Interestingly, this boost effect was enhanced when the LPMOs were bound to the designer cellulosome, in close proximity to the other cellulases. The origin of this proximity effect in cellulosomes is not yet well understood, but previous studies have suggested that it may be linked to limited diffusion of products between complementary acting enzymes (20, 23).
The successful incorporation of LPMOs into cellulosomes for enhanced cellulose degradation presented here offers interesting prospects for future plant-biomass degradation applications. Although mixtures of free cellulases supplemented with LPMOs have been commercially available for industrial applications (38), these systems do not benefit from the advantages offered by the cellulosomal architecture. Reciprocally, the cellulosomal complexes purified from anaerobic bacteria do not benefit from enhanced access to the microcrystalline cellulose warranted by LPMOs (20). In recent years, several studies have aimed at developing a consolidated bio-process based on host microbes like yeast for expressing designer cellulosome (39, 40), to efficiently combine saccharification and fermentation of cellulose to ethanol. These studies propose the expression of divalent and trivalent chimeric scaffoldins on the cell surface of Saccharomyces cerevisiae, and the formation of designer cellulosomes by incorporation of chimeric endo-cellulases and exo-cellulases (optionally coupled to a β-glucosidase). Based on the results presented here, the addition of a LPMO to these systems could lead to highly efficient consolidated processes. A large number of LPMOs from fungi have been expressed in yeast (29) and, given their similarities with AA10s (29), the conversion of AA9s to the cellulosomal mode should be possible by following the principles described here.
Further efforts should also be made to incorporate a wider array of enzymatic activities into cellulosomes. Indeed, the efficient degradation of lignocellulose also requires the breakdown of lignin components or their separation from the polysaccharide backbone. An earlier study successfully converted a feruloyl esterase from the fungus Aspergillus niger (41), and other types of “auxiliary activities” could be explored. Recently, the secretion by T. fusca of two copper-containing oxidases exhibiting laccase-like activities has been highlighted (42). Both enzymes appeared to be active on lignin and, when coupled to cellulases or hemicellulases, warranted enhanced release of soluble sugars from lignocellulosic biomass. After conversion to the cellulosomal mode, these enzymes could be integrated into designer cellulosomes bearing cellulases, hemicellulases, and LPMOs, potentially leading to the creation of synthetic, highly efficient, lignocellulolytic complexes.
Materials and Methods
Cloning.
All recombinant LPMOs were cloned by using a two-step restriction-free procedure. E7 and E8 coding sequences were amplified from T. fusca genomic DNA and cloned into the pET27b(+) vector (Novagen). E7-a, E7lnk-a, E8- a, E8∆-a, and E8∆∆-a were cloned by inserting a sequence coding for the DocA dockerin module at the desired position. All enzyme constructs were designed to contain a His tag for subsequent purification. Plasmids and primers used for restriction-free cloning are available in Table S1. Additional details for this and subsequent methods can be found in SI Materials and Methods.
Expression and Purification.
All recombinant proteins were expressed in E. coli BL21(DE3), and grown in autoinduction media at 20 °C. Protein purification was performed by immobilized metal-ion affinity chromatography on a Nickel-NTA column (Qiagen). The purified proteins were concentrated by ultrafiltration (Vivaspin, 10 kDa molecular mass cutoff, PES membrane; Sartorius) and buffer exchanged against Tris-buffered saline to achieve a 104-fold dilution. Purified proteins were stored in 50% (vol/vol) glycerol at −20 °C.
Cohesin–Dockerin Interaction.
The cohesin–dockerin interaction was analyzed by using an ELISA. The dockerin-bearing enzyme was first coated (0.1 µg) on MaxiSorp plate wells (Nunc), followed by incubation with a CBM-bearing cohesin. The CBM cohesins bound to the plate were detected by using an α-CBM primary antibody and a peroxidase-labeled secondary antibody. Antibody detection was performed by using 3,3′,5,5′-tetramethylbenzidine (ThermoScientific) and quantified spectrophotometrically (OD450).
Designer Cellulosome Assembly.
Equimolar mixtures of the different proteins constitutive of the designer cellulosome were prepared and incubated for 2 h at 37 °C. The electrophoretic mobility of the proteins was analyzed by PAGE under native and denaturing conditions.
Cellulose Degradation Assays.
Cellulose degradation was assayed by incubating the enzymes with 10 mg⋅mL−1 of microcrystalline cellulose (Avicel), at 50 °C for 72 h in 50 mM sodium acetate buffer at pH 5.9 with 2 mM ascorbic acid. Enzyme concentrations were 1 µM for the wild-type and chimeric LPMOs activity assay, 1 µM for the CBM-restoration assays (coupled to 1 µM Scaf-A), and 0.5 µM for the designer cellulosomes assays (coupled to 1 µM of each cellulase and 1 µM Scaf-ABT). In the case of the designer cellulosomes, an equimolar mixture of the different components was first allowed to interact for 2 h at 37 °C.
Soluble Sugar Analysis.
Soluble sugars released during the enzymatic degradation of cellulose were analyzed by HPAEC coupled to pulsed amperometric detection by using a Dionex ICS3000 system equipped with a CarboPac PA1 column.
Supplementary Material
Acknowledgments
We thank David Wilson for providing T. fusca gDNA, Dr. Jean-Guy Berrin for his advice regarding sugar analysis, Dr. Yuval Shoham for granting us access to his equipment, Drs. Sergey Malitsky and Asaph Aharoni for their assistance in setting-up the HPAEC equipment, and Lara Claite-Séchouaite for critically reading an early draft of the manuscript. This research was supported by Israel Science Foundation Grant 1349 (to E.A.B.). Additional support was received by the establishment of an Israeli Center of Research Excellence (I-CORE Center No. 152/11) managed by the Israel Science Foundation, by the Weizmann Institute of Science Alternative Energy Research Initiative, and the Helmsley Foundation (the Leona M. and Harry B. Helmsley Charitable Trust), and grants from the United States-Israel Binational Science Foundation, Jerusalem, Israel. E.A.B. is the incumbent of The Maynard I. and Elaine Wishner Chair of Bio-organic Chemistry.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1404148111/-/DCSupplemental.
References
- 1.Lynd LR, et al. How biotech can transform biofuels. Nat Biotechnol. 2008;26(2):169–172. doi: 10.1038/nbt0208-169. [DOI] [PubMed] [Google Scholar]
- 2.Ragauskas AJ, et al. The path forward for biofuels and biomaterials. Science. 2006;311(5760):484–489. doi: 10.1126/science.1114736. [DOI] [PubMed] [Google Scholar]
- 3.Himmel ME, Bayer EA. Lignocellulose conversion to biofuels: Current challenges, global perspectives. Curr Opin Biotechnol. 2009;20(3):316–317. doi: 10.1016/j.copbio.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 4.Sims REH, Mabee W, Saddler JN, Taylor M. An overview of second generation biofuel technologies. Bioresour Technol. 2010;101(6):1570–1580. doi: 10.1016/j.biortech.2009.11.046. [DOI] [PubMed] [Google Scholar]
- 5.Mba Medie F, Davies GJ, Drancourt M, Henrissat B. Genome analyses highlight the different biological roles of cellulases. Nat Rev Microbiol. 2012;10(3):227–234. doi: 10.1038/nrmicro2729. [DOI] [PubMed] [Google Scholar]
- 6.Henrissat B, Driguez H, Viet C, Schülein M. Synergism of cellulases from Trichoderma reesei in the degradation of cellulose. Bio/Technology. 1985;3:722–726. [Google Scholar]
- 7.Reese ET. A microbiological process report; enzymatic hydrolysis of cellulose. Appl Microbiol. 1956;4(1):39–45. doi: 10.1128/am.4.1.39-45.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vaaje-Kolstad G, et al. An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science. 2010;330(6001):219–222. doi: 10.1126/science.1192231. [DOI] [PubMed] [Google Scholar]
- 9.Forsberg Z, et al. Cleavage of cellulose by a CBM33 protein. Protein Sci. 2011;20(9):1479–1483. doi: 10.1002/pro.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Westereng B, et al. The putative endoglucanase PcGH61D from Phanerochaete chrysosporium is a metal-dependent oxidative enzyme that cleaves cellulose. PLoS ONE. 2011;6(11):e27807. doi: 10.1371/journal.pone.0027807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quinlan RJ, et al. Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc Natl Acad Sci USA. 2011;108(37):15079–15084. doi: 10.1073/pnas.1105776108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Phillips CM, Beeson WT, Cate JH, Marletta MA. Cellobiose dehydrogenase and a copper-dependent polysaccharide monooxygenase potentiate cellulose degradation by Neurospora crassa. ACS Chem Biol. 2011;6(12):1399–1406. doi: 10.1021/cb200351y. [DOI] [PubMed] [Google Scholar]
- 13.Harris PV, et al. Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: Structure and function of a large, enigmatic family. Biochemistry. 2010;49(15):3305–3316. doi: 10.1021/bi100009p. [DOI] [PubMed] [Google Scholar]
- 14.Cantarel BL, et al. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 2009;37(Database issue):D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42(Database issue):D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hemsworth GR, Henrissat B, Davies GJ, Walton PH. Discovery and characterization of a new family of lytic polysaccharide monooxygenases. Nat Chem Biol. 2014;10(2):122–126. doi: 10.1038/nchembio.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Isaksen T, et al. A C4-oxidizing lytic polysaccharide monooxygenase cleaving both cellulose and cello-oligosaccharides. J Biol Chem. 2014;289(5):2632–2642. doi: 10.1074/jbc.M113.530196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bayer EA, Lamed R, White BA, Flint HJ. From cellulosomes to cellulosomics. Chem Rec. 2008;8(6):364–377. doi: 10.1002/tcr.20160. [DOI] [PubMed] [Google Scholar]
- 19.Stahl SW, et al. Single-molecule dissection of the high-affinity cohesin-dockerin complex. Proc Natl Acad Sci USA. 2012;109(50):20431–20436. doi: 10.1073/pnas.1211929109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gefen G, Anbar M, Morag E, Lamed R, Bayer EA. Enhanced cellulose degradation by targeted integration of a cohesin-fused β-glucosidase into the Clostridium thermocellum cellulosome. Proc Natl Acad Sci USA. 2012;109(26):10298–10303. doi: 10.1073/pnas.1202747109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moraïs S, et al. Assembly of xylanases into designer cellulosomes promotes efficient hydrolysis of the xylan component of a natural recalcitrant cellulosic substrate. MBio. 2011;2(6):1–11. doi: 10.1128/mBio.00233-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bayer EA, Morag E, Lamed R. The cellulosome—a treasure-trove for biotechnology. Trends Biotechnol. 1994;12(9):379–386. doi: 10.1016/0167-7799(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 23.Mingardon F, Chanal A, Tardif C, Bayer EA, Fierobe H-P. Exploration of new geometries in cellulosome-like chimeras. Appl Environ Microbiol. 2007;73(22):7138–7149. doi: 10.1128/AEM.01306-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moraïs S, et al. Paradigmatic status of an endo- and exoglucanase and its effect on crystalline cellulose degradation. Biotechnol Biofuels. 2012;5(1):78. doi: 10.1186/1754-6834-5-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gomez Del Pulgar EM, Saadeddin A. The cellulolytic system of Thermobifida fusca. Crit Rev Microbiol. 2014;40(3):236–247. doi: 10.3109/1040841X.2013.776512. [DOI] [PubMed] [Google Scholar]
- 26.Barak Y, et al. Matching fusion protein systems for affinity analysis of two interacting families of proteins: The cohesin-dockerin interaction. J Mol Recognit. 2005;18(6):491–501. doi: 10.1002/jmr.749. [DOI] [PubMed] [Google Scholar]
- 27.Moraïs S, et al. Deconstruction of lignocellulose into soluble sugars by native and designer cellulosomes. MBio. 2012;3(6):1–11. doi: 10.1128/mBio.00508-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fierobe H-P, et al. Degradation of cellulose substrates by cellulosome chimeras. Substrate targeting versus proximity of enzyme components. J Biol Chem. 2002;277(51):49621–49630. doi: 10.1074/jbc.M207672200. [DOI] [PubMed] [Google Scholar]
- 29.Horn SJ, Vaaje-Kolstad G, Westereng B, Eijsink VG. Novel enzymes for the degradation of cellulose. Biotechnol Biofuels. 2012;5(1):45. doi: 10.1186/1754-6834-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caspi J, et al. Effect of linker length and dockerin position on conversion of a Thermobifida fusca endoglucanase to the cellulosomal mode. Appl Environ Microbiol. 2009;75(23):7335–7342. doi: 10.1128/AEM.01241-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caspi J, et al. Thermobifida fusca exoglucanase Cel6B is incompatible with the cellulosomal mode in contrast to endoglucanase Cel6A. Syst Synth Biol. 2010;4(3):193–201. doi: 10.1007/s11693-010-9056-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moser F, Irwin D, Chen S, Wilson DB. Regulation and characterization of Thermobifida fusca carbohydrate-binding module proteins E7 and E8. Biotechnol Bioeng. 2008;100(6):1066–1077. doi: 10.1002/bit.21856. [DOI] [PubMed] [Google Scholar]
- 33.Chen S, Wilson DB. Proteomic and transcriptomic analysis of extracellular proteins and mRNA levels in Thermobifida fusca grown on cellobiose and glucose. J Bacteriol. 2007;189(17):6260–6265. doi: 10.1128/JB.00584-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kostylev M, Wilson D. Two-parameter kinetic model based on a time-dependent activity coefficient accurately describes enzymatic cellulose digestion. Biochemistry. 2013;52(33):5656–5664. doi: 10.1021/bi400358v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Forsberg Z, et al. Comparative study of two chitin-active and two cellulose-active AA10-type lytic polysaccharide monooxygenases. Biochemistry. 2014;53(10):1647–1656. doi: 10.1021/bi5000433. [DOI] [PubMed] [Google Scholar]
- 36.Alahuhta M, et al. Structure of a fibronectin type III-like module from Clostridium thermocellum. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66(Pt 8):878–880. doi: 10.1107/S1744309110022529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jalak J, Kurašin M, Teugjas H, Väljamäe P. Endo-exo synergism in cellulose hydrolysis revisited. J Biol Chem. 2012;287(34):28802–28815. doi: 10.1074/jbc.M112.381624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cannella D, Hsieh C-WC, Felby C, Jørgensen H. Production and effect of aldonic acids during enzymatic hydrolysis of lignocellulose at high dry matter content. Biotechnol Biofuels. 2012;5(1):26. doi: 10.1186/1754-6834-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fan L-H, Zhang Z-J, Yu X-Y, Xue Y-X, Tan T-W. Self-surface assembly of cellulosomes with two miniscaffoldins on Saccharomyces cerevisiae for cellulosic ethanol production. Proc Natl Acad Sci USA. 2012;109(33):13260–13265. doi: 10.1073/pnas.1209856109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim S, Baek S-H, Lee K, Hahn J-S. Cellulosic ethanol production using a yeast consortium displaying a minicellulosome and β-glucosidase. Microb Cell Fact. 2013;12:14. doi: 10.1186/1475-2859-12-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levasseur A, et al. Design and production in Aspergillus niger of a chimeric protein associating a fungal feruloyl esterase and a clostridial dockerin domain. Appl Environ Microbiol. 2004;70(12):6984–6991. doi: 10.1128/AEM.70.12.6984-6991.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen C-Y, Hsieh Z-S, Cheepudom J, Yang C-H, Meng M. A 24.7-kDa copper-containing oxidase, secreted by Thermobifida fusca, significantly increasing the xylanase/cellulase-catalyzed hydrolysis of sugarcane bagasse. Appl Microbiol Biotechnol. 2013;97(20):8977–8986. doi: 10.1007/s00253-013-4727-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.