Abstract
A diverse array of tumor targeting agents ranging in size from peptides to nanoparticles is currently under development for applications in cancer imaging and therapy. However, it remains largely unclear how size differences among these molecules influence their targeting properties. Here we develop a simple, mechanistic model that can be used to understand and predict the complex interplay between molecular size, affinity, and tumor uptake. Empirical relationships between molecular radius and capillary permeability, interstitial diffusivity, available volume fraction, and plasma clearance were obtained using data in the literature. These relationships were incorporated into a compartmental model of tumor targeting using MATLAB to predict the magnitude, specificity, time dependence, and affinity dependence of tumor uptake for molecules across a broad size spectrum. In the typical size range for proteins, the model uncovers a complex trend in which intermediate sized targeting agents (MW ~ 25 kDa) have the lowest tumor uptake, while higher tumor uptake levels are achieved by smaller and larger agents. Small peptides accumulate rapidly in the tumor but require high affinity to be retained, while larger proteins can achieve similar retention with >100 fold weaker binding. For molecules in the size range of liposomes, the model predicts that antigen targeting will not significantly increase tumor uptake relative to untargeted molecules. All model predictions are shown to be consistent with experimental observations from published targeting studies. The results and techniques have implications for drug development, imaging, and therapeutic dosing.
Keywords: Tumor targeting, molecular size, affinity, antibodies, liposomes
Introduction
Traditional small molecule cancer therapeutics suffer from limited selectivity between tumor and healthy tissues, leading to marginal therapeutic indices. One approach for improving diagnostic or therapeutic specificity is to utilize targeting molecules capable of selectively binding antigens overexpressed in the tumor (1). Such agents can deliver payloads such as toxins, radiometals, or imaging agents to the tumor, or may induce cell death in a more direct manner by blocking proliferative signaling or recruiting immune effectors.
Traditionally, tumor targeting has focused on the use of IgG monoclonal antibodies due to their high functional affinity for the target antigen and favorable pharmacokinetic profile. However, effective antibody treatment of many solid tumors remains limited by an inability of the drugs to completely penetrate the tumor tissue leaving regions of untargeted cells (2,3). Additionally, slow plasma clearance of IgGs due to active recycling by the neonatal Fc receptor maintains high drug levels in the blood that complicate imaging or therapy.
With these limitations in mind, a number of alternative tumor targeting molecules have been developed with unique physical properties. Initial development of novel targeting agents focused on making smaller fragments of the full IgG that retain antigen binding properties, including 27 kDa scFvs, 50 kDa Fabs, 80 kDa minibodies, and various scFv and Fab based multimers (4). More recently, alternative binding scaffolds including 14 kDa DARPins and 7 kDa affibodies have been engineered that bind antigens with high affinity despite their small size (5,6). At the other end of the size spectrum, nanoparticles and liposomes with molecular radii ranging from 10–100 nm have been developed that incorporate targeting, imaging, and therapeutic functionalities (7). Chemical conjugation approaches such as PEGylation have allowed for further tailoring of molecular size (8).
While these diverse molecules vary in a number of properties including valency, geometry, stability, and surface charge, the most obvious difference is a wide range of molecular radii. Despite several experimental comparisons, however, the exact effects of these size differences on tumor targeting remain unclear. This confusion arises largely from the fact that size influences several distinct transport parameters relevant to tumor targeting including permeability across the tumor capillary wall (P), diffusivity within the tumor interstitium (D), available volume fraction in the tumor (ε), and rate of plasma clearance (kclear) (8–11). These parameters counteract each other in a manner that makes predicting the effects of size difficult a priori. For instance, small molecules have increased rates of transport across the capillary wall and within the tumor but are also rapidly cleared from the plasma, eliminating the diffusive gradient into the tumor. In contrast, large molecules are cleared from the blood more slowly, but simultaneously suffer from slower rates of transcapillary and interstitial transport.
Due to the complexity of these tradeoffs, computational tools are needed to accurately predict the effects of molecular size on tumor transport. Such models have previously been used to predict antibody microdistribution and macrodistribution within tumors and examine the effect of parameters such as tumor physiology, dose, binding affinity, and antigen turnover (12,13). Here we extend these models to predict the magnitude and specificity of tumor uptake for molecules covering the continuum of sizes from small peptides to liposomes by incorporating derived empirical relationships for the effect of hydrodynamic radius on the parameters P, D, ε, and kclear. Our predictions are consistent with published biodistribution studies targeting HER2 expressing xenografts in mice, as well as clinical data from targeting CEA expressing tumors in humans, and shed light on the complex interplay of size and binding affinity in tumor targeting.
Methods
Previously reported experimental measurements of capillary permeability (P), interstitial diffusivity (D), available volume fraction (ε), and plasma clearance (kclear) for molecules of various sizes in tumor tissues were collected from the literature (Supplemental Tables 1–4). The data sets include studies on proteins, small molecule tracers, dextrans, PEG chains, and liposomes primarily in mouse xenograft models. Mathematical equations describing the relationship between these parameters and molecular radius (Rmol) over a broad continuum of sizes were derived by fitting structural and empirical models of the capillary wall, tumor interstitial space, and renal and non-renal routes of plasma clearance. Implicit in all of these descriptions is the assumption that these are hydrophilic molecules that are not sequestered in membranes or fatty tissue. Fitting was performed using the non-linear least squares method in MATLAB.
The effect of molecular radius on diffusivity (D) and available volume fraction (ε) within the tumor can be described by modeling the tumor interstitial space as a series of small and large right circular cylindrical pores. Using this framework, the molecular diffusivity within each pore can be described as Dpore = Dfree * (Dpore/Dfree), where Dfree is the diffusivity of the molecule in solution (cm2/s) and Dpore/Dfree is the fractional reduction in free diffusion within the pores (14). Dfree can be estimated using the relationship Dfree = (3×10−6 cm2/s)/Rmol, where Rmol is the molecular radius in nm, while Dpore/Dfree can be solved as:
| (1) |
for values of λ < 0.6 where λ is defined as the ratio of molecular radius (Rmol) to pore radius (Rpore) (14). For 0.6 < λ < 1, numerical values of Dpore/Dfree were determined from previously described lookup tables (15). For λ > 1, Dpore/Dfree = 0. To account for diffusion through small and large pores in the tumor, diffusivity over the entire tumor space was defined as D = (A*Dpore_small + B*Dpore_large) where Dpore_small and Dpore_large are the diffusivities in the small and large pores respectively, and A and B are the relative amounts of diffusion that take place through each pore size (A + B = 1).
Using the same self-consistent two pore representation of the tumor interstitial space, the available volume fraction can be described using the equation:
| (2) |
where Vi is the interstitial fluid volume fraction, A and B are the ratios of small and large pores, and φpore_small and φpore_large are the partition coefficients of molecules in each pore size defined as φ = (1 − λ)2 for λ < 1, and φ = 0 for λ > 1 (16). From small molecule tracer studies, Vi was approximated as 0.5 (10). Since both the interstitial diffusivity and void fraction are described by the same model of the interstitial space, data sets describing each parameter (Supplemental Tables 1–2) were simultaneously fit to the respective equations to determine values for Rpore_small, Rpore_large, A, and B.
The effect of molecular radius on vascular permeability was similarly modeled using a two-pore representation of the tumor capillary wall. Transport was assumed to be primarily diffusive in nature due to high interstitial fluid pressure in tumors (17), such that permeability across a pore (cm2/s) can be modeled as:
| (3) |
where Dfree, (Dpore/Dfree), and φ are defined as above (16). Total permeability through small and large pores was defined as P = Acap*Pcap_pore_small + Bcap*Pcap_pore_large where Acap and Bcap are the fractional capillary pore areas per unit membrane thickness (cm−1) for small and large pores respectively. As above, the model was fit to experimentally determined P values (Supplemental Table 3) to estimate Acap, Bcap, Rcap_pore_small, and Rcap_pore_large. While these parameters are similar to those used to describe D and ε, here they are describing pores in the capillary wall versus pores in the interstitial space of the tumor.
Due to the various routes and complexities inherent in plasma clearance, there are no simple structural models to describe the size dependency of the clearance term kclear. Instead a largely empirical model was utilized for the renal and non-renal routes of clearance. For renal clearance, macromolecular filtration can be described as ClR = GFR*Θ, where ClR is the renal clearance in mL/hr, GFR is the rate of fluid filtration across the glomerular wall estimated at 10 mL/hr in female mice (18), and Θ is the macromolecular sieving coefficient. The sieving coefficient depends on molecular size and can be described as (19):
| (4) |
where Φ is the equilibrium partition coefficient, σ is a correction term for the geometry of the glomerular slits approximately equal to 2 for baseline glomeruli, Kconv is the solute hindrance factor for convection, and Pe is the Péclet number defined as:
| (5) |
In this description, v is the fluid velocity vector estimated at 0.001 cm/s, L is the membrane thickness approximated at 100 nm in mice (20), Dfree is the diffusivity in solution discussed above, and Kdiff is the diffusive hindrance factor. Since there are limited mechanistic models for the effect of size on the hindrance factors Kconv and Kdiff, they, along with the partition coefficient, are defined using empirical terms as reported previously (21):
| (6) |
| (7) |
where Rmol is the molecular radius of the targeting agent and α and β are empirical constants fit to the data (units nm−1).
Non-renal clearance was incorporated to account for plasma loss of molecules above the cutoff size for glomerular filtration. With several route of clearance and no structural models a fully empirical model was used with the form:
| (8) |
where ClNR,0 is the non-renal clearance for small molecule tracers (mL/hr), and δ (mL/hr) and γ (nm) are empirical constants fit to the data. While this equation has no physiological significance, it is consistent with experimental observations of decreasing non-renal clearance with increasing molecular size down to a constant level for large molecules (22). ClNR,0 was arbitrarily set to 2 mL/hr to account for the dominance of renal clearance in the size range of small peptides.
The single exponential plasma clearance term kclear (units hr−1) was then defined as:
| (9) |
where Vplasma is the plasma volume estimated in mice as 2 mL (23). For predictions of tumor uptake in human patients, the plasma volume was increased to 3 L. This equation was fit to experimental measurements of kclear for molecules of various sizes (Supplemental Table 4) to determine the constants α, β, δ, and γ. Although a biexponential description of plasma clearance is more physiologically accurate, the single exponential term is a reasonable approximation that allows us to better describe the broad features of size dependent clearance over the entire continuum of molecular radii using a single parameter.
Tumor uptake of targeting molecules was simulated using a mechanistic compartmental model of antibody uptake in tumors1 (Supplemental Figure 1), in which tumor concentration following a sub-saturating bolus iv injection can be described as:
| (10) |
| (11) |
where [Ab]plasma,0 is the initial plasma concentration of the targeting agent (%ID/mL), t is the time, [Ag] is the target antigen concentration (M), ke is the rate of endocytic clearance (sec−1), Kd is the targeting molecule’s affinity for the antigen (M), Rcap is the capillary radius (μm), and RKrogh is the average radius of tissue surrounding each blood vessel (μm). P, ε, and kclear represent permeability, available volume fraction, and plasma clearance rate, respectively, with values for each sized molecule taken from the fits described above. The diffusivity term (D) does not appear in the above equations as Thurber et al. demonstrated in the model derivation that antibody uptake from the tumor surface is negligible relative to vascular uptake for most experimentally or clinically relevant tumor types and sizes, and that the permeability term is the rate limiting step in vascular uptake. The diffusivity term is still included in the methods for fitting interstitial pore size, however, as it provides a check for self-consistency with the size-dependent trends in available volume fraction. Size-independent parameter values were estimated from the literature or were varied as described in each simulation. For figures plotted as a function of effective molecular weight, estimates were made from radius using the relationship MW = 1.32*Rmol3 (fit from data in reference (24)).
Using the above model, the time of peak tumor uptake following bolus injection can be defined as:
| (12) |
where Ω is defined in Equation 11.
Results
Relationship of size to transport parameters
The molecular size of a tumor targeting agent influences four parameters involved in tumor uptake: plasma clearance (kclear), capillary permeability (P), interstitial diffusivity (D), and available volume fraction in the tumor (ε). Values of these parameters for molecules of different sizes were collected from experimental studies reported in the literature (Supplemental Tables 1–4) and used to fit models of the capillary wall, tumor interstitial space, and renal and non-renal routes of plasma clearance. While these models may not fully represent the physiological phenomena behind each parameter, they provide a reasonable framework for describing experimental trends in these parameters over the range of targeting agent sizes.
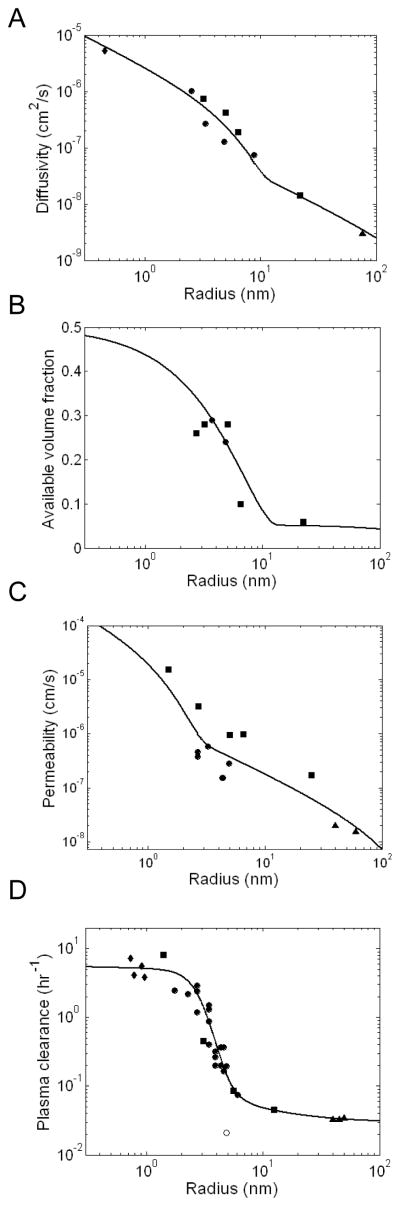
The interstitial diffusivity and available volume fraction data sets are best described by a two-pore model of the tumor interstitial space with pore radii of 13.8 nm and 1 μm at a ratio of 9 to 1 (Figure 1A,B). The small pore size is consistent with previous descriptions of size dependent transport in the tumor interstitial space (14), while the large pores are necessary to account for the observed diffusion of 2 MDa dextran and liposomes. Similarly, the relationship between molecular radius and transcapillary permeability was well fit by a two-pore model of the capillary wall with 4.5 nm and 500 nm radius pores with fractional area to thickness ratios of 17.6 cm−1 and 0.65 cm−1, respectively (Figure 1C). These values are again physiologically reasonable as ~5 nm pores are typical of healthy vasculature (16), and larger pores with 500 nm radii have been observed in leaky tumor vessels due to overexpression of VEGF and other hyperpermeability factors (24).
Figure 1.
Size dependent transport parameters. A, B. Relationship between molecular radius and effective diffusivity (D) and available volume fraction (ε) in the tumor. Data points were simultaneously fit to a two pore model of the tumor interstitial space. C. Relationship between molecular radius (R) and effective molecular permeability across the tumor vasculature (P). Data points were fit to a two pore model of the capillary wall. D. Relationship between molecular radius and plasma clearance rate (kclear). Data points were fit to an empirical model of renal and non-renal clearance. IgG clearance is denoted by an open circle and was not included in the fit. All data fitting was performed using a non-linear least squares method. Data points were collected from experimental results reported in the literature and include measurements of proteins (circles), dextran and PEG polymers (squares), small molecule tracers (diamonds), and liposomes (triangles). Additional descriptions of the experimental data are presented in Supplemental Tables 1–4.
The effect of size on plasma clearance is difficult to model as it is influenced by both renal and non-renal routes of clearance. As such, a largely empirical model of plasma clearance was derived with resulting parameters of α = 1.6 nm−1, β = 0.95 nm−1, δ = 1.94 mL/hr, and γ = 0.20 nm (Figure 1D). While these parameters have no physiological significance, they produce a fit that closely resembles the trend in the data and is similar to previous empirical descriptions of size dependent clearance (25). Plasma clearance data for IgGs are displayed separately and not included in the fit as the Fc domain significantly reduces clearance through interactions with endothelial FcRn receptors (26).
Predicted maximum tumor uptake
A previously described compartmental model of tumor uptake was used to predict the peak tumor concentration achieved for radiolabeled HER2 targeting molecules of various sizes following a bolus iv injection. Values for P, ε, and kclear were determined for each size using the relationships derived above. Size-independent model parameters were estimated for a well-vascularized HER2 expressing xenograft model from values in the literature and are presented in Table 1. While molecules bound to HER2 typically undergo net cellular internalization with a half time of ~6–8 hours ( ref (27) and unpublished results) endocytic clearance in the context of measured total tumor uptake of radiolabel depends also on the rate at which degraded label is cleared from the cell. Therefore we simulated two different radiolabels, faster-clearing 125I and residualizing 99mTc (28).
Table 1.
Size independent parameter values
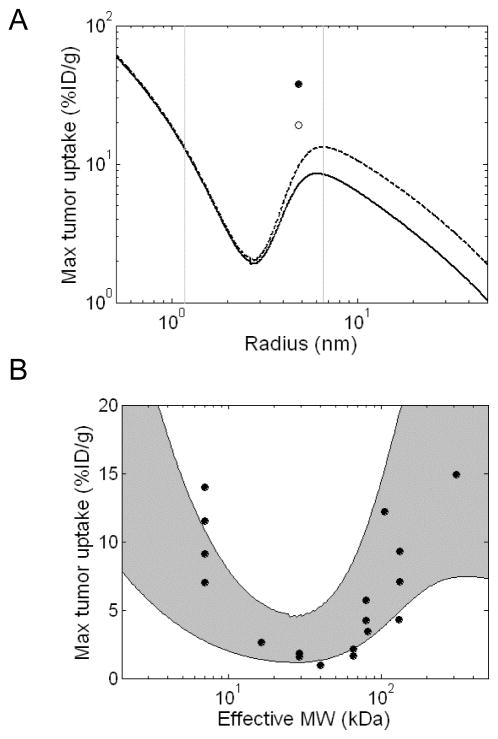
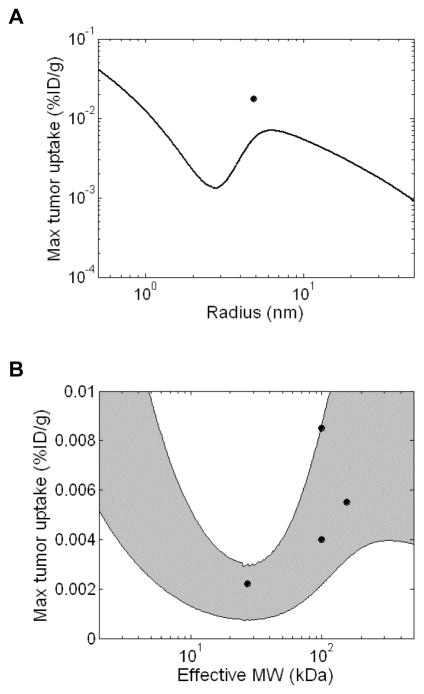
The simulations predict a complex relationship between peak tumor accumulation and size (Figure 2A). In general, predicted tumor uptake is highest for small targeting agents and decreases with increasing molecular radius due to the size-dependent decrease in tumor capillary permeability and available volume fraction reflected in Figures 1B and 1C. However, this trend briefly reverses in the size range typical of proteins producing a local minimum for tumor uptake at a radius of ~2.8 nm and a local maximum at ~6.5 nm. The existence of the minimum and maximum in this curve can be attributed primarily to the sigmoid dependence of renal clearance on size (Figure 1D). Molecules in this size range start to become larger than the kidney filtration cutoff leading to sustained circulation in the plasma which provides increased chances for extravasation into the tumor. Although capillary permeability and available volume fraction are still decreasing in this size range, the decrease in systemic clearance is greater, producing a net increase in tumor uptake. IgGs are predicted to achieve significantly higher tumor uptake than other molecules of equivalent size as a result of their slower plasma clearance due to size-independent FcRn-mediated salvage. The use of residualizing 99mTc is predicted to increase peak tumor uptake relative to 125I labeled molecules when used with large molecules.
Figure 2.
Predicted effect of molecular size on maximum tumor uptake. Simulations were performed using a compartmental model of tumor transport and size-dependent values of P, ε, and kclear. All size-independent parameters are reported in Table 1. Tumor concentrations are reported as %ID/g. A. Predicted peak tumor concentrations of HER2 binding molecules (Kd = 1 nM) labeled with 125I (solid line) or residualizing 99mTc (dashed line). IgG uptake was simulated independently (open circle - 125I, solid circle – 99mTc) and is predicted to be higher due to FcRn mediated reduction in plasma clearance. The vertical grey lines highlight the size range typical of protein therapeutics that is further analyzed in Figure 2B. B. Comparison to experimental data. Peak uptake simulations were performed as above and plotted as a function of effective molecular weight. The predicted uptake trends for RKrogh = 50 μm and RKrogh = 100 μm form the upper and lower bounds respectively of the shaded grey area. Data points were collected from HER2 targeting experiments in the literature including 99mTc, 111In, and 64Cu labeled molecules of various sizes. References and additional details for each experimental data point are presented in Supplemental Table 5. The units of radius and effective MW used in panel A and B, respectively, can be related using the relationship MW = 1.32*Rmol3 (for example 7 kDa affibodies, 27 kDa scFvs, 50 kDa Fabs, and 150 kDa IgGs have radii of 1.74 nm, 2.74 nm, 3.47 nm, and 4.86 nm, respectively.)
Model predictions for 99mTc labeled targeting agents in the size range typical of proteins (2–500 kDa) were compared to data from experimental HER2 targeting studies in the literature (Figure 2B). Each data point represents the highest tumor concentration achieved by a given targeting agent over an experimental time course (Supplemental Table 5). Since the RKrogh value for intercapillary spacing is the least well-characterized parameter in the model and depends on the extent of vascularization and necrosis within the tumor, simulations were performed for a range of values between 50 and 100 μm. While the computational predictions differ quantitatively from the experimental uptake for several molecules in this size range, the overall trends are consistent. In both the simulations and experimental precedents, intermediate sized proteins (~25 kDa) have the lowest tumor uptake while higher levels of targeting are achieved by smaller or larger agents. Higher predicted IgG uptake in the range of 30–40 %ID/g is also consistent with experimental precedents (Supplemental Table 5).
Time dependence of tumor uptake
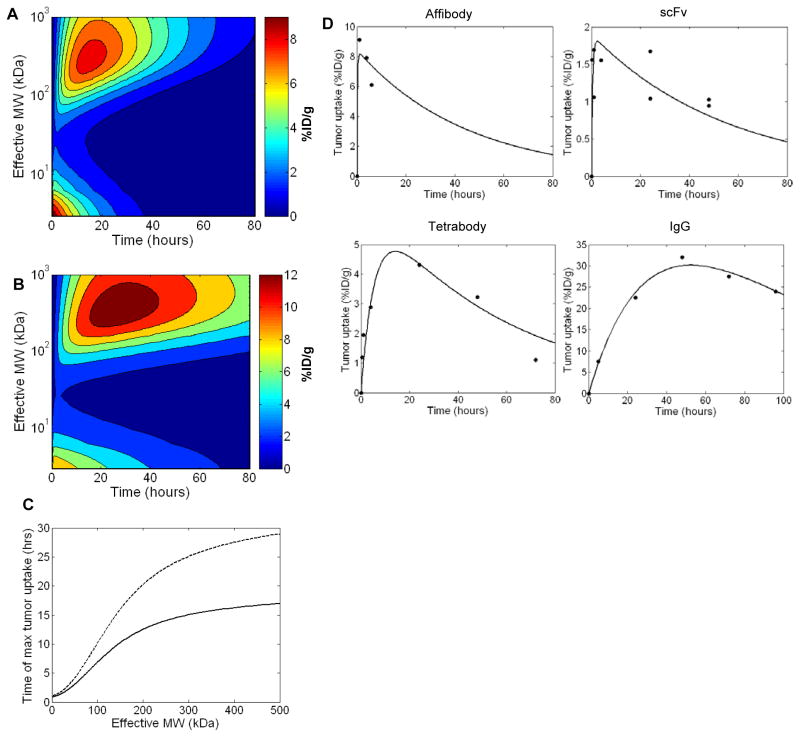
The time of peak tumor uptake and length of tumor retention also have important implications for imaging and therapy. Therefore, full time course simulations were performed for 125I or 99mTc labeled HER2 targeting molecules (Kd = 1 nM) ranging in size from 2–1000 kDa. As reported above, peak tumor levels are similar for proteins at the small and large ends of this size range with a local uptake minimum in between (Figure 3A,B). The time at which peak uptake occurs, however, differs significantly as small macromolecules reach their maximum tumor level within minutes, while uptake of larger molecules occurs on the time scale of hours to days (Figure 3C). Targeting agents labeled with residualizing 99mTc are predicted to achieve peak tumor uptake at later time points and display significantly greater tumor retention than those labeled with 125I as expected given their differences in cellular clearance. The predictions in Figures 3A–C are valid only for molecules lacking Fc domains or other active trafficking as FcRn mediated salvage increases the time until peak tumor uptake by increasing serum persistence.
Figure 3.
Predicted effect of molecular size on time course of tumor uptake. Tumor uptake over time was simulated for 125I (A) or 99mTc (B) labeled non-Fc domain containing HER2 binding molecules (Kd = 1 nM) ranging in size from 2–1000 kDa. C. Effect of molecular size on the time of maximum tumor uptake for 125I (solid line) or 99mTc (dashed line) labeled molecules. D. Comparison to experimental data. Tumor uptake simulations were performed for affibodies (MW = 7 kDa), scFvs (27 kDa), tetrabodies (130 kDa), and IgGs (150 kDa) and compared to experimentally measured time courses for 99mTc labeled HER2 targeting molecules (29–32). RKrogh values were fit to the experimental data for each molecule using the least squares method with results of 57, 80, 101, and 84 μm for the affibody, scFv, tetrabody, and IgG data sets respectively. These values are all in a physiologically reasonable range.
Computational predictions were compared to published biodistribution time courses for anti-HER2 proteins labeled with 99mTc (Figure 3D) (29–32). In order to more directly compare the time-dependency of the predicted uptake, the magnitude of uptake (%ID/g) was adjusted in each case by fitting the RKrogh value to the experimental data. For high affinity targeting molecules (Kd ≪ [Ag]) as is the case here, the RKrogh term impacts the height of the curve but has no influence on the shape or time of peak uptake. In all cases, the computationally predicted time course of tumor loading and retention matches the experimental results well. Affibodies and scFvs achieve peak uptake within the first few hours while larger tetramer and IgG molecules achieve tumor uptake more slowly. IgGs in particular have very slow tumor accumulation with peak uptake after days due to the slow rate of plasma clearance driven by FcRn mediated salvage.
Affinity dependence of tumor uptake
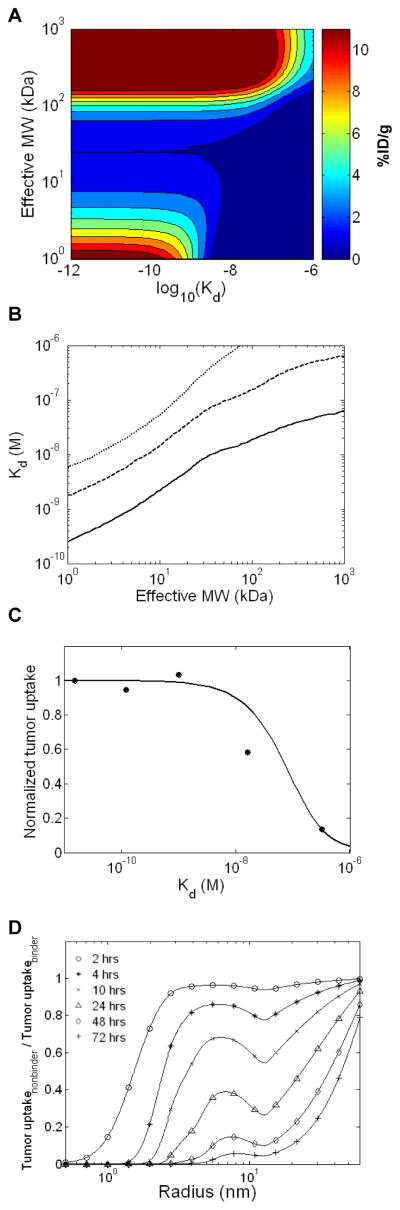
Experimental and theoretical analyses suggest that increasing the affinity of a targeting molecule for its antigen will increase tumor localization up to a point at which tumor levels plateau (12,33). However, the precise affinity at which maximum tumor uptake is achieved depends on the targeting molecule’s size. To examine this relationship in more detail, calculations were performed to predict the tumor uptake at 24 hours for macromolecules varying in both molecular size and affinity. For all sizes in the range of 1–1000 kDa, the expected improvement in tumor uptake with increasing affinity was observed up to a plateau at high affinities (Figure 4A). The threshold affinity of this plateau was size dependent, however, as smaller proteins require tighter binding on the order of 10−10 to 10−8 M Kd values to maintain significant tumor uptake, while large molecules are able to achieve similar uptake levels at much lower affinities in the 10−8 to 10−6 M Kd range (Figure 4A, B). IgGs, for instance, require only a 6×10−7 M affinity to achieve 50% of their peak tumor uptake at 24 hours. The differences in affinity dependence are due to the fact that small, unbound molecules are cleared rapidly from the tumor through vascular intravasation due to their high capillary permeability. As such, small proteins must be anchored to the antigen through high affinity interactions to be retained. In contrast, large, unbound molecules intravasate slowly such that moderate affinity molecules are able to rebind repeatedly and remain in the tumor.
Figure 4.
Binding and affinity dependence. A. Predicted tumor uptake at 24 hours for 99mTc labeled HER2 targeting molecules varying in both size and affinity for the target antigen. B. Affinity necessary to achieve 10% (small dashes), 50% (large dashes), or 90% (solid line) of the maximum tumor uptake at 24 hours as a function of molecular size. C. Comparison to experimental data. The predicted 24 hour tumor concentration for HER2 targeting scFvs (MW = 27 kDa) of various affinities were compared to experimental uptake measurements for affinity variants of the C6.5 scFv (33). Model predictions and experimental data are normalized by their respective uptake values for the highest affinity case. D. EPR mediated non-specific uptake. Predicted tumor concentrations of non-targeted molecules (Kd = 1 M) ranging in radii from 0.5–60 nm were calculated for various times and normalized by the predicted uptake of size matched antigen binding molecules with a Kd of 1 nM (untargeted to targeted uptake ratio). A value of 0 represents fully binding mediated tumor retention, while a value of 1 represents equivalent uptake of targeted and non-targeted molecules. RKrogh = 100 μm.
Computational predictions were compared to experimentally reported tumor uptake data for anti-HER2 scFvs ranging in affinity from 15 pM to 320 nM (33). The model accurately predicts the experimental trend in which the three highest affinity scFvs have similar uptake while lower tumor uptake levels are observed for the 16 nM and 320 nM Kd molecules (Figure 4C).
Non-binding mediated uptake: size dependence of the enhanced permeability and retention (EPR) effect
Experimental studies have suggested that significant tumor accumulation of large macromolecules may occur even in the absence of tumor-specific binding due to the EPR effect (34). We therefore calculated the uptake of untargeted macromolecules relative to the tumor levels of size-matched molecules that bind the target antigen with a 1 nM Kd (Figure 4D). The simulations demonstrate that at early time points uptake is similar for non-targeted and targeted molecules for all but the smallest peptides. Following this initial uptake phase, unbound molecules are cleared rapidly from the tumor while bound molecules are retained, producing a high level of specificity of targeting at later time points for molecules in the size range of most proteins (radius < 10 nm). In contrast, larger molecules in the size range of liposomes (~50 nm) are predicted to have similar tumor levels of targeted and non-targeted molecules even at later time points, as uptake is dominated by EPR effects. This situation arises as the slow clearance of large, unbound molecules by vascular intravasation occurs at the same rate as clearance of antigen-bound molecules by cellular internalization and degradation. These model predictions are consistent with experimentally measured values of tumor uptake specificity (Supplemental Figure 2).
Predicted uptake in human tumors
While comparisons to mouse xenograft studies are a useful validation for model predictions of size dependent trends, the true utility of a model depends on its ability to predict tumor uptake in human patients. Therefore, simulations were performed for tumor uptake of targeting agents of various sizes in human subjects and compared to clinical data for uptake of anti-carcinoembryonic antigen (CEA) scFv, F(ab′)2, DFM, and IgG molecules labeled with 131I (35). [Ab]plasma,0 was reduced from 50 %ID/mL to 0.033 %ID/mL due to the increase in plasma volume from 2 mL to 3 L, and the [Ag] and ke values were changed to 300 nM and 9.6E-6 sec−1 to reflect the different expression and trafficking properties of CEA (36). All other parameter values were left the same as used in the mouse studies as they should be relatively independent of animal species or body weight in their stated form and few measured values are available for human patients. The predicted max tumor level for molecules ranging in size from peptides to liposomes is presented in Figure 5A. The size-dependent trends are identical to those observed in the mouse simulations, while the absolute values are significantly reduced due to the increased plasma volume. The predicted uptake levels in the size range of proteins match closely with the clinically measured tumor concentrations (Figure 5B).
Figure 5.
Predicted tumor uptake in humans. Simulations were performed as described in Figure 2 except with Vplasma = 3L, and [Ag] and ke adjusted for targeting CEA. A. Predicted peak tumor concentrations in humans of CEA binding molecules (Kd = 1 nM) labeled with 125I. IgG uptake was simulated independently and denoted by the solid circle. B. Comparison to clinical data. Peak uptake simulations were performed as above and plotted as a function of effective molecular weight in the size range typical of proteins (2–500 kDa). The predicted uptake trends for RKrogh = 50 μm and RKrogh = 100 μm form the upper and lower bounds respectively of the shaded grey area. The data points represent clinically measured tumor concentrations for scFv, F(ab′)2, DFM, and IgG molecules targeting CEA expressing tumors in humans (35).
Discussion
The increased development of novel tumor binding agents for applications in cancer therapy and imaging has raised the question of how size differences among these molecules impact their targeting properties. Here we incorporate derived relationships between molecular radius and the transport parameters permeability, available volume, and plasma clearance into a compartmental model of tumor uptake to quantitatively assess the effect of molecular size on the magnitude and specificity of tumor localization. Despite the simplicity of the model, we are able to accurately predict several experimental trends for HER2 targeting molecules in mice and CEA targeting molecules in humans, suggesting that size and affinity alone can largely account for the targeting properties of most macromolecules. The modeling framework presented here can also be applied to other tumor types and antigens by simply altering the relevant parameters, which can be independently measured.
While several groups have experimentally or computationally compared tumor uptake for small sets of different sized molecules (30,37,38), here we compare molecules across a broad continuum of molecular radii uncovering complex trends of size dependency. In particular, the model predicts that in the size range of most protein agents, there is a local uptake minimum at ~25 kDa while larger and smaller agents achieve higher tumor levels. This prediction is consistent with experimental measurements of HER2 targeting molecules and suggests that small proteins such as affibodies and DARPins, along with larger molecules including multimers and PEGylated proteins should be superior targeting agents compared to scFvs. For large molecules, uptake can be further increased by incorporation of Fc or albumin binding domains to actively reduce plasma clearance (26,39).
Although small and large proteins are predicted to have similar peak tumor levels, they differ significantly in the time and affinity dependence of uptake. Small proteins achieve high tumor levels rapidly but require high affinity to be retained, as unbound molecules clear from the tumor rapidly. The rapid uptake of small proteins combined with their efficient clearance from the plasma and normal tissues may make them ideally suited for imaging applications (6). In contrast, large molecules can achieve high uptake at comparatively low affinities but accumulate in the tumor on a much slower time scale. These molecules may be best suited for multi-step pretargeting strategies in which the slow clearance from the plasma can be augmented by clearing agents (40).
Outside the size range of typical protein agents, the model predicts very high tumor uptake for small, hydrophilic, high affinity peptides. While high affinity peptides of this size are rare, there are some experimental precedents supporting the prediction. Somatostatin antagonists and glucagon-like peptide analogues have been shown to achieve tumor levels of 61 and 287% ID/g, respectively, in mouse tumor models (41,42). While these high tumor levels may be partially attributable to differences among the tumor models or target antigens, they provide enticing precedents that may motivate future development of targeting agents in this size range. At the other end of the size spectrum, the model predicts lower tumor uptake levels on the order of 1–2 %ID/g for liposomes and other large targeting agents. There are several caveats for making general predictions about liposome and nanoparticle uptake including molecular radii close to the capillary cut-off and greater variation in molecular geometry and chemical structure relative to proteins. Nevertheless, the model predictions are consistent with experimentally measured uptake values of 1–8 %ID/g for a majority of liposomes and nanoparticles (43–45).
One of the more intriguing predictions from the model is that for molecules beyond a certain size there is little to no increase in tumor uptake from antigen targeting. The precise size at which this targeting independent uptake dominates depends on several parameters, but is generally predicted to occur in the ~50 nm size range typical of liposomes and nanoparticles. These predictions are consistent with several experimental reports of insignificant differences in tumor levels of liposomes and nanoparticles with or without targeting ligands (43,44). In contrast, antigen specific targeting may be observed with smaller particles, in tumors with high vascular permeability, or by targeting antigens on the luminal side of the tumor vasculature. For vascular targeting agents in particular, the entire analysis of extravasation and diffusion presented here is irrelevant. It is also important to note that antigen targeting may still improve the therapeutic efficacy of liposomes and nanoparticles even without altering the total tumor concentration by increasing the amount of drug internalized by cells within the tumor (44).
Given the large number of parameters used in the model and the inherent variability in these parameter values due to tumor heterogeneity and experimental error, it is inevitable that there will be some variation or error in the tumor uptake predictions presented here for specific molecules in a given tumor. We believe, however, that the overall trends predicted by the model including predominance of EPR in liposome targeting, and the greater importance of high affinity for small binding molecules should be relatively consistent as they are well supported by published experimental evidence. Better understanding of these trends through the modeling framework presented here should aid in the future design of targeting agents with improved uptake and specificity.
Supplementary Material
Acknowledgments
Funding - National Cancer Institute (CA101830); NCI/MIT/NIGMS Center of Cancer Nanotechnology Excellence; MIT/NIGMS Biotechnology Training Grant
We thank Greg Thurber and John Rhoden for critical comments on the manuscript.
Appendix
Definition of parameter values
- D
Diffusion coefficient in tumor (cm2/s)
- P
Tumor capillary permeability (cm/s)
- ε
Available volume fraction in the tumor
- kclear
Single exponential plasma clearance rate (hr−1)
- Rcap
Capillary radius (μm)
- RKrogh
Radius of tissue surrounding capillary (μm)
- [Ab]plasma0
Initial plasma antibody concentration (%ID/mL)
- [Ab]tumor
Average concentration of total antibody (bound + free) in tumor (%ID/g)
- [Ag]
Antigen concentration in tumor (M)
- Kd
Antibody dissociation constant (M) = koff/kon
- ke
Internalization/degradation rate of bound antibody (sec−1)
- Dfree
Diffusion coefficient in solution (cm2/s)
- Dpore
Diffusion coefficient in cylindrical pore (cm2/s)
- Rmol
Hydrodynamic radius of the targeting molecule (nm)
- Rpore
Radius of the pore (nm)
- λ
Ratio of molecular radius to pore radius
- Vi
Interstitial fluid volume fraction in tumor
- φ
Partition coefficient in pore
- A, B
Relative amounts of diffusion through small and large pores, respectively
- Acap, Bcap
Fractional capillary pore areas per unit membrane thickness through small and large pores, respectively (cm−1)
- ClR
Renal clearance (mL/hr)
- GFR
Glomerular filtration rate (mL/hr)
- Θ
Macromolecular sieving coefficient
- Φ
Equilibrium partition coefficient at glomerular wall
- σ
Correction term for geometry of glomerular slits
- Kconv
Solute hindrance factor for convection
- Kdiff
Diffusive hindrance factor
- Pe
Péclet number, ratio of convection to diffusion
- v
Fluid velocity vector (cm/s)
- L
Membrane thickness (nm)
- α, β
Empirical constants for describing kidney filtration (nm−1)
- ClNR
Non-renal clearance (mL/hr)
- δ, γ
Empirical constants for non-renal clearance (units mL/hr and nm, respectively)
- Vplasma
Plasma volume (mL)
Footnotes
Thurber GM, Wittrup KD. A mechanistic compartmental model for antibody uptake in tumors. Submitted.
References
- 1.Allen TM. Ligand-targeted therapeutics in anticancer therapy. Nat Rev Cancer. 2002;2:750–63. doi: 10.1038/nrc903. [DOI] [PubMed] [Google Scholar]
- 2.Jain RK. Delivery of molecular and cellular medicine to solid tumors. Adv Drug Deliv Rev. 2001;46:149–68. doi: 10.1016/s0169-409x(00)00131-9. [DOI] [PubMed] [Google Scholar]
- 3.Thurber GM, Schmidt MM, Wittrup KD. Factors determining antibody distribution in tumors. Trends Pharmacol Sci. 2008;29:57–61. doi: 10.1016/j.tips.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holliger P, Hudson PJ. Engineered antibody fragments and the rise of single domains. Nat Biotechnol. 2005;23:1126–36. doi: 10.1038/nbt1142. [DOI] [PubMed] [Google Scholar]
- 5.Zahnd C, Wyler E, Schwenk JM, et al. A designed ankyrin repeat protein evolved to picomolar affinity to Her2. J Mol Biol. 2007;369:1015–28. doi: 10.1016/j.jmb.2007.03.028. [DOI] [PubMed] [Google Scholar]
- 6.Orlova A, Magnusson M, Eriksson TL, et al. Tumor imaging using a picomolar affinity HER2 binding affibody molecule. Cancer Res. 2006;66:4339–48. doi: 10.1158/0008-5472.CAN-05-3521. [DOI] [PubMed] [Google Scholar]
- 7.Ferrari M. Cancer nanotechnology: opportunities and challenges. Nat Rev Cancer. 2005;5:161–71. doi: 10.1038/nrc1566. [DOI] [PubMed] [Google Scholar]
- 8.Caliceti P, Veronese FM. Pharmacokinetic and biodistribution properties of poly(ethylene glycol)-protein conjugates. Adv Drug Deliv Rev. 2003;55:1261–77. doi: 10.1016/s0169-409x(03)00108-x. [DOI] [PubMed] [Google Scholar]
- 9.Dreher MR, Liu W, Michelich CR, Dewhirst MW, Yuan F, Chilkoti A. Tumor vascular permeability, accumulation, and penetration of macromolecular drug carriers. J Natl Cancer Inst. 2006;98:335–44. doi: 10.1093/jnci/djj070. [DOI] [PubMed] [Google Scholar]
- 10.Krol A, Maresca J, Dewhirst MW, Yuan F. Available volume fraction of macromolecules in the extravascular space of a fibrosarcoma: implications for drug delivery. Cancer Res. 1999;59:4136–41. [PubMed] [Google Scholar]
- 11.Pluen A, Boucher Y, Ramanujan S, et al. Role of tumor-host interactions in interstitial diffusion of macromolecules: cranial vs. subcutaneous tumors. Proc Natl Acad Sci U S A. 2001;98:4628–33. doi: 10.1073/pnas.081626898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graff CP, Wittrup KD. Theoretical analysis of antibody targeting of tumor spheroids: importance of dosage for penetration, and affinity for retention. Cancer Res. 2003;63:1288–96. [PubMed] [Google Scholar]
- 13.Thurber GM, Zajic SC, Wittrup KD. Theoretic criteria for antibody penetration into solid tumors and micrometastases. J Nucl Med. 2007;48:995–9. doi: 10.2967/jnumed.106.037069. [DOI] [PubMed] [Google Scholar]
- 14.Nugent LJ, Jain RK. Pore and fiber-matrix models for diffusive transport in normal and neoplastic tissues. Microvasc Res. 1984;28:270–4. doi: 10.1016/0026-2862(84)90022-0. [DOI] [PubMed] [Google Scholar]
- 15.Paine PL, Scherr P. Drag coefficients for the movement of rigid spheres through liquid-filled cylindrical pores. Biophys J. 1975;15:1087–91. doi: 10.1016/S0006-3495(75)85884-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michel CC, Curry FE. Microvascular permeability. Physiol Rev. 1999;79:703–61. doi: 10.1152/physrev.1999.79.3.703. [DOI] [PubMed] [Google Scholar]
- 17.Baxter LT, Jain RK. Transport of fluid and macromolecules in tumors. I. Role of interstitial pressure and convection. Microvasc Res. 1989;37:77–104. doi: 10.1016/0026-2862(89)90074-5. [DOI] [PubMed] [Google Scholar]
- 18.Qi Z, Whitt I, Mehta A, et al. Serial determination of glomerular filtration rate in conscious mice using FITC-inulin clearance. Am J Physiol Renal Physiol. 2004;286:F590–6. doi: 10.1152/ajprenal.00324.2003. [DOI] [PubMed] [Google Scholar]
- 19.Deen WM, Lazzara MJ, Myers BD. Structural determinants of glomerular permeability. Am J Physiol Renal Physiol. 2001;281:F579–96. doi: 10.1152/ajprenal.2001.281.4.F579. [DOI] [PubMed] [Google Scholar]
- 20.Yamada E. The fine structure of the renal glomerulus of the mouse. J Biophys Biochem Cytol. 1955;1:551–66. doi: 10.1083/jcb.1.6.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lazzara MJ, Deen WM. Effects of plasma proteins on sieving of tracer macromolecules in glomerular basement membrane. Am J Physiol Renal Physiol. 2001;281:F860–8. doi: 10.1152/ajprenal.2001.281.5.F860. [DOI] [PubMed] [Google Scholar]
- 22.Kaneo Y, Uemura T, Tanaka T, Kanoh S. Polysaccharides as drug carriers: biodisposition of fluorescein-labeled dextrans in mice. Biol Pharm Bull. 1997;20:181–7. doi: 10.1248/bpb.20.181. [DOI] [PubMed] [Google Scholar]
- 23.Kaliss N, Pressman D. Plasma and blood volumes of mouse organs, as determined with radioactive iodoproteins. Proc Soc Exp Biol Med. 1950;75:16–20. doi: 10.3181/00379727-75-18083. [DOI] [PubMed] [Google Scholar]
- 24.Yuan F, Dellian M, Fukumura D, et al. Vascular permeability in a human tumor xenograft: molecular size dependence and cutoff size. Cancer Res. 1995;55:3752–6. [PubMed] [Google Scholar]
- 25.Nakaoka R, Tabata Y, Yamaoka T, Ikada Y. Prolongation of ther serum half-life period of superoxide dismutase by poly(ethylene glycol) modification. Journal of controlled release. 1997;46:253–61. [Google Scholar]
- 26.Ghetie V, Ward ES. Transcytosis and catabolism of antibody. Immunol Res. 2002;25:97–113. doi: 10.1385/IR:25:2:097. [DOI] [PubMed] [Google Scholar]
- 27.Austin CD, De Maziere AM, Pisacane PI, et al. Endocytosis and sorting of ErbB2 and the site of action of cancer therapeutics trastuzumab and geldanamycin. Mol Biol Cell. 2004;15:5268–82. doi: 10.1091/mbc.E04-07-0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karacay H, Ong GL, Hansen HJ, Griffiths GL, Goldenberg DM, Mattes MJ. Intracellular processing of 99Tcm-antibody conjugates. Nucl Med Commun. 1998;19:971–9. doi: 10.1097/00006231-199810000-00007. [DOI] [PubMed] [Google Scholar]
- 29.Willuda J, Kubetzko S, Waibel R, Schubiger PA, Zangemeister-Wittke U, Pluckthun A. Tumor targeting of mono-, di-, and tetravalent anti-p185(HER-2) miniantibodies multimerized by self-associating peptides. J Biol Chem. 2001;276:14385–92. doi: 10.1074/jbc.M011669200. [DOI] [PubMed] [Google Scholar]
- 30.Kubetzko S, Balic E, Waibel R, Zangemeister-Wittke U, Pluckthun A. PEGylation and multimerization of the anti-p185HER-2 single chain Fv fragment 4D5: effects on tumor targeting. J Biol Chem. 2006;281:35186–201. doi: 10.1074/jbc.M604127200. [DOI] [PubMed] [Google Scholar]
- 31.Tran T, Engfeldt T, Orlova A, et al. (99m)Tc-maEEE-Z(HER2:342), an Affibody molecule-based tracer for the detection of HER2 expression in malignant tumors. Bioconjug Chem. 2007;18:1956–64. doi: 10.1021/bc7002617. [DOI] [PubMed] [Google Scholar]
- 32.Tsai SW, Sun Y, Williams LE, Raubitschek AA, Wu AM, Shively JE. Biodistribution and radioimmunotherapy of human breast cancer xenografts with radiometal-labeled DOTA conjugated anti-HER2/neu antibody 4D5. Bioconjug Chem. 2000;11:327–34. doi: 10.1021/bc9901292. [DOI] [PubMed] [Google Scholar]
- 33.Adams GP, Schier R, McCall AM, et al. High affinity restricts the localization and tumor penetration of single-chain fv antibody molecules. Cancer Res. 2001;61:4750–5. [PubMed] [Google Scholar]
- 34.Iyer AK, Khaled G, Fang J, Maeda H. Exploiting the enhanced permeability and retention effect for tumor targeting. Drug Discov Today. 2006;11:812–8. doi: 10.1016/j.drudis.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 35.Green AJ, Johnson CJ, Adamson KL, Begent RH. Mathematical model of antibody targeting: important parameters defined using clinical data. Phys Med Biol. 2001;46:1679–93. doi: 10.1088/0031-9155/46/6/307. [DOI] [PubMed] [Google Scholar]
- 36.Schmidt MM, Thurber GM, Wittrup KD. Kinetics of anti-carcinoembryonic antigen antibody internalization: effects of affinity, bivalency, and stability. Cancer Immunol Immunother. 2008;57:1879–90. doi: 10.1007/s00262-008-0518-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams LE, Wu AM, Yazaki PJ, et al. Numerical selection of optimal tumor imaging agents with application to engineered antibodies. Cancer Biother Radiopharm. 2001;16:25–35. doi: 10.1089/108497801750095989. [DOI] [PubMed] [Google Scholar]
- 38.Thomas GD, Chappell MJ, Dykes PW, et al. Effect of dose, molecular size, affinity, and protein binding on tumor uptake of antibody or ligand: a biomathematical model. Cancer Res. 1989;49:3290–6. [PubMed] [Google Scholar]
- 39.Dennis MS, Jin H, Dugger D, et al. Imaging tumors with an albumin-binding Fab, a novel tumor-targeting agent. Cancer Res. 2007;67:254–61. doi: 10.1158/0008-5472.CAN-06-2531. [DOI] [PubMed] [Google Scholar]
- 40.Goldenberg DM, Sharkey RM, Paganelli G, Barbet J, Chatal JF. Antibody pretargeting advances cancer radioimmunodetection and radioimmunotherapy. J Clin Oncol. 2006;24:823–34. doi: 10.1200/JCO.2005.03.8471. [DOI] [PubMed] [Google Scholar]
- 41.Ginj M, Zhang H, Waser B, et al. Radiolabeled somatostatin receptor antagonists are preferable to agonists for in vivo peptide receptor targeting of tumors. Proc Natl Acad Sci U S A. 2006;103:16436–41. doi: 10.1073/pnas.0607761103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wicki A, Wild D, Storch D, et al. [Lys40(Ahx-DTPA-111In)NH2]-Exendin-4 is a highly efficient radiotherapeutic for glucagon-like peptide-1 receptor-targeted therapy for insulinoma. lin Cancer Res. 2007;13:3696–705. doi: 10.1158/1078-0432.CCR-06-2965. [DOI] [PubMed] [Google Scholar]
- 43.Bartlett DW, Su H, Hildebrandt IJ, Weber WA, Davis ME. Impact of tumor-specific targeting on the biodistribution and efficacy of siRNA nanoparticles measured by multimodality in vivo imaging. Proc Natl Acad Sci U S A. 2007;104:15549–54. doi: 10.1073/pnas.0707461104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kirpotin DB, Drummond DC, Shao Y, et al. Antibody targeting of long-circulating lipidic nanoparticles does not increase tumor localization but does increase internalization in animal models. Cancer Res. 2006;66:6732–40. doi: 10.1158/0008-5472.CAN-05-4199. [DOI] [PubMed] [Google Scholar]
- 45.Gu F, Zhang L, Teply BA, et al. Precise engineering of targeted nanoparticles by using self-assembled biointegrated block copolymers. Proc Natl Acad Sci U S A. 2008;105:2586–91. doi: 10.1073/pnas.0711714105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hilmas DE, Gillette EL. Morphometric analyses of the microvasculature of tumors during growth and after x-irradiation. Cancer. 1974;33:103–10. doi: 10.1002/1097-0142(197401)33:1<103::aid-cncr2820330116>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 47.Jackson TL, Lubkin SR, Siemers NO, Kerr DE, Senter PD, Murray JD. Mathematical and experimental analysis of localization of anti-tumour antibody-enzyme conjugates. Br J Cancer. 1999;80:1747–53. doi: 10.1038/sj.bjc.6690592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baker JH, Lindquist KE, Huxham LA, Kyle AH, Sy JT, Minchinton AI. Direct visualization of heterogeneous extravascular distribution of trastuzumab in human epidermal growth factor receptor type 2 overexpressing xenografts. Clin Cancer Res. 2008;14:2171–9. doi: 10.1158/1078-0432.CCR-07-4465. [DOI] [PubMed] [Google Scholar]
- 49.Press OW, Shan D, Howell-Clark J, et al. Comparative metabolism and retention of iodine-125, yttrium-90, and indium-111 radioimmunoconjugates by cancer cells. Cancer Res. 1996;56:2123–9. [PubMed] [Google Scholar]
- 50.Engfeldt T, Tran T, Orlova A, et al. (99m)Tc-chelator engineering to improve tumour targeting properties of a HER2-specific Affibody molecule. Eur J Nucl Med Mol Imaging. 2007;34:1843–53. doi: 10.1007/s00259-007-0474-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.