Abstract
A negative transcriptional feedback loop generates circadian rhythms in Drosophila. PERIOD (PER) is a critical state-variable in this mechanism, and its abundance is tightly regulated. We found that the Drosophila homolog of Ataxin-2 (ATX2) – an RNA binding protein implicated in human neurodegenerative diseases - was required for circadian locomotor behavior. ATX2 was necessary for PER accumulation in circadian pacemaker neurons, and thus determined period length of circadian behavior. ATX2 was required for the function of TWENTY-FOUR (TYF), a crucial activator of PER translation. Indeed, ATX2 formed a complex with TYF, and promoted its interaction with Poly-A binding protein (PABP). Our work uncovers a role for ATX2 in circadian timing, and reveals that this protein functions as an activator of PER translation in circadian neurons.
In animals, circadian pacemakers synchronize a wide range of processes with the day-night cycle, from basic cellular metabolism to complex behaviors. These molecular clocks are composed of highly conserved transcriptional feedback loops (1). In Drosophila, PERIOD (PER) is a critical component of the circadian pacemaker, and is under tight transcriptional and post-translational control (1). PER, with the help of TIMELESS (TIM), negatively regulates its own gene expression by displacing the dimeric transactivator CLOCK-CYCLE from its promoter (2). PER phosphorylation - regulated by several kinases (NEMO, DOUBLETIME and Casein Kinase II) and by the phosphatase PP2A (3, 4) - controls PER stability, activity and nuclear entry. per mRNA stability and translation are also regulated (5–9). The protein TWENTY-FOUR (TYF) (9) promotes PER translation in the Pigment-Dispersing Factor (PDF)-containing small ventral lateral neurons (sLNvs), which play a particularly important role in the control of circadian behavior (10, 11). TYF binds both Poly-A Binding Protein (PABP) and eukaryotic translation initiation factor 4F (eIF4F), thus presumably promoting per mRNA circularization and translation. However, TYF does not appear to bind directly per mRNA (9).
Ataxin-2 (ATX2) is an RNA binding protein that is proposed to regulate translation. It interacts with PABP, is found in stress granules, and in Drosophila has an important role in miRNA silencing (12–14). In an RNA interference (RNAi) screen aimed at identifying novel regulators of circadian behavior, Atx2 was among the genes linked to miRNA silencing that were downregulated with long or short dsRNAs. The expression of these dsRNAs is controlled by UAS binding sites (15, 16) and can thus be activated with tissue-specific GAL4 transgenes. When Atx2 dsRNAs (Atx2RNAi-1; Fig. S1A) were expressed in all circadian neurons with tim-GAL4 (17), the period of circadian locomotor behavior under constant darkness (DD) was lengthened to about 26.7 hr (Table S1, Fig. 1A). We also noticed increased arrhythmicity, and lower amplitude of rhythms (see “power” in Table S1). These data indicate a crucial role for ATX2 in the circadian molecular pacemaker. Depletion of GW182, a protein essential for miRNA silencing (18), resulted in a distinct circadian phenotype (19), indicating that ATX2 may regulate circadian behavior independently of miRNA silencing.
Fig. 1.
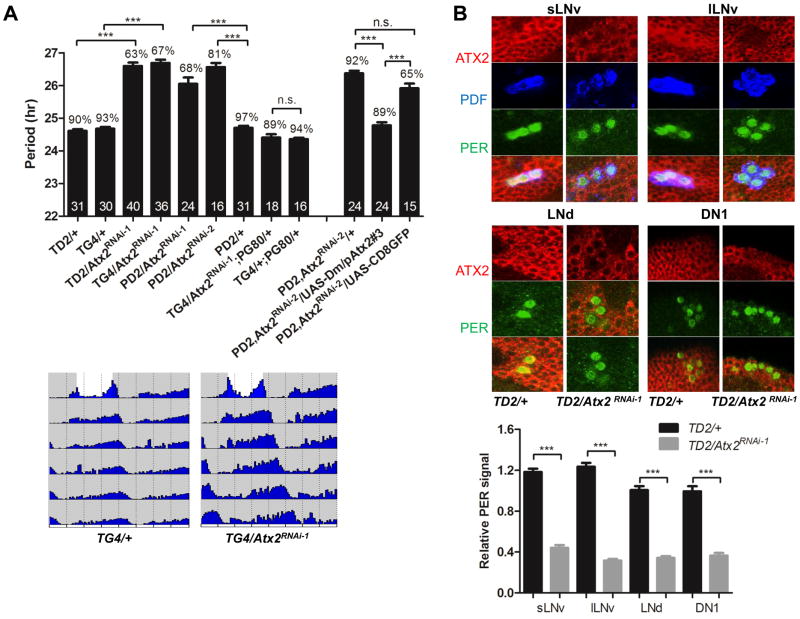
Requirement of ATX2 in LNvs for normal circadian behavior. (A) Effects of depleting ATX2 in pacemaker neurons on period length of circadian behavior. (Upper) Bar graph showing period length (Y axis), percentage of rhythmic flies and number of flies tested (in the bars). Error bars = SEM. TG4 is short for tim-GAL4, TD2 for tim-GAL4, UAS-dicer2, PD2 for Pdf-GAL4, UAS-dicer2 and PG80 for Pdf-GAL80. Dicer2 increases RNAi effects (15). (Lower) Double-plotted actograms of control and TG4/Atx2RNAi-1 flies showing the last day of the light-dark (LD) cycle and 5 days of DD. (B) (Upper) ATX2 is severely downregulated in circadian neurons expressing Atx2 dsRNA. Fly brains were dissected at Zeitgeber Time (ZT) 0 (ZT0 corresponds to the beginning of the light phase of the LD cycle) and immunostained with anti-ATX2, anti-PER and anti-PDF antibodies. (Lower) Quantification of ATX2 levels in sLNvs, lLNvs, LNds and DN1s. Between 11 to 20 neurons were quantified per data point. *** = P< 0.001 as determined by Tukey’s multiple comparison test after one-way ANOVA, n.s. = not significant at the 0.05 level, in both panel A and B.
Because the PDF-containing sLNvs control circadian behavior in DD (10, 11), we restricted expression of Atx2 dsRNAs with the pdf-GAL4 driver. The phenotype in DD was as severe as that observed with tim-GAL4 (Fig. 1A, Table S1). Furthermore, no phenotype was observed when we restricted expression of Atx2 dsRNAs by combining tim-GAL4 with Pdf-GAL80, which blocks GAL4 mediated transcription in PDF cells (10) (Fig. 1A, Table S1). To exclude off-target effects, we expressed a second dsRNA in PDF-containing circadian neurons, which targets a different region of the Atx2 mRNA (Atx2RNAi-2: Fig. S1A). A similar long-period phenotype was observed (Fig. 1A, Table S1). Moreover, when dsRNAs were expressed with tim-GAL4, ATX2 was depleted in all circadian neurons examined (sLNvs, large LNvs [lLNvs], dorsal Lateral Neurons [LNds] and Dorsal Neurons 1 [DN1s], Fig. 1B). Importantly, the long-period phenotype was prevented by expression of a chimeric UAS-Atx2 (UAS-Dm/pAtx2) in which the region targeted by one of the dsRNAs was from D. pseudoobscura and thus sufficiently divergent to be resistant to RNAi (20) (Fig. 1A, Fig. S1A and Table S1). We therefore conclude that ATX2 is required in PDF-containing sLNvs for normal circadian behavior in DD.
No obvious anatomical defects were observed in sLNvs when ATX2 was depleted (Fig. S2A), but subtle developmental defects cannot be excluded. We therefore restricted expression of the dsRNAs either to development or to adulthood with GAL80ts, a temperature sensitive repressor of GAL4 (21). Flies that developed at 29°C and were transferred after eclosion to 18°C to prevent further production of Atx2 dsRNAs behaved like control flies in DD. (Fig. S2B, Table S1). Thus, developmental expression of Atx2 dsRNAs does not appear to affect circadian behavior in adult flies. On the contrary, flies that developed at 18°C and were transferred to 29°C after eclosion showed a ca. 1hr increase in period (Fig. S2B, Table S1). The depletion of ATX2 in adults thus appears to be sufficient to lengthen circadian behavioral rhythms. The weaker effect on period compared to that of life-long ATX2 downregulation (Fig. 1A, Table S1) may reflect less effective depletion of ATX2 in the presence of GAL80ts, either because this protein still weakly represses GAL4 even at high temperature, or because less time has elapsed for the sLNvs to accumulate dsRNAs. We conclude that ATX2 is required acutely in adult sLNvs.
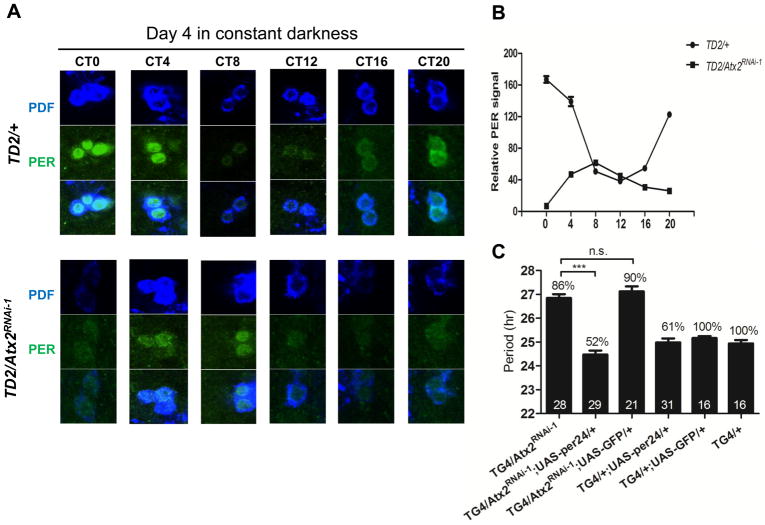
To understand the consequences of ATX2 depletion on the circadian pacemaker, we measured abundance of PER in circadian neurons on the 4th day of DD. PER rhythms were delayed by about 8 hours in the sLNvs (Fig. 2) and in the LNds (Fig. S3A) of flies with low ATX2 amounts, which is consistent with the long-period behavioral phenotype. Unexpectedly, in the DN1s, peak PER concentrations were not delayed, although its minimum concentrations were (Fig. S3A). The reason for this partial delay in DN1s is not clear. Amounts of PER in the sLNvs were low (Fig. 2), but close to normal in the DN1s and the LNds. Amounts of TIM were not affected in any of these neurons (Figure S3B). Low abundance of PER appeared to be sufficient to explain the long period phenotype, because overexpression of PER restored normal period length in flies with depleted ATX2 (Fig. 2C).
Fig. 2.
Regulation of PER accumulation in sLNvs by ATX2. (A) sLNvs of brains from wild-type and Atx2 RNAi flies, dissected at different time points (Circadian Time, CT) during the 4th day of DD and stained with anti-PDF and anti-PER antibodies. (B) Quantification of PER staining in 12–16 sLNvs per data point. (C) PER overexpression corrects the period lengthening induced by Atx2 knock-down. Statistics and abbreviations as in Fig. 1.
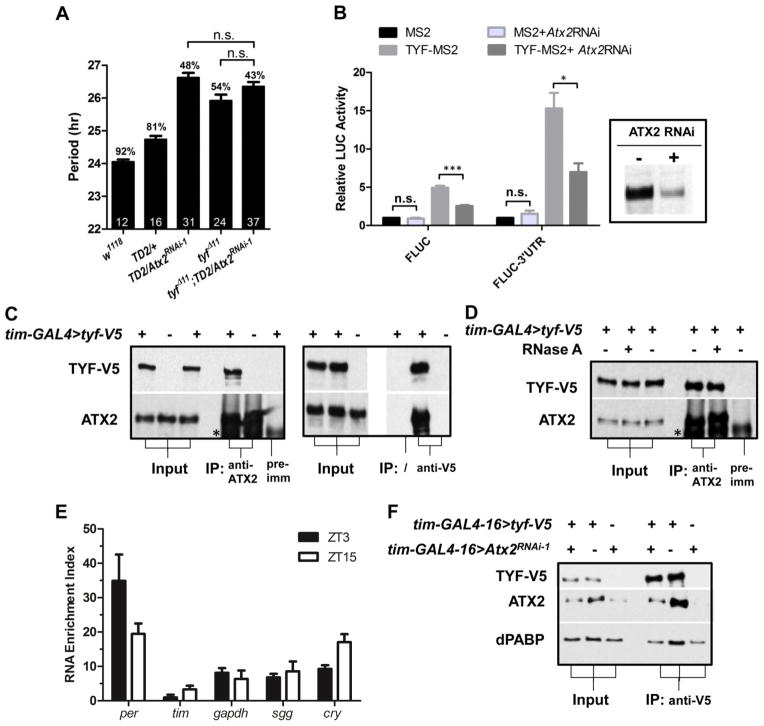
The long period phenotype, increased behavioral arrhythmicity, and low abundance of PER in the sLNvs are reminiscent of the phenotypes observed in tyf mutant flies (9). To determine whether ATX2 and TYF might work together in the circadian cycle, we tested double loss-of-function mutants. The double-mutants showed no additive effects compared to the single mutants (Fig. 3A), which indicates that ATX2 and TYF regulate the same step of the circadian cycle: PER translation.
Fig. 3.
Functional interaction between Atx2 and tyf. (A) Effects of Atx2 RNAi and tyf mutations on circadian period. Statistics and abbreviations as in Fig. 1. (B) Requirement of ATX2 for TYF to promote per translation. MS2 and TYF-MS2 expression plasmids were cotransfected in Drosophila S2R+ cells with firefly luciferase reporter plasmids (FLUC) controlled by MS2 binding sites, in the presence or absence of the per 3′-UTR. Luciferase activity with MS2 was set to 1. dsRNAs targeting Atx2 were added two days before transfection and strongly reduced ATX2 levels (see insertion). N=3. Error bars = SEM. * = P< 0.05, *** = P<0.001, n.s. = not significant (P>0.05) as determined by Student’s t test with Bonferroni correction. (C) Physical association of ATX2 with TYF. Protein from head extracts of flies expressing (+) or not (−) TYF-V5 were immunoprecipitated (IP) with an antibody to ATX2 (left panel) or V5 (right panel). A serum obtained prior to immunization with ATX2 antigens was used as negative control (pre-imm). * = cross-reacting band. (D) The association between ATX2 and TYF is resistant to RNase A treatment, and thus independent of RNA. (E) Association of ATX2 with per and other transcripts. The RNA enrichment index (Y axis) was calculated by subtracting the relative amount of RNA found “immunoprecipitated” with the pre-immunization control from the relative amount found in the immunoprecipitation with the antibody to ATX2, N=4. (F) ATX2 promotes TYF-PABP interactions. Much less PABP is co-immunoprecipitated with TYF-V5 when ATX2 is downregulated by RNAi.
TYF does not bind RNA on its own. Thus, to assay TYF’s translational activity in cell culture, TYF was fused to the MS2 RNA binding protein and MS2 binding sites were placed in the 3′ untranslated region (UTR) of a luciferase reporter gene (9). MS2-TYF promoted luciferase expression, and this transactivation was increased in the presence of per 3′UTR sequences (Fig. 3B), as previously reported (9). If ATX2 was depleted with dsRNAs, TYF transactivation was decreased (Fig. 3B). However, basal luciferase expression was unaffected. We therefore conclude that TYF requires ATX2 to promote PER translation.
To further test cooperative action of ATX2 and TYF to regulate PER translation, we determined whether they form a complex in vivo. We overexpressed a V5-tagged TYF and either isolated TYF complexes with an antibody to V5, or immunoprecipitated endogenous ATX2 with a polyclonal antibody to this protein. In both cases, ATX2 and TYF co-immunoprecipitated (Fig. 3C). Moreover, the ATX2-TYF complexes were resistant to RNase A treatment (Fig. 3D). Because TYF is associated with per mRNA, we tested whether ATX2 is also associated with per mRNA. We immunoprecipitated ATX2 and found per mRNA to be enriched in ATX2 immunoprecipitates compared to control (Fig. 3E, S4A). Although TYF appears to bind per mRNAs preferentially (9), ATX2 also associated with other mRNAs. Thus, how TYF is specifically recruited to per mRNAs remains unclear. A rhythm in ATX2-per mRNA complex formation was observed. It appears to be largely driven by mRNA abundance rhythms (Fig. S4B–C).
TYF interacts with PABP and eIF4F (9). Because ATX2 also binds to PABP (12), we tested whether ATX2 contributes to the formation of a stable TYF complex that bridges eIF4F and PABP and thus stimulates PER translation through mRNA circularization (22). We immunoprecipitated TYF-V5 with an antibody to V5 from whole head extracts of control flies, or from heads of flies expressing Atx2 dsRNAs (Fig. 3F). Depletion of ATX2 reduced the amount of both ATX2 and PABP in the V5-TYF immunoprecipitate. Thus, ATX2 is necessary for the formation of a stable complex between TYF and PABP (Fig. S5).
Our results implicate ATX2 in the control of circadian rhythms as a critical activator of PER translation. They support the emerging notion that translational control is critical for the function of circadian pacemaker neurons (8, 9). Our results also emphasize that ATX2 can function as a translational activator, as well as a translational repressor (12–14). Whether ATX2 functions as an activator or repressor may depend on the combination of proteins associated with a specific mRNA. In the case of per, ATX2’s association with TYF might explain its positive effect on PER translation.
In humans, extension of Ataxin-2’s Poly-Q stretch to pathological lengths is responsible for Spinocerebellar Ataxia and is associated with increased risk of Amyotrophic Lateral Sclerosis (23, 24). Patients affected with Spinocerebellar Ataxia, and even asymptomatic carriers of a pathological Atx2 allele, suffer from sleep disruption, particularly disruption of REM sleep (25–28), which is under circadian regulation (29, 30). Decreased ATX2 activity in the circadian clock perhaps contributes to these sleep disorders.
Supplementary Material
Acknowledgments
We thank D. Wentworth and D. Szydlik for technical assistance, the Allada lab for communicating results prior to publication, and for fly stocks and plasmids; M. Moore and her lab, as well as M. Ramaswami for helpful discussions and reagents, and D. Weaver for critical reading of our manuscript; M. Rosbash for PER and PDF antibodies and N. Sonenberg for PABP antibody; the Vienna Drosophila RNAi Center (VDRC) for Atx2 RNAi lines, R. Stanewsky for fly stocks, the Drosophila Genomics Research Center for the Atx2 cDNA, and the Developmental Studies Hybridoma Bank for PDF antibodies. This work was supported by R01 grants GM66777, GM79182 and GM100091 to P.E.
Footnotes
References and Notes
- 1.Yu W, Hardin PE. J Cell Sci. 2006;119:4793. doi: 10.1242/jcs.03174. [DOI] [PubMed] [Google Scholar]
- 2.Menet JS, Abruzzi KC, Desrochers J, Rodriguez J, Rosbash M. Genes Dev. 2010;24:358. doi: 10.1101/gad.1883910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ko HW, et al. J Neurosci. 2010;30:12664. doi: 10.1523/JNEUROSCI.1586-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sathyanarayanan S, Zheng X, Xiao R, Sehgal A. Cell. 2004;116:603. doi: 10.1016/s0092-8674(04)00128-x. [DOI] [PubMed] [Google Scholar]
- 5.So WV, Rosbash M. EMBO J. 1997;16:7146. doi: 10.1093/emboj/16.23.7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suri V, Lanjuin A, Rosbash M. EMBO J. 1999;18:675. doi: 10.1093/emboj/18.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stanewsky R, Lynch KS, Brandes C, Hall JC. J Biol Rhythms. 2002;17:293. doi: 10.1177/074873002129002609. [DOI] [PubMed] [Google Scholar]
- 8.Bradley S, Narayanan S, Rosbash M. Genetics. 2012;192:943. doi: 10.1534/genetics.112.143248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lim C, et al. Nature. 2011;470:399. doi: 10.1038/nature09728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stoleru D, Peng Y, Nawathean P, Rosbash M. Nature. 2005;438:238. doi: 10.1038/nature04192. [DOI] [PubMed] [Google Scholar]
- 11.Renn SCP, Park JH, Rosbash M, Hall JC, Taghert PH. Cell. 1999;99:791. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- 12.Satterfield TF, Pallanck LJ. Hum Mol Genet. 2006;15:2523. doi: 10.1093/hmg/ddl173. [DOI] [PubMed] [Google Scholar]
- 13.McCann C, et al. Proc Natl Acad Sci U S A. 2011;108:E655. doi: 10.1073/pnas.1107198108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nonhoff U, et al. Mol Biol Cell. 2007;18:1385. doi: 10.1091/mbc.E06-12-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dietzl G, et al. Nature. 2007;448:151. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 16.Ni JQ, et al. Nat Methods. 2011;8:405. doi: 10.1038/nmeth.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaneko M, Park JH, Cheng Y, Hardin PE, Hall JC. J Neurobiol. 2000;43:207. doi: 10.1002/(sici)1097-4695(20000605)43:3<207::aid-neu1>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 18.Eulalio A, Tritschler F, Izaurralde E. RNA. 2009;15:1433. doi: 10.1261/rna.1703809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Emery P. Neuron. 2013 doi: 10.1016/j.neuron.2013.01.035. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langer CC, Ejsmont RK, Schonbauer C, Schnorrer F, Tomancak P. PLoS One. 2010;5:e8928. doi: 10.1371/journal.pone.0008928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGuire SE, Mao Z, Davis RL. Sci STKE. 2004;2004:pl6. doi: 10.1126/stke.2202004pl6. [DOI] [PubMed] [Google Scholar]
- 22.Sonenberg N, Hinnebusch AG. Cell. 2009;136:731. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lastres-Becker I, Rub U, Auburger G. Cerebellum. 2008;7:115. doi: 10.1007/s12311-008-0019-y. [DOI] [PubMed] [Google Scholar]
- 24.Elden AC, et al. Nature. 2010;466:1069. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Velazquez-Perez L, et al. 2011;8:447. [Google Scholar]
- 26.Rodriguez-Labrada R, et al. Movement disorders. 2011;26:347. doi: 10.1002/mds.23409. [DOI] [PubMed] [Google Scholar]
- 27.Tuin I, et al. Neurology. 2006;67:1966. doi: 10.1212/01.wnl.0000247054.90322.14. [DOI] [PubMed] [Google Scholar]
- 28.Boesch SM, et al. Movement disorders. 2006;21:1751. doi: 10.1002/mds.21036. [DOI] [PubMed] [Google Scholar]
- 29.Dijk DJ, Czeisler CA. J Neurosci. 1995;15:3526. doi: 10.1523/JNEUROSCI.15-05-03526.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee ML, Swanson BE, de la Iglesia HO. Curr Biol. 2009;19:848. doi: 10.1016/j.cub.2009.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.