Abstract
In the search for biomarkers of metastasis, attention has been largely placed on ensemble-averaged measurements that screen for molecules or genes. However, individual molecular changes do not always result in disease, and population-based measurements can mask the molecular signatures of the cells responsible for disease. Here, we describe a device that selects for cells based on chemotactic behavior rather than based on molecular differences, enabling the most aggressive cells to be studied independently from the heterogeneous population.
INNOVATION
Cell migratory phenotype drives numerous disease processes, including metastasis, but little is known about the mechanisms that drive the heterogeneity of migratory phenotypes observed within cell populations. While numerous devices have been developed that permit the creation and maintenance of chemotactic gradients, our specific innovation is the ability to then select for sub-populations of cells within the device as a function of their chemotactic response. Sorting cells based on behavior rather than the more conventional approach of sorting cells based on the expression of a particular receptor or molecule may prove to be a valuable method to uncovering the molecular underpinnings of disease.
NARRATIVE
Metastasis is often attributed to the behaviors of individual cells that successfully move from the primary tumor to secondary sites. The traditional approach to identifying molecular biomarkers of metastasis has been through the screening of large populations of cells and tissues. Western blotting, RT-PCR, and microarray analysis (for example) each involve lysing thousands of cells to assay protein or gene expression within cells. However, this approach has met with limited success. One potential reason for this may be that population-based assays mask the behaviors of individual cells and small cell sub-populations. To truly understand the cellular properties that lead to metastasis, the cells that exhibit the most metastatic behaviors should be studied separately from the heterogeneity found in the overall cell population1.
Chemotaxis has been suggested to play a role in metastasis. Both in vivo and in vitro experiments have shown that chemokines and growth factors within the tumor microenvironment can promote invasion and directed migration during metastasis2. Notably, cells, whether from a primary source or a clonal cell line, can exhibit significant variation in their response to chemotactic gradients. Some cells may move directly towards the source, whereas others may take a more indirect path, may not move at all, or may even move away from the source. As such, screening large populations may mask the differences seen only in the small subpopulations that successfully metastasize.
To unmask the heterogeneous responses of cells to chemotactic gradients, we developed a novel microfluidic device that selects for cells based on their chemotactic response to gradients of Epidermal Growth Factor (EGF). We focus on EGF because Epidermal Growth Factor receptor (EGFR) is overexpressed in most human cancers3 and is tightly linked to poor clinical prognosis4. Moreover, EGFR is one of the most common drug targets on the market5. We show that our device can successfully sort cells based on chemotactic response and that these cells can then be isolated for further molecular-based analysis. Further analysis of chemotactic cells relative to metastatic potential may reveal key insights to the molecular drivers of metastatic migration. We expect that this device will be of interest not only to the cancer community but also the broader biomedical community interested in isolating and studying cells based on their migratory properties for application related to tissue engineering, inflammation, and wound healing, to name a few.
RESULTS
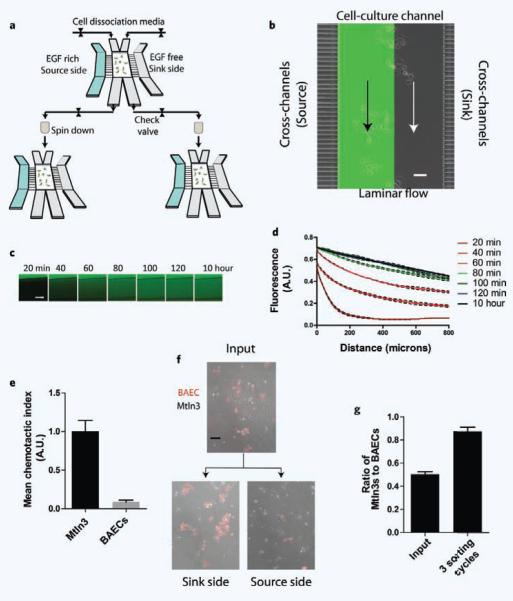
To achieve chemotaxis-based sorting, we fabricated a microfluidic device capable of generating a stable chemotactic gradient of EGF. The device is comprised of three sections (Fig. 1a,b): a central-channel (width: 800 μm, height: 150 μm) used for cell culture, two flow-through side channels that act as a steady source and a steady sink for EGF that diffuses into the central channel through cross-channels (square cross-section, 16 μm2), and finally, inlet and outlet ports into the central channel that allow two laminar flows to be introduced simultaneously into the central channel (Fig. 1a). These inlet and outlet ports into the central channel permit the segregation of cells in each half of the central channel into different populations: one closest to the source and one closest to the sink. After imposing a chemotactic gradient, cells are disassociated from the device using balanced laminar flow through the inlets ports of the central channel, and re-plated in different devices, thereby completing one sorting stage (Fig. 1b; FITC-Dextran solution was used to visualize the formation of laminar flow). To validate and characterize the chemotactic gradient, we used a 5 kDa fluorescein isothiocyanate (FITC)-tagged dextran (approximately the molecular weight of EGF) as a fluorescent marker (Fig. 1c). The gradient stabilized in approximately 2 hours (Fig. 1d) and was linear throughout the channel.
Figure 1.
Design of microfluidic device for chemotaxis (phenotype) based sorting. (a) Schematic showing the device design and sorting scheme. The EGF-rich/free side channels, cross-channels, and central channel. (b) Overlay image of the device showing the three channels, and establishment of laminar flow (visualized by 50 mM of 40 kDa FITC-Dextran). Scale bar: 100 μm. (c) Formation of a chemotactic gradient, as imaged based on the diffusion of FITC-tagged 5 kDa dextran. Scale bar: 200 μm. (d) Dynamics of gradient formation in the central channel over time. The gradient stabilizes in approx. 2 hours. (e) BAECs exhibit negligible mean CI as compared to Mtln3 cells. N = 2 experiments, n = 78 cells. ***p < 0.0001 in Student’s unpaired t-test. (f) Micrographs showing the change in composition of the cell population during sorting. EGF-responsive Mtln3 cells (untagged) dominate the source side and BAEC cells (tagged red) dominate the sink side after three sorting stages. Scale bar: 100 μm. (g) Quantification of sorting efficiency. A 50/50 co-culture of Mtln3 cells and BAECs is refined into an approximately 88% Mtln3-rich cell population at the source side after three sorting cycles. All bar graphs show mean ± SEM.
To validate that the device can sort cells based on chemotactic response, we mixed an EGF-responsive cell line (Mtln3 cells) with an EGF non-responsive cell type (bovine aortic endothelial cells, BAEC). Mtln3 rat mammary adenocarcinoma cells overexpressing EGFR were chosen because of their robust chemotactic response to EGF gradients6. Under identical EGF gradient conditions, the mean chemotactic index (CI, as defined in methods section) of BAECs was negligible as compared to mean CI of Mtln3 cells (Fig. 1e). To separate the non-responsive BAECs from the responsive Mtln3 cells, the cell mixture was subjected to a gradient of EGF, and using laminar flow of cell dissociation solution through the two parallel ports in the culture channel, cells were isolated from the source-side and sink-side of the device. The cells were then separately re-plated into new devices. With each additional segregation stage, there was an increase in the percentage of BAECs on the sink side. After three segregation steps, the cell population collected at the source-side was approximately 85% Mtln3s (Fig. 1f,g), demonstrating the ability of the device to sort the most chemotactic cells from the least chemotactic cells.
To demonstrate the ability of our device to sort cells within one given cell type based on chemotactic behavior, we plated Mtln3 cells in the central channel of our device containing a steady EGF gradient and imaged the cells in real-time for 12 hours at 10 minute intervals. To characterize their response to EGF, the start-to-end trajectory of the cells was categorized based on whether it terminated in the “forward”, “reverse”, or “neutral” sectors of a polar plot. As expected, the cells exhibited a wide range of chemotactic responses (Fig. 2a). Approximately 43% of cells were actively chemotactic towards the source, whereas approximately 41% of cells did not exhibit any significant chemotactic response (n = 73 cells). To sort the cells based on this heterogeneous response, we subjected Mtln3 cells to three stages (S1, S2, and S3) of sorting as described above (Fig. 2b). Upon quantification of all the cell trajectories, we found that the mean CI of cells collected from the source side was consistently and significantly higher than the mean CI of cells collected from the sink side of a device (Fig. 2c). The CI of source-side cells is lower than the CI of cells from the original, unsorted population likely due to de-sensitization effects from prolonged exposure to EGF. Also, we observed no significant change in the mean speed of the cells, irrespective of whether they were collected from the source side or the sink side (Fig. 2d), indicating there is no chemokinesis effect.
Figure 2.
Sorting of cells based on chemotactic phenotype. (a) The heterogeneity of chemotactic response of Mtln3 cells exposed to EGF gradient. n = 73 cells, N = 4 experiments. Scale: 50 μm. (b) Schematic for multi-staged sorting of Mtln3 cells, yielding two distinct cell populations at each stage (S1, S2, and S3): Source-side and sink-side cell population. (c) The cell population collected from source side (S1-, S2-, and S3-source) has a significantly higher mean CI at each stage as compared to cells collected from sink-side (S1-, S2-, and S3-sink), n = 89 cells minimum, N = 4 independent experiments. ***p < 0.0001 in Students’ unpaired t-test. (d) Mtln3 cells collected at different stages of sorting exhibit no significant change in mean speed. (e) Flow cytometry analysis of EGFR surface expression of source and sink sides of the device indicates that EGFR expression is higher in the sink cells than in the source cells (N = 3). (f) Representative flow cytometry analysis of cells collected from the source-side (S1-, S2-, and S3-source) and sink-side (S1-, S2-, and S3-sink) of the device indicating the sink-side cells consistently have fewer EGFRs on the cell surface, as compared to cells on the source-side. Negative control (no antibody) is shown in red. 10,000 cells were analyzed for each condition. (g) CI of the same population of cells when subjected to both forward and reverse gradients over equal periods of time in the same device (without re-plating). Source (marked +, Side A) and sink (marked −, Side B) were interchanged at the end of 12 hours. (h) Mean speed of Mtln3 cells as a function of EGF concentration in the absence of a gradient. (i) EGFR inhibition with PD153035 (75 nM, 15 min incubation) induced a decrease of CI over a 4-hour imaging period. Error bars on bar graphs depict ± SEM.
Having established that the device successfully segregates cells into two distinct chemotactic populations, we examined if differences in the chemotactic phenotype of sorted cells correlate with differences at a molecular level. Since response to EGF is mediated by EGFR expression on the surface of the cells, we focused here on the surface expression of EGFR, however it is also possible to investigate other molecular differences through approaches such as RNA-seq. Indeed, flow cytometry analysis of cells indicates that there was significant difference in the EGFR levels of the sink and source side populations (Fig. 2e,f), with cells at the source side (S1-, S2-, and S3-source) displaying increased levels of EGFR compared to sink side cells. The two-port design of the device also permits the reversal of the gradient and sorting of cells without re-plating the cells as would be required in a transwell assay. We analyzed the CI of cells from the source and sink side of the device before and after the direction of the gradient is reversed (Fig. 2g) and find that the cells on the source side both before and after gradient reversal exhibit a higher chemotactic index. As controls, we also investigated the speed of cells in response to EGF in the absence of a gradient (Fig. 2h) and the chemotactic response in the presence of an inhibitor to the EGFR (Fig. 2i). Our data indicate that the cells are responsive to EGF in the range of concentrations used in our experiments and the chemotactic response we observe is specifically mediated by EGFR. Together, these results show that our multi-staged sorting device segregates cells based on functional phenotype. While our focus was EGFR, cells isolated using our device could also be analyzed for other behaviors (invasion, adhesion, proliferation) or other transcriptional changes using sequencing.
DISCUSSION
Here, we introduce the use of a microdevice to sort cells based on chemotactic response. This approach may offer the opportunity to discover new biomarkers by screening the most aggressive behaviors in smaller sub-populations of cells that may be masked by screening full, heterogeneous populations. Using a microfluidic-based platform offers the advantages of being able to establish well-defined, stable gradients, observe cells in real-time, and isolate cells based on response. Approaches such as this are critical in focusing on the most aggressive cells and unmasking the heterogeneities that exist within cell populations to gain new understanding of the molecular drivers of chemotaxis.
Using microfluidics and microdevices to sort cells has received significant attention in recent literature. However, the primary focus of this pioneering work has been on the isolation and study of rare cells types from complex mixtures of cells like those that occur in blood7–9. Our approach builds from this prior work by focusing on sorting based on cell behavior. Since behaviors are the drivers of disease, this tool and the concept of sorting based on behavior may become a powerful approach in understanding the individual genetic and molecular traits within sub-populations that promote disease. As single cell sequencing technologies move forward, investigating genetic changes in these populations should uncover new insights into the mechanisms that drive the most efficient chemotaxis.
The sorting scheme we report here is based on chemotactic phenotype and could potentially be used in various tissue engineering approaches. For instance, inducing cells to populate a scaffold is essential to the formation and integration of a tissue-engineered construct. Sorting cells based on migratory phenotype opens up the possibility of deriving populations of “super-migrating” cells that could be used for faster production of functional constructs. Additionally, this type of sorting approach could be used to understand drug resistance in cancer treatments, where some cells survive identical drug treatments that arrest other similar cells within the same tumor or in the same clonal population of cells10. Behavior-based selection is a powerful tool towards both understanding disease and controlling for desired cell behaviors.
METHODS SUMMARY
Chemotaxis device fabrication
A polyimide-based mold of the device was exposed on N-type silicon wafers. Briefly, SU-8 2100 was spin-coated on silicon wafers for a target photoresist thickness of 200 μm, using a 5-second, 100 rpm/sec acceleration phase followed by steady phase at 1500 rpm for 45 seconds. After edge-bead removal and two-staged soft-baking at 65 °C and 95 °C for 5 minutes and 45 minutes respectively, the wafer was cooled to room temperature before being exposed to 300 mJ/cm2 using a long-pass filtered UV light (SUSS-ABM©) contact aligner.
The quartz mask was designed for two-staged exposure alignment. The first stage was designed to create the cross channels — 4 μm in both thickness and width and sufficiently long to connect the side channels and the central channel. The second stage was designed for both the central and side channels, which were 200 μm high, 800 μm wide and spaced 50 μm apart. The central channel had inlet and outlet flow-guides at both ends at an emergence angle of 20° each, facilitating the efficient laminar flushing of either sides of the central channel without inducing cross-mixing.
The entire device was cast in PDMS, plasma-cleaned, and bound immediately to the glass substrate to ensure a leak-proof chemotaxis device. To ensure efficient adhesion of cells to the substrate, the whole volume of the device was filled with DI water immediately, and the device was used within 4 hours. For some measurements, PDMS-cast devices were sequentially soaked in triethylamine, ethyl acetate, and acetone for 6 days for long-term usage11 and to make the device bubble-proof during cell culture.
Imaging and analysis
The microfluidic device was assembled using standard tubing and fittings. Approximately 7 nl/min of flow was maintained in both the side-channels and equalized using a hydraulic setup augmented with a peristaltic pump. Bubble traps were introduced at the inlet ports, so as to allow efficient, undisturbed development of gradient in the central channel.
Cells in the central channel were imaged using a Zeiss 10X objective (N.A. 0.3) in real-time for 12 hours at 10-minute intervals, beginning 2 hours after initiating the formation of the gradient to ensure that the recorded images correspond only to the stable phase of the gradient. Individual cell trajectories were tracked manually. The chemotactic index (CI) of a cell was defined as the product of the direction index — reflecting the cosine of the angle between the direction of maximum gradient (a line perpendicular to the length of the source channel) and the net direction of cellular translation — and the speed index, defined as the start-to-end speed of the cell normalized by the mean start-to-end speed of the input population. Each reported CI is normalized by the maximum CI obtained to have a common frame of reference for all experiments.
Segregation scheme
To induce and control segregation of cells with different chemotactic potential, the cells were exposed to twin, balanced, laminar flow (150 μm/sec) of pre-warmed cell dissociation media (50% Trypsin/EDTA in phosphate buffered saline (PBS)), with a residence time of approximately 130 sec, for a duration of 5 minutes. The detached cells were collected at each port on the outlet into a 1.5 ml Eppendorf tube. After washing three times with cell culture media, the two cell subpopulations were plated in different, new devices. The same scheme was repeated for each stage of sorting. We estimate a near 30% loss of cells between one device and the next.
Cell culture and plating
Mtln3 rat mammary adenocarcinoma cells were cultured in α-MEM (Life Technologies Corp., Carlsbad, CA) supplemented with 5% fetal bovine serum (FBS), and 1% penicillin/streptomycin (Gibco, Life Technologies, Carlsbad, CA). Co-culture of bovine aortic endothelial cells (BAECs) and Mtln3 cells was done in Mtln3 media. For experiments requiring tagging cells, CellTracker™ (Molecular Probes, OR) was used per the manufacturer’s protocol. Cells were cultured at 37 °C and 5% CO2 during cell culture and imaging. All cells used were between passages 6 and 12, with a passage ratio of 1:20. For plating cells inside the microfluidic device, the central channel of the device was quenched with de-ionized water and incubated in 1% fibronectin solution in PBS for 30 minutes. Cells were plated in the central channel at a density not less than 5,000 cells/ml.
Flow cytometry
Cells harvested from either the source-side or the sink-side of each sorting stage were spun-down and resuspended in 500 μl ice-cold staining buffer (1X PBS, 0.1% sodium azide, 2% BSA). Using an Alexa-fluor 647 pre-conjugated anti-EGFR antibody (sc-120 AF647, Santa Cruz Bio-technology, CA), cells were stained in suspension at 1:20 and incubated for 30 minutes. Excess antibody was removed by washing the cells three times in ice-cold staining buffer. Cells were immediately mounted on a CyFlow Space™ (Partec, NJ) flow cytometer at a concentration of 40,000 cells/ml and probed at the far red channel. As negative control, unstained Mtln3 cells were used.
ACKNOWLEDGEMENTS
This work was funded by an NSF CAREER Award (CBET 1055502) and a grant from the National Cancer Institute through the Cornell Center on Microenvironment and Metastasis (award number U54CA143876).
Th is work was performed in part at the Cornell NanoScale Facility, a member of the National Nanotechnology Infrastructure Network, which is supported by the National Science Foundation (Grant ECCS-0335765). Mtln3 cells were kindly gifted by Dr. JeffSegall, Albert Einstein College of Medicine.
REFERENCES
- 1.Altschuler SJ, Wu LF. Cellular heterogeneity: Do differences make a difference? Cell. 2010;141(4):559–563. doi: 10.1016/j.cell.2010.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Condeelis J, Singer RH, Segall JE. The Great Escape: When cancer cells hijack the genes for chemotaxis and motility. Annu. Rev. Cell Dev. Biol. 2005;21(1):695–718. doi: 10.1146/annurev.cellbio.21.122303.120306. [DOI] [PubMed] [Google Scholar]
- 3.Salomon DS, Brandt R, Ciardiello F, Normanno N. Epidermal growth factor-related peptides and their receptors in human malignancies. Crit. Rev. Oncol. 1995;19(3):183–232. doi: 10.1016/1040-8428(94)00144-i. [DOI] [PubMed] [Google Scholar]
- 4.Fox S, Smith K, Hollyer J, Greenall M, Hastrich D, Harris A. The epidermal growth factor receptor as a prognostic marker: Results of 370 patients and review of 3009 patients. Breast Cancer Res. Treat. 1994;29(1):41–49. doi: 10.1007/BF00666180. [DOI] [PubMed] [Google Scholar]
- 5.Tortora G, Ciardiello F, Gasparini G. Combined targeting of EGFR-dependent and VEGF-dependent pathways: Rationale, preclinical studies and clinical applications. Nat. Clin. Pract. Oncol. 2008;5(9):521–530. doi: 10.1038/ncponc1161. [DOI] [PubMed] [Google Scholar]
- 6.Bailly M, Condeelis JS, Segall JE. Chemoattractant-induced lamellipod extension. Microsc. Res. Tech. 1998;43(5):433–443. doi: 10.1002/(SICI)1097-0029(19981201)43:5<433::AID-JEMT9>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 7.Ozkumur E, Shah AM, Ciciliano JC, et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci. Transl. Med. 2013;5(179):179ra47–179ra47. doi: 10.1126/scitranslmed.3005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hughes AD, Mattison J, Western LT, Powderly JD, Greene BT, King MR. Microtube device for selectin-mediated capture of viable circulating tumor cells from blood. Clin. Chem. 2012;58(5):846–853. doi: 10.1373/clinchem.2011.176669. [DOI] [PubMed] [Google Scholar]
- 9.Nagrath S, Sequist LV, Maheswaran S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450(7173):1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell LL, Polyak K. Breast tumor heterogeneity: Cancer stem cells or clonal evolution? Cell Cycle. 2007;6(19):2332–2338. doi: 10.4161/cc.6.19.4914. [DOI] [PubMed] [Google Scholar]
- 11.Zhou J, Ellis AV, Voelcker NH. Recent developments in PDMS surface modification for microfluidic devices. Electrophoresis. 2010;31(1):2–16. doi: 10.1002/elps.200900475. [DOI] [PubMed] [Google Scholar]