Abstract
The generation of cellular diversity is dependent on the precise spatiotemporal regulation of gene expression by both cis- and trans-acting mechanisms. The developmental principles regulating expression of specific gene subsets in individual cell types are not fully understood. Here we define the cis-regulatory mechanisms driving expression of cell-selective and broadly expressed genes in vivo in the AWB olfactory neuron subtype in C. elegans. We identify an element that is necessary to drive expression of neuron-selective chemoreceptor genes in the AWB neurons, and show that this element functions in a context-dependent manner. We find that the expression of broadly expressed sensory neuronal genes in the AWB neurons is regulated by diverse cis- and trans-regulatory mechanisms that act partly in parallel to the pathways governing expression of AWB-selective genes. We further demonstrate that cis-acting mechanisms driving gene expression in the AWB neurons appear to have diverged in related nematode species. Our results provide insights into the cis-regulatory logic driving cell-specific gene expression, and suggest that variations in this logic contribute to the generation of functional diversity.
Keywords: cis-regulatory motifs, chemoreceptors, olfactory neuron, C. elegans
INTRODUCTION
Cellular functional diversity is generated by the precise spatiotemporal expression of specific gene subsets. Gene expression in turn, is dictated by cis-regulatory sequences which interact with ensembles of transcription factors (TFs) to drive activation or repression of transcription (Yuh et al., 1998; Howard and Davidson, 2004). Genome-wide searches for genes sharing cis-acting sequences have led to the identification of predicted co-regulated genes (Halfon et al., 2002; Markstein et al., 2002; Efimenko et al., 2005). In a complementary approach, the cis-regulatory sequences of co-expressed genes have been analyzed to identify motifs required for co-regulation (Roth et al., 1998; Gaudet and Mango, 2002; Zhang et al., 2002; Wenick and Hobert, 2004; Etchberger et al., 2007; McGhee et al., 2007). However, experimental verification of spatiotemporal expression patterns has shown that the presence of a sequence motif may not be predictive of the expression pattern of the corresponding gene (Harbison et al., 2004; Efimenko et al., 2005; Etchberger et al., 2007), and conversely, that co-expressed genes may not necessarily be co-regulated (Wenick and Hobert, 2004; Zhao et al., 2005; Etchberger et al., 2009). Thus, bioinformatics-based analyses of cis-regulatory sequence information, coupled with experimental validation of identified motifs, may be a more effective approach towards defining the cis-regulatory logic driving gene expression.
The availability of genome sequences, and the ability to rapidly verify predicted gene expression patterns in model organisms such as S. cerevisiae, C. elegans and D. melanogaster, have led to a description of the cis-acting sequences driving gene expression in specific cell or tissue types, or under defined environmental conditions (eg. Gaudet and Mango, 2002; Ao et al., 2004; Markstein et al., 2004; McGhee et al., 2007; Harbison et al., 2004). Identification and comparison of cis-regulatory sequences in phylogenetically related species have also enabled analyses of evolutionary constraint and divergence in gene expression patterns (Kellis et al., 2003; Grad et al., 2004; Wittkopp et al., 2008). These experiments have shown that similar to mutations in coding regions, variations in cis-regulatory sequences are also critically important for the generation of phenotypic diversity (Wray, 2007).
Cell fate diversification is especially critical in the nervous system which consists of thousands of different specialized cell types. For instance, in the chemosensory system, each chemosensory neuron type must not only express ‘generic’ neuronal molecules such as channels required to confer cellular excitability, but also express genes required for its unique chemosensory profile. Expression of all genes in a given cell type may be regulated by a shared motif, such that multiple cell-specific motifs act in a piecemeal fashion to drive the expression of broadly expressed genes in different neuron types. Alternatively, multiple pathways may act in parallel to drive gene expression in a neuron-specific manner.
C. elegans provides an ideal system in which to dissect the cis- and trans-regulatory principles driving neuron-specific gene expression due to its well-defined nervous system, and the ability to monitor and manipulate gene expression in single cell types. Analyses of a limited number of neuron types in C. elegans have suggested that the unique properties of a given neuron type are regulated by TFs termed ‘terminal selector TFs’ acting via defined cis-regulatory ‘terminal selector motifs’ upstream of terminal differentiation genes (Hobert, 2008). However, analyses of additional neurons, and particularly of diverse neuronal subtypes, is essential to determine whether these principles are generalizable across all neuronal types, or whether distinct mechanisms are employed on a cell-by-cell basis.
Here we describe the cis-regulatory logic driving gene expression in the AWB olfactory neuron subtype in C. elegans. We find that AWB-expressed chemoreceptor (CR) genes are co-regulated via partly shared cis-regulatory motifs. Interestingly, these motifs are poorly conserved in their orthologs in related nematode species, and expression pattern analyses suggest divergence of both cis- and trans-regulatory mechanisms regulating CR gene expression across species. We also find that the expression of more broadly expressed sensory neuronal genes in the AWB neurons is regulated via diverse mechanisms acting in parallel to the pathways regulating cell-specific aspects of AWB identity. While a subset of these genes appears to be co-regulated in all expressing neurons, other genes may be regulated via neuron-specific mechanisms. Our results indicate that gene regulatory mechanisms in the AWB olfactory neuron type are complex, and suggest that this complexity may maximize flexibility in neuronal functions, while constraining neuron-specific properties.
RESULTS
Identification of AWB-expressed genes
Genes defining AWB neuron properties can represent members of three categories: AWB-specific or -selective genes which are expressed only in the two AWB neurons, or also in a small subset of additional cell types; sensory genes which are expressed in most or all sensory neuron types including in the AWB neurons; and pan-neuronal genes which are expressed broadly in the nervous system. Since the expression of genes in each category may be regulated via different mechanisms, we included both AWB-specific or –selective, as well as more broadly expressed sensory neuronal genes, in our study. Pan-neuronally expressed genes, a subset of which has been reported to be regulated via the N1 motif in C. elegans (Ruvinsky et al., 2007) were excluded from this analysis.
To identify AWB-specific or -selective genes, we mined published literature and databases for reported expression patterns. In particular, CR genes are expected to exhibit selective expression in one or a few chemosensory neuron types (Troemel et al., 1995). The str-1 CR gene was previously shown to be expressed specifically in the AWB neurons (Troemel et al., 1997). Expression profiling of isolated populations of embryonic AWB neurons identified the sru-38, srd-23, str-220 and srsx-3 CR genes, which were confirmed to be expressed in the AWB neurons, and a few other chemosensory cell types (Colosimo et al., 2004). Examination of the genomic location of str-1 and str-220 revealed the str-44 CR gene located in close proximity, which was also expressed in the AWB neurons (C. Bargmann, pers. comm.). Three additional AWB-expressed CR genes (srab-16, srab-24 and str-163) were identified via expression pattern analyses of gfp reporter fusion genes generated by the C. elegans gene expression project (www.wormbase.org; C. Bargmann, pers. comm.).
A number of genes have been shown to be expressed more broadly in sensory neuron types, including in the AWB neurons. These include the tax-2 and tax-4 cyclic nucleotide-gated channel subunit genes which are expressed in many, but not all, chemosensory neurons (Coburn and Bargmann, 1996; Komatsu et al., 1996), the odr-4 chaperone protein gene which is also expressed in multiple chemosensory neurons (Dwyer et al., 1998), as well as ciliogenic genes which are expressed in all ciliated neurons and are required for cilia assembly and function (Scholey, 2003). Although the expression of many ciliogenic genes is regulated by the DAF-19 RFX transcription factor (Swoboda et al., 2000; Blacque et al., 2005; Efimenko et al., 2005), additional ciliogenic genes such as the osm-3 and kap-1 kinesin genes are regulated via a DAF-19-independent mechanism (Swoboda et al., 2000; Mukhopadhyay et al., 2007), and were, therefore, included in our analysis. We also analyzed upstream cis-regulatory sequences driving expression of the lim-4 LIM-homeobox gene in the AWB neurons and a subset of non-sensory neuron types (Sagasti et al., 1999; Zheng et al., 2005). LIM-4 has previously been shown to be required for fate specification of the AWB neurons (Sagasti et al., 1999). A summary of these genes and their expression patterns is shown in Table 1.
Table 1.
Expression patterns of genes described in this study.
| Gene name | Sequence name | Encoded protein | Expression patterna |
|---|---|---|---|
| AWB-specific or –selective genes | |||
| str-1 | C42D4.5 | Chemoreceptor | AWB |
| str-220 | C42D4.9 | Chemoreceptor | AWB (weak) |
| str-44 | C42D4.4 | Chemoreceptor | AWB (weak) |
| sru-38 | R07B5.4 | Chemoreceptor | AWB, ASH, PHA/B |
| srd-23 | Y40H7A.5 | Chemoreceptor | AWB, other head neurons |
| srsx-3 | Y97E10B.9 | Chemoreceptor | AWB, AWC |
| srab-16 | R13D11.6 | Chemoreceptor | AWB, AWC |
| srab-24 | Y64G10A.8 | Chemoreceptor | AWB, ASH/I (variable) |
| str-163 | Y9C9A.3 | Chemoreceptor | AWB, other head neurons |
| lim-4 | ZC64.4 | LIM homeodomain transcription factor | AWB, other motor and interneurons |
| Genes broadly expressed in chemosensory neurons | |||
| tax-2 | F36F2.5 | Cyclic nucleotide-gated channel | AWB, many sensory neurons |
| tax-4 | ZC84.2 | Cyclic nucleotide-gated channel | AWB, many sensory neurons |
| odr-4 | Y102E9.1 | Chaperone protein required for chemoreceptor localization | AWB, many sensory neurons |
| kap-1 | F08F8.3 | Kinesin motor | All ciliated neurons, including AWB |
| osm-3 | M02B7.3 | Kinesin motor | All ciliated neurons, including AWB |
Described expression patterns are from Wormbase (www.wormbase.org), references in the text, and personal communication from C. I. Bargmann.
Deletion analyses reveal regions required to drive expression in the AWB neurons
To determine whether genes expressed in the AWB neurons contain a shared regulatory motif, we first used bioinformatic tools to align their upstream regulatory sequences (see Experimental Procedures). No significantly overrepresented shared motifs were identified likely due to the large genomic space. We thus used deletion analyses to narrow the regions required to drive expression in the AWB neurons. Regulatory regions were fused to gfp coding sequences, and expression of gfp was examined in animals transgenic for these constructs. Expression levels of str-44 and str-220 were too weak to be analyzed with confidence, and were excluded from further analysis. For simplicity, we henceforth assume that the expression pattern of gfp driven by the regulatory sequences of a gene reflects the spatiotemporal expression patterns of the endogenous gene, although it is possible that these expression patterns are not identical.
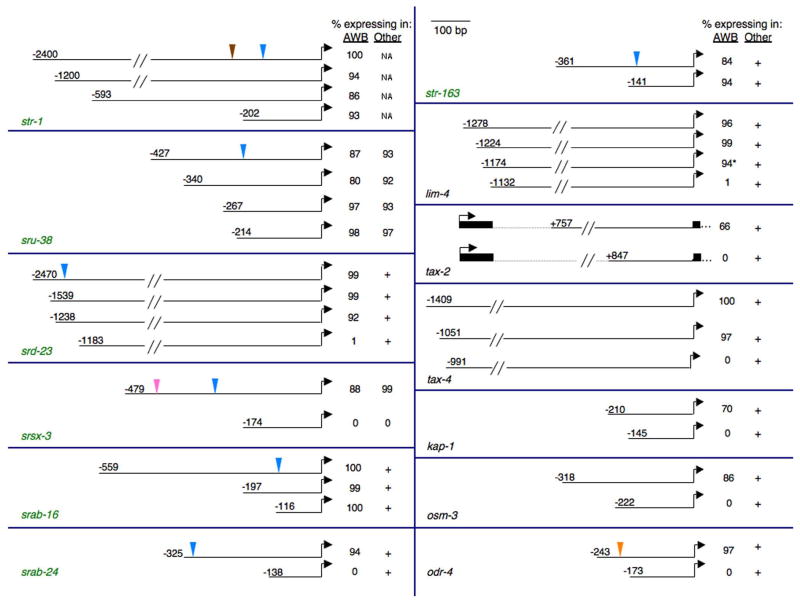
As has been previously reported for many C. elegans genes (eg. Gaudet and Mango, 2002; Efimenko et al., 2005; Etchberger et al., 2007), sequences required for expression in the AWB neurons were located within ~400 bp upstream of the initiator ATG for the majority of examined genes (Figure 1). However, sequences driving expression of srd-23, lim-4 and tax-4 in the AWB neurons were located >900 bp upstream of the ATG (Figure 1). In addition, as reported previously (Coburn and Bargmann, 1996), regulatory sequences for expression of tax-2 in the AWB neurons were located within its first intron.
Figure 1. Deletion analyses identify the minimal regulatory regions required to drive expression in the AWB chemosensory neurons.
CR genes selectively expressed in the AWB neurons are indicated in green. Numbers indicate the length of the regulatory region (in bp) relative to the translational start site (indicated by an arrowhead). For osm-3, the translational start site represents that of the osm-3b isoform (www.wormbase.org). Cis-regulatory sequences were inserted upstream of gfp coding sequences via the PCR fusion technique (Hobert, 2002). Colored inverted triangles represent the locations of the AWB motif (blue; see Figure 3); the motif recognized by MEF-2 in str-1 regulatory sequences (brown; van der Linden et al., 2007); the motif shown to drive srsx-3 expression in the AWC chemosensory neurons (pink; Lesch et al., 2009); and the X box motif in odr-4 regulatory sequences (orange; Efimenko et al., 2005). Expression in at least one AWB neuron in one or more independent transgenic lines for each construct was examined, and the average of multiple lines is reported where applicable. Adult animals grown at 20°C were examined in all cases at 400X magnification. + indicates retained expression in all, or subsets of cell types other than the AWB neurons; NA – not applicable. n > 100 for each.* indicates weak expression.
We found that for genes expressed in multiple neuron types, deletions that abolished expression in the AWB neurons continued to drive expression in subsets of additional neuron types, indicating that individual motifs may drive expression in distinct subsets of cells. Consistent with this hypothesis, different cis-regulatory elements appear to drive expression of the srsx-3 CR gene in the AWC and AWB neurons (Figure 1) (Lesch et al., 2009). Moreover, sequences required to drive developmental expression of the str-1 CR gene in the AWB neurons are distinct from the previously identified motif that is recognized by the MEF-2 TF which modulates str-1 expression under specific conditions (Figure 1) (van der Linden et al., 2007). Expression of lim-4 in the AWB neurons is initiated by the CEH-37 Otx and the NHR-67 Tailless/TLX nuclear receptor transcription factors, and is maintained via autoregulation (Sagasti et al., 1999; Lanjuin et al., 2003; Sarin et al., 2009). We found that deletion of a 42 bp region abolished lim-4 expression in the AWB neurons in adults (Figure 1), as well as in older L1 larvae, suggesting that this region contains sequences required for maintenance of lim-4 expression. We were unable to determine whether sequences required for initiation of lim-4 expression were also present with this region, since transient lim-4 expression in the AWB neurons in late stage embryos or early L1 larvae could not be determined with confidence. We rarely observed ectopic expression in other cell types upon deletion of regulatory sequences of any gene examined, suggesting that these regulatory sequences do not contain repressive elements (Wenick and Hobert, 2004). These results indicate that defined sequences promote developmentally regulated gene expression in the AWB neurons, and that additional sequences drive expression in other cell types, or under alternate conditions.
A bipartite A/T-rich element(s) is necessary for the expression of CR genes in the AWB neurons
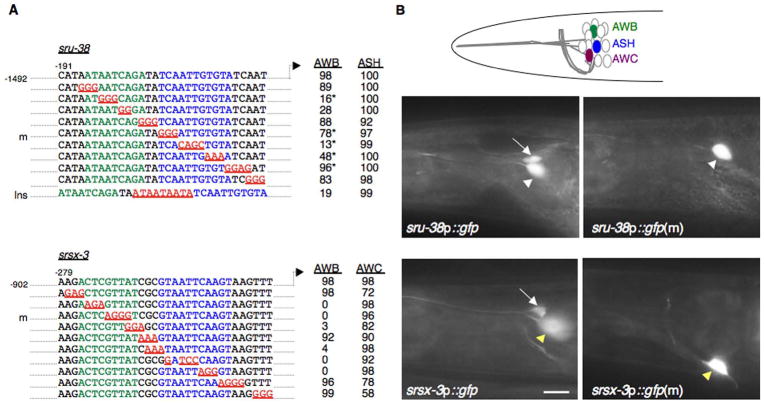
We next defined the necessary elements within the minimal sequences required to drive expression in the AWB neurons. Reasoning that AWB-expressed CR genes may be co-regulated, we first focused on the str-1, sru-38, srd-23, srsx-3, srab-16, srab-24 and str-163 CR genes. We performed scanning substitution mutageneses of the required minimal regulatory sequences of the sru-38 and srsx-3 genes, and identified two necessary sequences in each region (Figure 2A, B). Required elements upstream of both genes included A/T rich sequences which can comprise the core of homeodomain transcription factor binding sites (Desplan et al., 1988; Kalionis and O’Farrell, 1993). Replacement of these A/T sequences in each element abolished or significantly weakened expression of both sru-38 and srsx-3 in the AWB neurons without affecting expression in other neuron types (Figure 2A, B). For both genes, the required regulatory sequences may be viewed as a bipartite element in which each individual motif is separated by a 2–4 bp ‘linker’ sequence, mutations in which did not affect expression (Figure 2A), although we are unable to rule out the possibility that the particular base substitutions used were insufficient to abolish function. To determine whether the length of the linker is a critical parameter, we increased its length, and found that expression of sru-38 in the AWB neurons was partially affected (Figure 2A), suggesting that the relative spacing of the two motifs was important for correct regulation of gene expression.
Figure 2. Mutageneses of the regulatory regions of the sru-38 and srsx-3 CR genes reveal sequences required for AWB-specific expression.
A) The indicated lengths and positions of regulatory elements relative to the translational start site were fused to gfp in expression vectors (gift from A. Fire). Mutations generated in these sequences are indicated in red and underlined. Sequences in green and blue indicate the bipartite AWB motif (see Figure 3). The percentage of animals expressing gfp in at least one of each neuron type from one or two lines for each construct is shown. Adult animals were examined under 400X magnification. m- indicates the mutated construct whose expression pattern is shown in B; Ins – indicates an insertion to increase linker length. n > 50 for each line. * - indicates weak expression.
B) Representative expression patterns driven by wild-type and mutated (m) sru-38 and srsx-3 regulatory sequences in adult wild-type animals. Cartoon at top indicates relative cell body positions of the AWB, ASH and AWC chemosensory neurons. Arrow in each panel indicates the cell body of the AWB neurons. Arrowheads indicate cell body of the ASH (white) or AWC neuron (yellow). Anterior is at left; scale – 10 μm.
We next determined whether a similar bipartite element could be identified in the defined minimal regulatory sequences of the remaining five AWB-expressed CR genes. We identified a single copy of a related bipartite element with an intervening linker sequence of <5bp upstream of srd-23 and srab-24 (Figure 3A). However, although multiple copies of each individual motif could be identified upstream of srab-16, str-1 and str-163 (Supplementary Table 1), we did not identify a similar bipartite element in any of these sequences. We termed each of the individual motifs in the element, AWB CR Motif 1 and AWB CR Motif 2 (Motif 1 and Motif 2 for short). Each motif could be present in either orientation (Figure 3A).
Figure 3. Identification of motifs driving CR gene expression in the AWB neurons.
A) The sequences, positions of the first base in the motif on the forward strand relative to the ATG, strands, and POSSUM (see Experimental Procedures) score of Motifs 1 (green) and 2 (blue) are indicated for seven AWB-expressed CR genes. Linker lengths between the two motifs are indicated where applicable.
B) For each CR gene regulatory sequence, mutations (indicated in red and underlined) were generated in each motif, and expression was examined. Fusion genes were generated via site directed mutagenesis in a gfp expression vector (str-1), or PCR fusions (all other genes). Animals from at least two independent transgenic lines were examined for each construct. + – gfp expression was retained in one or more cells other than the AWB neurons. n > 100 for each. * indicates weak expression.
C) Shown is a sequence logo (Schneider and Stephens, 1990) for each of the motif sequences from A and B. Motif 1 and Motif 2 sequences were derived from five and six experimentally validated sequences, respectively.
To verify the necessity of the identified motifs in driving expression in the AWB neurons, we mutated the motifs in the regulatory sequences of each gene. Individually mutating each motif of the bipartite element significantly affected expression driven by srd-23 and srab-24 sequences specifically in the AWB neurons (Figure 3B), as observed for similar mutations in the motifs present upstream or sru-38 and srsx-3 (Figure 2A). Mutation of single motifs identified in the minimal regulatory sequences of srab-16, str-1, and str-163 also abolished expression in the AWB neurons (Figure 3B). Experimentally verified sequences from all analyzed CR genes were used to derive a position weight matrix (PWM) for each motif, confirming the requirement of an A/T-rich core sequence in each motif (Figure 3C). Taken together, these results suggest that subsets of AWB-expressed CR genes may be developmentally co-regulated via partly shared cis-regulatory elements.
Regulatory sequences driving CR gene expression in the AWB neurons are not conserved in related species
Regulatory motifs are frequently conserved among closely related species, and this conservation has been exploited to identify required elements (Kellis et al., 2003; Xie et al., 2005; Etchberger et al., 2007). Both conserved, as well as divergent gene expression patterns and cis-regulatory logic have been noted between C. elegans (Ce) and the related nematode species C. briggsae (Cb) and C. remanei (Cr) (Stein et al., 2003; Teng et al., 2004; Ortiz et al., 2006; Etchberger et al., 2007; Marri and Gupta, 2009), suggesting that evolutionary pressures may affect individual genes differently.
To determine whether the cis-regulatory sequences driving CR gene expression in the AWB neurons are conserved among related species, we examined the upstream regulatory sequences of the C. briggsae and C. remanei orthologs of the seven AWB-expressed CR genes. Overall sequence conservation across the regulatory sequences was low (Supplementary Figure 1), and alignment of the C. elegans sequences with their C. briggsae orthologs did not identify significantly overrepresented conserved motifs. We searched the upstream sequences using the PWMs derived for each motif from the C. elegans sequences. We restricted the search using three parameters: we searched only within the upstream 1.5kb sequences; we required that the score for each motif in the PWM be greater than a threshold; and we restricted the linker length to <5bp. Using these criteria, no bipartite elements could be identified in the regulatory sequences of the CR gene orthologs in C. briggsae or C. remanei. We next searched for the occurrence of each motif singly in the upstream regulatory sequences of CR gene orthologs, and identified several possible candidates (Supplementary Table 1). These candidate motifs were seldom located in a similar position as the required motifs in their C. elegans counterparts, and showed limited sequence conservation (data not shown).
Since the CR gene subfamilies in Caenorhabditis species are large, and have undergone species- and family-specific gene expansions and losses (Troemel et al., 1995; Thomas and Robertson, 2008), we considered the possibility that regulatory and coding sequences may have diverged independently. In this case, we might expect that conserved regulatory sequences may be present upstream of a homologous, but not orthologous C. briggsae CR gene. To address this possibility, we searched 1.5 kb of upstream sequences of all predicted C. briggsae genes using the PWMs derived from each motif with the above restrictions (see Experimental Procedures). However, only a single annotated CR gene was identified by this analysis from the str, sru, srsx, srd, or srab subfamilies (data not shown). Moreover, no conserved motifs were identified upon specifically searching the upstream regulatory sequences of members of the C. briggsae srsx and sru gene families with the bipartite motifs derived from the C. elegans srsx-3 and sru-38 CRs, respectively. These observations suggest that the cis-regulatory logic driving CR gene expression may be divergent among related species. However, it remains possible that sequence elements driving expression in the AWB neurons may be present further upstream, within introns, or downstream, in the C. briggsae or C. remanei CR orthologs or homologs.
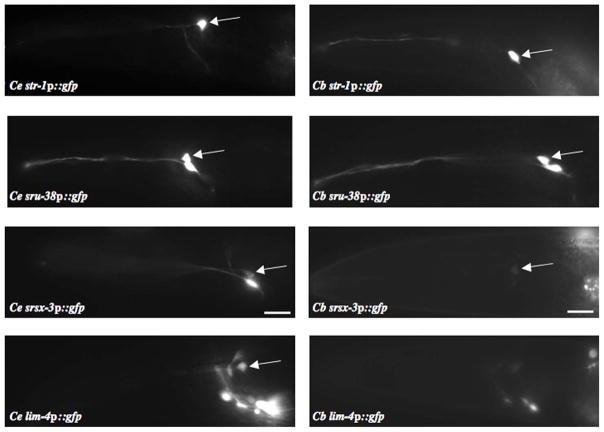
To determine whether the C. briggsae regulatory sequences could drive expression in the AWB neurons in C. elegans, we analyzed the expression patterns of the identified C. briggsae orthologs of str-1, sru-38 and srsx-3 in C. elegans. As shown in Figure 4, while Cb-str-1 and Cb-sru-38 exhibited the same expression pattern as their C. elegans orthologs, the expression pattern of Cb-srsx-3 was distinct. Unlike Ce-srsx-3 which is strongly expressed in both the AWB and AWC sensory neurons, Cb-srsx-3 was expressed only weakly in the AWB neurons (Figure 4). Since the LIM-4 LIM homeodomain protein has been shown to regulate expression of terminally differentiated genes, including CR genes, in the AWB neurons (Sagasti et al., 1999), we next determined whether expression of this TF was conserved. Cb-lim-4 was not expressed in the AWB neurons in C. elegans (Figure 4). These results suggest that divergence in CR expression patterns may arise from differences in both cis- and trans-regulatory mechanisms.
Figure 4. Regulatory sequences of a subset of C. briggsae orthologs drive expression in the AWB neurons in C. elegans.
Shown are the expression patterns of gfp driven by regulatory sequences of the C. elegans or C. briggsae orthologs of the indicated genes in C. elegans. Arrows indicate cell bodies of the AWB neurons. Adult animals; scale – 15 μm.
The AWB CR element drives expression in the AWB neurons in a context-dependent manner
We next asked whether the presence of the AWB CR bipartite element can be predictive of gene expression in the AWB neurons. Using the PWMs derived for each motif, we examined 1.5 kb of upstream regulatory sequences of all predicted C. elegans genes (www.wormbase.org; WS195) for the presence of the bipartite element. A large geneset was identified by this analysis, and as expected, contained the sru-38, srsx-3, srd-23 and srab-24 genes (Supplementary Table 2). However, other genes known to be selectively expressed in the AWB neurons were not identified by this analysis.
To examine whether any of the identified genes, and specifically CR genes, were expressed in the AWB neurons, we examined transgenic animals carrying fusion genes between gfp and regulatory sequences of three CR genes containing high scoring motifs (Supplementary Table 2). No expression was observed in the AWB neurons in adults (Supplementary Table 3), indicating that the AWB CR element may function together with additional context-dependent sequences to drive expression in the AWB neurons.
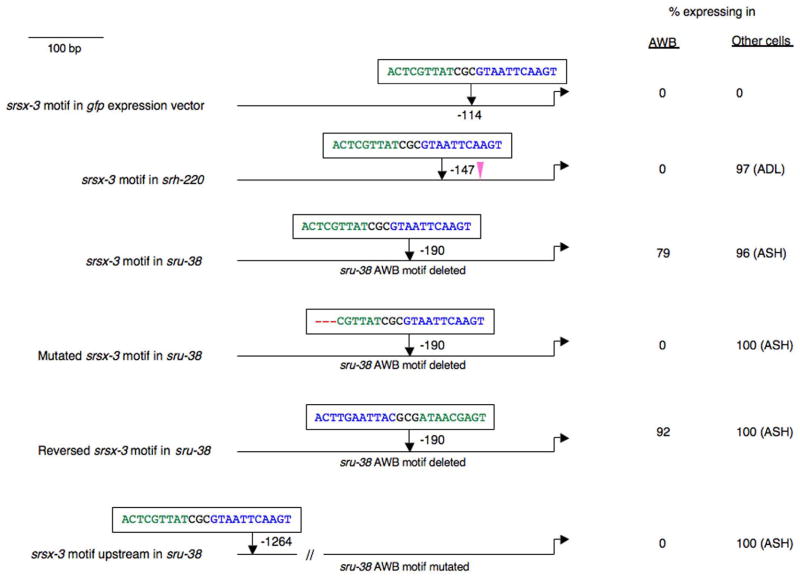
To further examine this hypothesis, we inserted a copy of the bipartite motif from srsx-3 into a minimal gfp expression vector (gift of A. Fire), and found that the motifs did not confer expression in the AWB or any other neuron type (Figure 5). Insertion of a single copy of the srsx-3 element into the upstream regulatory sequences of the ADL chemosensory neuron-expressed srh-220 gene (McCarroll et al., 2005) also did not drive expression in the AWB neurons, while expression in the ADL neurons remained unaffected (Figure 5).
Figure 5. The AWB CR element drives expression in a context-dependent manner.
A copy of a wild-type or mutated srsx-3 bipartite AWB element (Motif 1 – green; Motif 2 – blue) was inserted into the pPD95.75 gfp expression vector (gift of A. Fire); or the regulatory sequences of the srh-220 or sru-38 CR genes at the indicated sites. The E box that may drive expression of srh-220 in the ADL neurons is located at −99 (McCarroll et al., 2005), and is indicated by a pink triangle. Adult animals from one or two independent transgenic lines were examined in all cases. n = 50 – 100 for each.
If the genomic context is necessary for the function of the AWB CR element, we would predict that the element in one AWB-expressed CR gene could functionally replace the element from a second CR gene. Consistent with this prediction, replacing the sru-38 AWB element with a wild-type, but not mutated, srsx-3 AWB element resulted in efficient gfp expression in the AWB neurons (Figure 5), without affecting gfp expression in the ASH neurons. Insertion of the srsx-3 element in the reverse orientation in the sru-38 regulatory sequences was also sufficient to drive expression in the AWB neurons (Figure 5), further indicating that the AWB CR element can function in either orientation. The AWB elements in both the sru-38 and srsx-3 regulatory sequences are located within 200 bp of the translation start sites (Figure 1). To investigate whether the position of the AWB element relative to the ATG affects function, we inserted the srsx-3 AWB element at ~1.2kb upstream of the ATG in sru-38 regulatory sequences containing a mutated sru-38 AWB element (Figure 5). No expression was observed in the AWB neurons, suggesting that although the AWB CR element can be located in close proximity to the translational start site (as for sru-38 and srsx-3) or further upstream (as for srd-23), the position of the element relative to the ATG was important in the context of an individual regulatory sequence. Together, these results indicate that the genomic context of the AWB CR element, together with its relative position, plays an important role in the ability of the element to drive expression in the AWB neurons.
The LIM-4 LIM homeodomain protein does not bind the AWB CR motif in sru-38 regulatory sequences
LIM-4 has been proposed to act as a master regulator of the AWB fate, such that expression of AWB-specific genes is lost in lim-4 mutants (Sagasti et al., 1999). Although the expression of only the str-1 CR was previously examined, we confirmed that expression of sru-38 was also abolished in the AWB neurons in lim-4 mutants (Supplementary Figure 2). Since Motifs 1 and 2 may comprise homeodomain binding sites, we determined whether LIM-4 is able to directly bind these motifs in vitro. LIM-4 was expressed as a fusion protein in yeast (Vermeirssen et al., 2007), and tested for binding to the bipartite sequence motif from sru-38 via electrophoretic mobility shift assays (see Experimental Procedures). However, under the conditions tested, no binding was observed (data not shown), suggesting that LIM-4 may not bind these sequences directly on its own.
Distinct sequences drive expression of broadly expressed sensory neuronal genes
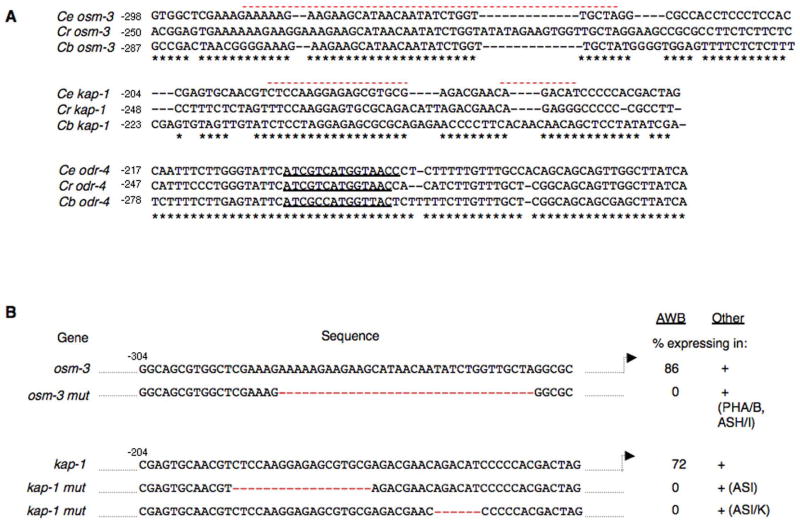
Deletion analyses identified the minimal regulatory sequences required to drive expression of the broadly expressed sensory neuronal genes kap-1, osm-3, odr-4, tax-2 and tax-4 in the AWB neurons (Figure 1). These molecules play roles in the formation of sensory neuron ciliary structures (osm-3, kap-1) (Signor et al., 1999; Snow et al., 2004), as a chaperone for seven transmembrane domain receptors (odr-4) (Dwyer et al., 1998), and sensory transduction (tax-2, tax-4) (Coburn and Bargmann, 1996; Komatsu et al., 1996). We reasoned that unlike CR genes whose expression may be regulated in a species-specific manner to optimize sensory responses, expression of genes required for sensory signal transduction, or for generation of sensory neuron-specific ciliary morphologies may be mediated via conserved mechanisms. Consistent with this notion, we found that unlike the regulatory sequences of the orthologous CR genes which were highly divergent, the regulatory sequences of kap-1 and to a lesser extent, osm-3 contained large stretches of identical sequences (Figure 6A; Supplementary Figure 1). Using these blocks of highly conserved sequences as guides, we deleted conserved nucleotides, and further defined the sequences required to drive expression of kap-1 and osm-3 in the AWB neurons of C. elegans (Figure 6B). These sequences were conserved in C. elegans, C. briggsae and C. remanei (Figure 6A). Deletions that abolished expression in the AWB neurons continued to drive expression in subsets of additional neuron types (Figure 6B), suggesting that the cis-regulatory sequences of these genes are modular, with multiple motifs driving expression in different cell types. Surprisingly, however, the regulatory sequence of tax-2, tax-4 and odr-4 were poorly conserved overall (Supplementary Figure 1), and we did not identify large segments of conserved sequences in the regions identified by deletion analyses to be required to drive expression in the AWB neurons (Figure 1).
Figure 6. Sensory neuron-expressed genes in the AWB neurons may be regulated by multiple mechanisms.
A) Alignment of the minimal required regulatory sequences of C. elegans (Ce) osm-3, kap-1 and odr-4, and their orthologs in C. briggsae (Cb), and C. remanei (Cr) showing highly conserved sequences. Nucleotides conserved in at least two orthologs are marked by asterisks at the bottom. The overall level of conservation across 1.5 kb of upstream sequences is shown in Supplementary Figure 1. The translational start site of osm-3 represents that of the osm-3b isoform. The X box motif in odr-4 is indicated by an underline. A red dashed overline indicates the sequences deleted in (B). The sequence of the X box motif was found to contain a one nucleotide mismatch with the previously reported sequence (Efimenko et al., 2005) (reported as ATCGTCATCGTAAC; direct sequencing of our wild-type strain indicates ATCGTCATGGTAAC).
B) Deletion of sequences abolishes expression in the AWB neurons. Deleted sequences are indicated by a red dashed line. gfp expression is retained in additional cell subsets. Adult animals from at least two independent transgenic lines were examined for each. n > 100 for each.
We next determined whether the AWB CR element could be identified in the minimal cis-regulatory sequences of these sensory neuron-expressed genes. We included the transcriptional regulator lim-4 in this analysis. Neither the bipartite element, nor either of the single motifs could be identified in any of the examined sequences. We also aligned the minimal required regulatory sequences of all six genes to determine whether a common motif suggestive of co-regulation could be defined. However, analyses using multiple algorithms failed to identify a robustly conserved motif in all of these genes. Moreover, mutating or deleting A/T-rich sequences which form the core of Motifs 1 and 2 singly in the required regulatory sequences of lim-4 had no effect on gfp expression in the AWB neurons (Supplementary Figure 3), although we cannot rule out the possibility that these motifs act redundantly to regulate expression. These results suggest that expression of these genes may be regulated by multiple trans-acting mechanisms.
In support of the above hypothesis, we noted that the minimal sequences driving expression of the odr-4, but not osm-3, kap-1, tax-2 or tax-4 genes in the AWB neurons contained a predicted X box motif which was conserved in related species (Figure 1; Figure 6A). The X-box motif has previously been shown to be recognized by the DAF-19 RFX transcription factor, and is present upstream of many ciliogenic, and other genes in C. elegans (Swoboda et al., 2000; Blacque et al., 2005; Efimenko et al., 2005; Chen et al., 2006). The X box in the odr-4 regulatory sequences was previously identified in a genome-wide search for genes containing X box motifs (Efimenko et al., 2005), and mutation of the X box was reported to affect odr-4 expression (Efimenko et al., 2005). In support of these findings, we also noted that sequences lacking the X box failed to drive expression in the AWB neurons, although expression in a subset of other cell types was retained (Figure 1). Moreover, we have previously shown that expression of the kap-1 kinesin-II subunit gene is regulated by the FKH-2 forkhead domain transcription factor in the AWB neurons, but not other neuron types (Mukhopadhyay et al., 2007). Together, these results suggest that mechanisms regulating expression of broadly expressed genes are distinct from those regulating expression of CR genes specifically in the AWB neurons, and that in addition, these mechanisms may differ in other neuron types.
DISCUSSION
We have shown that multiple cis-regulatory mechanisms drive gene expression in the AWB neurons, and contribute to functional diversification of this olfactory neuron type. The cis- and trans-regulatory principles driving gene expression have been described for a subset of neuron types in C. elegans (Hobert, 2008). A common principle that has emerged from these studies is the concept of ‘terminal selector’ TFs which are expressed throughout the life of the postmitotic neuron type, and which act directly on ‘terminal selector motifs’ in the regulatory sequences of terminal differentiation genes to govern neuronal identities (Hobert, 2008). Selector TFs may also act on, or together with, downstream TFs which in turn, specify additional neuronal differentiation programs. A key feature of selector TFs is that they govern the unique identity of the neuron type, whereas additional generic neuronal properties may be regulated via other parallel pathways (Swoboda et al., 2000; Ruvinsky et al., 2007).
Our analyses of the cis-regulatory mechanisms driving gene expression in the AWB neurons are consistent with the concept of a selector TF-driven differentiation program, and suggest that these TFs may act with multiple parallel pathways to dictate neuron-specific properties. The expression of seven AWB-expressed CR genes appears to be in part, co-regulated by a shared cis-regulatory motif. Four of seven CR genes contain a bipartite element, in which each sequence motif is separated by a linker of restricted length, and at least one of the two motifs was also identified upstream of the remaining three analyzed CR genes. Bipartite elements have been identified as binding sites for homo- or heterodimeric TFs. For instance, nuclear hormone receptors bind cooperatively as homodimers or heterodimers to bipartite sequences containing half-site motifs, in which the linker length determines binding specificity (Khorasanizadeh and Rastinejad, 2001). The binding specificity and affinity of the AP-1 complex consisting of Fos and Jun proteins is provided by cooperative interaction with other members of related and unrelated TF families, and binding to composite elements, individual motifs of which may or may not be located in immediate proximity (Chinenov and Kerppola, 2001; Macian et al., 2001). In C. elegans, LIM homeodomain proteins such as TTX-3 and MEC-3 have been shown to bind cooperatively with other homeodomain proteins to bipartite motifs to regulate transcription (Xue et al., 1993; Duggan et al., 1998; Wenick and Hobert, 2004). We suggest that CR genes may similarly be co-regulated by a heterodimeric TF terminal selector complex in the AWB neurons of which LIM-4 may be a member, and that LIM-4 may bind cooperatively with different TFs to regulate expression of different CR subsets. This combinatorial regulation of expression may allow for co-regulation, while retaining the ability to differentially modulate the expression of subsets of CR genes.
However, this cis-regulatory element is absent in more broadly expressed sensory neuronal genes. Moreover, the expression of genes such as odr-4 and kap-1 in the AWB neurons is regulated by alternate TFs such as DAF-19 and FKH-2 which have been suggested to act in parallel pathways to the pathway regulating fate specification of the AWB neurons (Mukhopadhyay et al., 2007). It is interesting to note that kinesin motors may be used differentially to generate and maintain ciliary structural diversity in C. elegans sensory neurons (Evans et al., 2006; Mukhopadhyay et al., 2007), and that moreover, TAX-2 and TAX-4 may mediate subsets of different functions (Ailion and Thomas, 2000; Satterlee et al., 2004). We suggest that independent regulation of the kinesin and channel genes in different neuron types, perhaps under different environmental conditions, may enable precise regulation of expression under different conditions in order to exquisitely modulate sensory neuronal function. It remains possible, however, that expression of a subset of these sensory neuronal genes may be regulated indirectly by the selector TF(s) for the AWB neurons acting via downstream TFs (Hwang and Lee, 2003; Uchida et al., 2003; Koga and Ohshima, 2004). We are unable to determine whether expression of these genes is regulated by LIM-4, the best identified candidate for an AWB selector TF, since expression of these genes is retained in lim-4 mutants due to transformation of the AWB neuron fate to that of the AWC sensory neurons (Sagasti et al., 1999). Further characterization of the regulatory mechanisms employed by the AWB neurons for fate specification will require determining whether LIM-4 is a bona fide selector TF, and identification of interacting TFs and binding sites.
Our experiments also indicate that both cis- and trans-regulatory mechanisms of gene expression in the AWB neurons may have diverged between C. elegans and related species. Independent evolution of cis- and trans-regulatory mechanisms has been shown to contribute to functional diversification via changes in gene expression in many species, including nematodes (King and Wilson, 1975; Yvert et al., 2003; Denver et al., 2005; Wittkopp et al., 2008). Indeed, cis-regulatory changes have been suggested to be a critical contributor to the ability of animals to adapt to dynamically changing environmental conditions (Wray, 2007). These changes may be particularly critical in the sensory system, allowing optimal adaptation to species-specific niches. CR genes in Caenorhabditis species represent a rapidly diverging gene family (Stewart et al., 2005; Thomas and Robertson, 2008); diversification of both cis-regulatory and coding sequences of CR genes may, therefore, be an important contributor to species-specific chemosensory properties. However, a recent analysis of sequence variation among related nematode species found a higher degree of divergence and polymorphisms in candidate chemosensory neuron TF selector genes than in downstream chemosensory signaling molecules or in TFs required for the development of other neuron clases (Jovelin, 2009; Jovelin et al., 2009), suggesting that employment of both divergent cis- and trans-acting mechanisms may contribute to evolution of functional diversity in the sensory nervous system. Investigations into, and comparisons of, the developmental principles employed by additional sensory neuron types may provide insights into the mechanisms by which both specificity and plasticity are maintained in sensory neuron functions.
EXPERIMENTAL PROCEDURES
Strains
Wild-type strains used were C. elegans Bristol N2 and C. briggsae AF16. Transgenic strains were generated by injecting fusion gene constructs at 50 ng/μl together with the unc-122::dsRed coninjection marker at 50 ng/μl. One or two independent transgenic lines were established for each construct.
Molecular biology
Promoter::gfp fusion genes were generated by amplifying upstream genomic sequences, which were then subcloned into the pPD95.77 gfp expression vector (gift of A Fire) or fused to gfp-coding sequences via the PCR fusion technique (Hobert, 2002). Mutagenesis was performed with the QuikChange site-directed mutagenesis kit (Stratagene), and/or by PCR fusion, and all mutations were confirmed by sequencing. Primer sequences are available upon request.
Vector sequences present in the standard C. elegans expression vectors have been reported to inhibit gfp expression driven by regulatory sequences subcloned into these vectors (Etchberger and Hobert, 2008). To minimize confounding effects of comparing expression patterns driven by regulatory sequences subcloned into vectors, or present in linear, vector-free PCR fusion constructs, comparisons were performed with wild-type sequences present correspondingly in either linear PCR fusions, or subcloned into the same expression vector.
To examine the sufficiency of the AWB CR element in driving expression in the AWB neurons, the srsx-3 AWB CR element either replaced the sru-38 AWB CR element, or was inserted upstream of a mutated sru-38 AWB CR element in the pPD95.77 gfp expression vector. The srsx-3 CR element was also inserted into ~1.6 kb of srh-220 upstream regulatory sequences.
Microscopy
Animals were grown at 20°C for at least two generations prior to analyses. GFP expression in the AWB neurons was examined under 400X magnification on a Zeiss Axioplan microscope. The AWB neurons were identified by their characteristic cell body positions, ciliary morphology, or by dye-filling with lipophilic dyes (Ward et al., 1975; Perkins et al., 1986). Any GFP expression in at least one AWB neuron was scored as positive for expression.
Bioinformatic analyses
Different lengths (500 bp, 1 kb, 1.5 kb) of upstream regulatory sequences of all or subsets of AWB-expressed genes in C. elegans and C. briggsae were aligned and analyzed using MEME (http://meme.sdsc.edu/meme4_1/cgi-bin/meme.cgi) or GLAM2 http://meme.sdsc.edu/meme4_1/cgi-bin/glam2.cgi). Motifs identified by mutational analyses in the regulatory sequences of sru-38 and srsx-3 were used to identify similar motifs in the regulatory sequences of additional AWB-expressed CR genes using the Improbizer algorithm (Ao et al., 2004). Sequences verified experimentally from all seven CR genes were then used to generate a position weight matrix (PWM) for each motif. Realignment of the identified motifs with the PWMs generated a score of >5.5 for each motif using POSSUM (http://zlab.bu.edu/~mfrith/possum/), a matrix-based motif search program. The presence of the bipartite element with a linker sequence of < 5bp, or individual motifs with scores of >5.5 in 1.5 kb of upstream regulatory sequences in C. elegans AWB-expressed CR genes and their orthologs in related species was determined by searching with the PWMs for each motif, and are shown in Supplementary Table 1. The PWMs for each motif were also used to search the C. elegans or the C. briggsae (WS195) genomes using POSSUM for the presence of the motifs in 1.5 kb of upstream regulatory sequences. A custom-written Perl script was used to further restrict the search to sequences containing both motifs, each with a score of >5.5, and separated by a linker sequence of ≤ 5 bp in the output from POSSUM. The 651 genes identified by this analysis in C. elegans are shown in Supplementary Table 2.
To identify possible shared motifs in the minimal regulatory regions required to drive expression of odr-4, osm-3, kap-1,lim-4, tax-2 and tax-4, the required sequences were analyzed together with, or without regulatory sequences from their C. briggsae, C. remanei and C. japonica orthologs using MEME, RSAT (http://rsat.ulb.ac.be/rsat/), GLAM2, PhyloCon(PMID: 14668220), Improbizer, YMF (http://wingless.cs.washington.edu/YMF/YMFWeb/YMFInput.pl) CLOVER(PMID: 14988425) and MatInspector (PUBMED: 8532532). No significant conserved motifs were found to be overrepresented in these sequences.
Electrophoretic mobility shift assay
A yeast strain constitutively expressing a LIM-4::AD fusion protein (Vermeirssen et al., 2007) was generously provided by J. Reece-Hoyes and M. Walhout (University of Massachusetts Medical School). Electrophoretic mobility shift assays with total yeast nuclear extract were carried out essentially as described (Kim et al., 2005). Probes used were the following: wild type: 5′-AATAATCAGATATCAATTGTGTAT -3′; mutant: 5′-AAGGGGCAGATATCGGGGGTGTAT-3′; mutated bases are underlined.
Supplementary Material
Acknowledgments
We thank the Caenorhabditis Genetics Center for strains, Andy Fire for expression vectors, Cori Bargmann for noting the expression patterns of additional AWB-expressed CR genes, Joe Rodriguez, Pengyu Hong, Xiaoyun Sun, and Hennady Shulha for assistance with bioinformatic analyses, John Reece-Hoyes and Marian Walhout for an yeast strain expressing a LIM-4::AD fusion protein, the Sengupta lab for discussions and technical assistance, and Susan Birren and Scott Neal for comments on the manuscript. This work was supported by the NIH (P.S. - R01 GM56223).
References
- Ailion M, Thomas JH. Dauer formation induced by high temperatures in Caenorhabditis elegans. Genetics. 2000;156:1047–1067. doi: 10.1093/genetics/156.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ao W, Gaudet J, Kent WJ, Muttumu S, Mango SE. Environmentally induced foregut remodeling by PHA-4/FoxA and DAF-12/NHR. Science. 2004;305:1743–1746. doi: 10.1126/science.1102216. [DOI] [PubMed] [Google Scholar]
- Blacque OE, Perens EA, Boroevich KA, Inglis PN, Li C, Warner A, Khattra J, Holt RA, Ou G, Mah AK, McKay SJ, Huang P, Swoboda P, Jones SJ, Marra MA, Baillie DL, Moerman DG, Shaham S, Leroux MR. Functional genomics of the cilium, a sensory organelle. Curr Biol. 2005;15:935–941. doi: 10.1016/j.cub.2005.04.059. [DOI] [PubMed] [Google Scholar]
- Chen N, Mah A, Blacque OE, Chu J, Phgora K, Bakhoum MW, Newbury CR, Khattra J, Chan S, Go A, Efimenko E, Johnsen R, Phirke P, Swoboda P, Marra M, Moerman DG, Leroux MR, Baillie DL, Stein LD. Identification of ciliary and ciliopathy genes in Caenorhabditis elegans through comparative genomics. Genome Biol. 2006;7:R126. doi: 10.1186/gb-2006-7-12-r126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinenov Y, Kerppola TK. Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity. Oncogene. 2001;20:2438–2452. doi: 10.1038/sj.onc.1204385. [DOI] [PubMed] [Google Scholar]
- Coburn C, Bargmann CI. A putative cyclic nucleotide-gated channel is required for sensory development and function in C. elegans. Neuron. 1996;17:695–706. doi: 10.1016/s0896-6273(00)80201-9. [DOI] [PubMed] [Google Scholar]
- Colosimo ME, Brown A, Mukhopadhyay S, Gabel C, Lanjuin AE, Samuel AD, Sengupta P. Identification of thermosensory and olfactory neuron-specific genes via expression profiling of single neuron types. Curr Biol. 2004;14:2245–2251. doi: 10.1016/j.cub.2004.12.030. [DOI] [PubMed] [Google Scholar]
- Denver DR, Morris K, Streelman JT, Kim SK, Lynch M, Thomas WK. The transcriptional consequences of mutation and natural selection in Caenorhabditis elegans. Nat Genet. 2005;37:544–548. doi: 10.1038/ng1554. [DOI] [PubMed] [Google Scholar]
- Desplan C, Theis J, O’Farrell PH. The sequence specificity of homeodomain-DNA interaction. Cell. 1988;54:1081–1090. doi: 10.1016/0092-8674(88)90123-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggan A, Ma C, Chalfie M. Regulation of touch receptor differentiation by the Caenorhabditis elegans mec-3 and unc-86 genes. Development. 1998;125:4107–4119. doi: 10.1242/dev.125.20.4107. [DOI] [PubMed] [Google Scholar]
- Dwyer ND, Troemel ER, Sengupta P, Bargmann CI. Odorant receptor localization to olfactory cilia is mediated by ODR-4, a novel membrane-associated protein. Cell. 1998;93:455–466. doi: 10.1016/s0092-8674(00)81173-3. [DOI] [PubMed] [Google Scholar]
- Efimenko E, Bubb K, Mak HY, Holzman T, Leroux MR, Ruvkun G, Thomas JH, Swoboda P. Analysis of xbx genes in C. elegans. Development. 2005;132:1923–1934. doi: 10.1242/dev.01775. [DOI] [PubMed] [Google Scholar]
- Etchberger JF, Flowers EB, Poole RJ, Bashllari E, Hobert O. Cis-regulatory mechanisms of left/right asymmetric neuron-subtype specification in C. elegans. Development. 2009;136:147–160. doi: 10.1242/dev.030064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchberger JF, Hobert O. Vector-free DNA constructs improve transgene expression in C. elegans. Nat Methods. 2008;5:3. doi: 10.1038/nmeth0108-3. [DOI] [PubMed] [Google Scholar]
- Etchberger JF, Lorch A, Sleumer MC, Zapf R, Jones SJ, Marra MA, Holt RA, Moerman DG, Hobert O. The molecular signature and cis-regulatory architecture of a C. elegans gustatory neuron. Genes Dev. 2007;21:1653–1674. doi: 10.1101/gad.1560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JE, Snow JJ, Gunnarson AL, Ou G, Stahlberg H, McDonald KL, Scholey JM. Functional modulation of IFT kinesins extends the sensory repertoire of ciliated neurons in Caenorhabditis elegans. J Cell Biol. 2006;172:663–669. doi: 10.1083/jcb.200509115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet J, Mango SE. Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science. 2002;295:821–825. doi: 10.1126/science.1065175. [DOI] [PubMed] [Google Scholar]
- Grad YH, Roth FP, Halfon MS, Church GM. Prediction of similarly acting cis-regulatory modules by subsequence profiling and comparative genomics in Drosophila melanogaster and D. pseudoobscura. Bioinformatics. 2004;20:2738–2750. doi: 10.1093/bioinformatics/bth320. [DOI] [PubMed] [Google Scholar]
- Halfon MS, Grad Y, Church GM, Michelson AM. Computation-based discovery of related transcriptional regulatory modules and motifs using an experimentally validated combinatorial model. Genome Res. 2002;12:1019–1028. doi: 10.1101/gr.228902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, Jennings EG, Zeitlinger J, Pokholok DK, Kellis M, Rolfe PA, Takusagawa KT, Lander ES, Gifford DK, Fraenkel E, Young RA. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques. 2002;32:728–730. doi: 10.2144/02324bm01. [DOI] [PubMed] [Google Scholar]
- Hobert O. Regulatory logic of neuronal diversity: terminal selector genes and selector motifs. Proc Natl Acad Sci USA. 2008;105:20067–20071. doi: 10.1073/pnas.0806070105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard ML, Davidson EH. cis-Regulatory control circuits in development. Dev Biol. 2004;271:109–118. doi: 10.1016/j.ydbio.2004.03.031. [DOI] [PubMed] [Google Scholar]
- Hwang SB, Lee J. Neuron cell type-specific SNAP-25 expression driven by multiple regulatory elements in the nematode Caenorhabditis elegans. J Mol Biol. 2003;333:237–247. doi: 10.1016/j.jmb.2003.08.055. [DOI] [PubMed] [Google Scholar]
- Jovelin R. Rapid sequence evolution of transcription factors controlling neuron differentiation in Caenorhabditis. Mol Biol Evol. 2009 doi: 10.1093/molbev/msp142. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovelin R, Dunham JP, Sung FS, Phillips PC. High nucleotide divergence in developmental regulatory genes contrasts with the structural elements of olfactory pathways in caenorhabditis. Genetics. 2009;181:1387–1397. doi: 10.1534/genetics.107.082651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalionis B, O’Farrell PH. A universal target sequence is bound in vitro by diverse homeodomains. Mech Dev. 1993;43:57–70. doi: 10.1016/0925-4773(93)90023-q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- Khorasanizadeh S, Rastinejad F. Nuclear-receptor interactions on DNA-response elements. Trends Biochem Sci. 2001;26:384–390. doi: 10.1016/s0968-0004(01)01800-x. [DOI] [PubMed] [Google Scholar]
- Kim K, Colosimo ME, Yeung H, Sengupta P. The UNC-3 Olf/EBF protein represses alternate neuronal programs to specify chemosensory neuron identity. Dev Biol. 2005;286:136–148. doi: 10.1016/j.ydbio.2005.07.024. [DOI] [PubMed] [Google Scholar]
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- Koga M, Ohshima Y. The C. elegans ceh-36 gene encodes a putative homemodomain transcription factor involved in chemosensory functions of ASE and AWC neurons. J Mol Biol. 2004;336:579–587. doi: 10.1016/j.jmb.2003.12.037. [DOI] [PubMed] [Google Scholar]
- Komatsu H, Mori I, Ohshima Y. Mutations in a cyclic nucleotide-gated channel lead to abnormal thermosensation and chemosensation in C. elegans. Neuron. 1996;17:707–718. doi: 10.1016/s0896-6273(00)80202-0. [DOI] [PubMed] [Google Scholar]
- Lanjuin A, VanHoven MK, Bargmann CI, Thompson JK, Sengupta P. Otx/otd homeobox genes specify distinct sensory neuron identities in C. elegans. Dev Cell. 2003;5:621–633. doi: 10.1016/s1534-5807(03)00293-4. [DOI] [PubMed] [Google Scholar]
- Lesch BJ, Gehrke AR, Bulyk ML, Bargmann CI. Transcriptional regulation and stabilization of left-right neuronal identity in C. elegans. Genes Dev. 2009;23:345–358. doi: 10.1101/gad.1763509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macian F, Lopez-Rodriguez C, Rao A. Partners in transcription: NFAT and AP-1. Oncogene. 2001;20:2476–2489. doi: 10.1038/sj.onc.1204386. [DOI] [PubMed] [Google Scholar]
- Markstein M, Markstein P, Markstein V, Levine MS. Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc Natl Acad Sci USA. 2002;99:763–768. doi: 10.1073/pnas.012591199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M, Zinzen R, Markstein P, Yee KP, Erives A, Stathopoulos A, Levine M. A regulatory code for neurogenic gene expression in the Drosophila embryo. Development. 2004;131:2387–2394. doi: 10.1242/dev.01124. [DOI] [PubMed] [Google Scholar]
- Marri S, Gupta BP. Dissection of lin-11 enhancer regions in Caenorhabditis elegans and other nematodes. Dev Biol. 2009;325:402–411. doi: 10.1016/j.ydbio.2008.09.026. [DOI] [PubMed] [Google Scholar]
- McCarroll SA, Li H, Bargmann CI. Identification of transcriptional regulatory elements in chemosensory receptor genes by probabilistic segmentation. Curr Biol. 2005;15:347–352. doi: 10.1016/j.cub.2005.02.023. [DOI] [PubMed] [Google Scholar]
- McGhee JD, Sleumer MC, Bilenky M, Wong K, McKay SJ, Goszczynski B, Tian H, Krich ND, Khattra J, Holt RA, Baillie DL, Kohara Y, Marra MA, Jones SJ, Moerman DG, Robertson AG. The ELT-2 GATA-factor and the global regulation of transcription in the C. elegans intestine. Dev Biol. 2007;302:627–645. doi: 10.1016/j.ydbio.2006.10.024. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S, Lu Y, Qin H, Lanjuin A, Shaham S, Sengupta P. Distinct IFT mechanisms contribute to the generation of ciliary structural diversity in C. elegans. EMBO J. 2007;26:2966–2980. doi: 10.1038/sj.emboj.7601717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz CO, Etchberger JF, Posy SL, Frokjaer-Jensen C, Lockery S, Honig B, Hobert O. Searching for neuronal left/right asymmetry: genomewide analysis of nematode receptor-type guanylyl cyclases. Genetics. 2006;173:131–149. doi: 10.1534/genetics.106.055749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins LA, Hedgecock EM, Thomson JN, Culotti JG. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev Biol. 1986;117:456–487. doi: 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
- Roth FP, Hughes JD, Estep PW, Church GM. Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nat Biotechnol. 1998;16:939–945. doi: 10.1038/nbt1098-939. [DOI] [PubMed] [Google Scholar]
- Ruvinsky I, Ohler U, Burge CB, Ruvkun G. Detection of broadly expressed neuronal genes in C. elegans. Dev Biol. 2007;302:617–626. doi: 10.1016/j.ydbio.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Sagasti A, Hobert O, Troemel ER, Ruvkun G, Bargmann CI. Alternative olfactory neuron fates are specified by the LIM homeobox gene lim-4. Genes Dev. 1999;13:1794–1806. doi: 10.1101/gad.13.14.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarin S, Antonio C, Tursun B, Hobert O. The C. elegans Tailless/TLX transcription factor nhr-67 controls neuronal identity and left/right asymmetric fate diversification. Development. 2009;136:2933–2944. doi: 10.1242/dev.040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterlee JS, Ryu WS, Sengupta P. The CMK-1 CaMKI and the TAX-4 Cyclic nucleotide-gated channel regulate thermosensory neuron gene expression and function in C. elegans. Curr Biol. 2004;14:62–68. doi: 10.1016/j.cub.2003.12.030. [DOI] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey JM. Intraflagellar transport. Annu Rev Cell Dev Biol. 2003;19:423–443. doi: 10.1146/annurev.cellbio.19.111401.091318. [DOI] [PubMed] [Google Scholar]
- Signor D, Wedaman KP, Rose LS, Scholey JM. Two heteromeric kinesin complexes in chemosensory neurons and sensory cilia of Caenorhabditis elegans. Mol Biol Cell. 1999;10:345–360. doi: 10.1091/mbc.10.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow JJ, Ou G, Gunnarson AL, Walker MR, Zhou HM, Brust-Mascher I, Scholey JM. Two anterograde intraflagellar transport motors cooperate to build sensory cilia on C. elegans neurons. Nat Cell Biol. 2004;6:1109–1113. doi: 10.1038/ncb1186. [DOI] [PubMed] [Google Scholar]
- Stein LD, Bao Z, Blasiar D, Blumenthal T, Brent MR, Chen N, Chinwalla A, Clarke L, Clee C, Coghlan A, Coulson A, D’Eustachio P, Fitch DH, Fulton LA, Fulton RE, Griffiths-Jones S, Harris TW, Hillier LW, Kamath R, Kuwabara PE, Mardis ER, Marra MA, Miner TL, Minx P, Mullikin JC, Plumb RW, Rogers J, Schein JE, Sohrmann M, Spieth J, Stajich JE, Wei C, Willey D, Wilson RK, Durbin R, Waterston RH. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 2003;1:E45. doi: 10.1371/journal.pbio.0000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart MK, Clark NL, Merrihew G, Galloway EM, Thomas JH. High genetic diversity in the chemoreceptor superfamily of Caenorhabditis elegans. Genetics. 2005;169:1985–1996. doi: 10.1534/genetics.104.035329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swoboda P, Adler HT, Thomas JH. The RFX-type transcription factor DAF-19 regulates sensory neuron cilium formation in C. elegans. Mol Cell. 2000;5:411–421. doi: 10.1016/s1097-2765(00)80436-0. [DOI] [PubMed] [Google Scholar]
- Teng Y, Girard L, Ferreira HB, Sternberg PW, Emmons SW. Dissection of cis-regulatory elements in the C. elegans Hox gene egl-5 promoter. Dev Biol. 2004;276:476–492. doi: 10.1016/j.ydbio.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Thomas JH, Robertson HR. The Caenorhabditis chemoreceptor gene families. BMC Biol. 2008;6:42. doi: 10.1186/1741-7007-6-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troemel ER, Chou JH, Dwyer ND, Colbert HA, Bargmann CI. Divergent seven transmembrane receptors are candidate chemosensory receptors in C. elegans. Cell. 1995;83:207–218. doi: 10.1016/0092-8674(95)90162-0. [DOI] [PubMed] [Google Scholar]
- Troemel ER, Kimmel BE, Bargmann CI. Reprogramming chemotaxis responses: sensory neurons define olfactory preferences in C. elegans. Cell. 1997;91:161–169. doi: 10.1016/s0092-8674(00)80399-2. [DOI] [PubMed] [Google Scholar]
- Uchida O, Nakano H, Koga M, Ohshima Y. The C. elegans che-1 gene encodes a zinc finger transcription factor required for specification of the ASE chemosensory neurons. Development. 2003;130:1215–1224. doi: 10.1242/dev.00341. [DOI] [PubMed] [Google Scholar]
- van der Linden AM, Nolan KM, Sengupta P. KIN-29 SIK regulates chemoreceptor gene expression via an MEF2 transcription factor and a class II HDAC. EMBO J. 2007;26:358–370. doi: 10.1038/sj.emboj.7601479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeirssen V, Deplancke B, Barrasa MI, Reece-Hoyes JS, Arda HE, Grove CA, Martinez NJ, Sequerra R, Doucette-Stamm L, Brent MR, Walhout AJ. Matrix and Steiner-triple-system smart pooling assays for high-performance transcription regulatory network mapping. Nat Methods. 2007;4:659–664. doi: 10.1038/nmeth1063. [DOI] [PubMed] [Google Scholar]
- Ward S, Thomson N, White JG, Brenner S. Electron microscopical reconstruction of the anterior sensory anatomy of the nematode Caenorhabditis elegans. J Comp Neurol. 1975;160:313–337. doi: 10.1002/cne.901600305. [DOI] [PubMed] [Google Scholar]
- Wenick AS, Hobert O. Genomic cis-regulatory architecture and trans-acting regulators of a single interneuron-specific gene battery in C. elegans. Dev Cell. 2004;6:757–770. doi: 10.1016/j.devcel.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG. Regulatory changes underlying expression differences within and between Drosophila species. Nat Genet. 2008;40:346–350. doi: 10.1038/ng.77. [DOI] [PubMed] [Google Scholar]
- Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue D, Tu Y, Chalfie M. Cooperative interaction between the Caenorhabditis elegans homeoproteins UNC-86 and MEC-3. Science. 1993;261:1324–1328. doi: 10.1126/science.8103239. [DOI] [PubMed] [Google Scholar]
- Yuh CH, Bolouri H, Davidson EH. Genomic cis-regulatory logic: experimental and computational analysis of a sea urchin gene. Science. 1998;279:1896–1902. doi: 10.1126/science.279.5358.1896. [DOI] [PubMed] [Google Scholar]
- Yvert G, Brem RB, Whittle J, Akey JM, Foss E, Smith EN, Mackelprang R, Kruglyak L. Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nat Genet. 2003;35:57–64. doi: 10.1038/ng1222. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Ma C, Delohery T, Nasipak B, Foat BC, Bounoutas A, Bussemaker HJ, Kim SK, Chalfie M. Identification of genes expressed in C. elegans touch receptor neurons. Nature. 2002;418:331–335. doi: 10.1038/nature00891. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Fang L, Chen N, Johnsen RC, Stein L, Baillie DL. Distinct regulatory elements mediate similar expression patterns in Caenorhabditis elegans’ excretory cell. J Biol Chem. 2005;280:38787–38794. doi: 10.1074/jbc.M505701200. [DOI] [PubMed] [Google Scholar]
- Zheng X, Chung S, Tanabe T, Sze JY. Cell-type specific regulation of serotonergic identity by the C. elegans LIM-homeodomoain factor LIM-4. Dev Biol. 2005;286:618–628. doi: 10.1016/j.ydbio.2005.08.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.